Abstract
Winter wheat (Triticum spp.) varieties require long exposures to low temperatures to flower, a process called vernalization. The VRN2 locus includes two completely linked zinc finger-CCT domain genes (ZCCT1 and ZCCT2) that act as flowering repressors down-regulated during vernalization. Deletions or mutations in these two genes result in the elimination of the vernalization requirement in diploid wheat (Triticum monococcum). However, natural allelic variation in these genes has not been described so far in polyploid wheat (tetraploid Triticum turgidum and hexaploid Triticum aestivum). A tetraploid wheat population segregating for both VRN-A2 and VRN-B2 loci facilitated the characterization of different alleles. Comparisons between functional and nonfunctional alleles revealed that both ZCCT1 and ZCCT2 genes are able to confer vernalization requirement and that different ZCCT genes are functional in different genomes. ZCCT1 and ZCCT2 proteins from nonfunctional vrn2 alleles have mutations at arginine amino acids at position 16, 35, or 39 of the CCT domain. These positions are conserved between CCT and HEME ACTIVATOR PROTEIN2 (HAP2) proteins, supporting a model in which the action of CCT domains is mediated by their interactions with HAP2/HAP3/HAP5 complexes. This study also revealed natural variation in gene copy number, including a duplication of the functional ZCCT-B2 gene and deletions or duplications of the complete VRN-B2 locus. Allelic variation at the VRN-B2 locus was associated with a partially dominant effect, which suggests that variation in the number of functional ZCCT genes can be used to expand allelic diversity for heading time in polyploid wheat and, hopefully, improve its adaptation to different environments.
Wheat (Triticum aestivum) is one of the major crop species and occupies a wide range of environments from 65°N to 45°S (Lantican et al., 2005). This wide adaptability is favored by diverse growth habits, which include winter and spring forms. Winter wheats are sown in autumn and require long exposures to cold temperatures (vernalization) to accelerate flowering. The vernalization requirement prevents flower development during winter, protecting sensitive floral organs from freezing temperatures. Spring wheats are planted in the spring or in the fall (in regions with mild winters) and do not have a vernalization requirement.
The three major genes responsible for natural variation in vernalization requirement in wheat (and also in barley [Hordeum vulgare]) are VRN1, VRN2, and VRN3. VRN1 is a homolog of the Arabidopsis (Arabidopsis thaliana) meristem identity gene APETALA1, which determines the transition between the production of leaves and flowers at the shoot apical meristem (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). Mutagenized plants of diploid wheat (Triticum monococcum; 2n = 14; Am genome similar to the A genome of polyploid wheat) with complete deletions of the VRN1 gene fail to flower (Shitsukawa et al., 2007), indicating that VRN1 is essential for the initiation of the reproductive phase in this species. Several natural mutations have been identified in regulatory regions of the VRN1 promoter or first intron, which are associated with the elimination or reduction of the vernalization requirement and consequently with spring growth habit (Yan et al., 2003, 2004a; Fu et al., 2005; vonZitzewitz et al., 2005).
VRN3 is a homolog of the Arabidopsis photoperiod gene FLOWERING LOCUS T, and in both species this gene up-regulates VRN1 transcription under long days (Yan et al., 2006; Hemming et al., 2008) through interactions with its promoter (Wigge et al., 2005; Li and Dubcovsky, 2008). Before vernalization, VRN3 is down-regulated by VRN2, preventing winter wheats from flowering during the fall. Vernalization results in the induction of VRN1 and the down-regulation of VRN2 (Loukoianov et al., 2005; Trevaskis et al., 2006), thereby releasing VRN3 to further induce VRN1 and initiate the reproductive phase during the long days of spring (reviewed by Trevaskis et al., 2007; Distelfeld et al., 2009). The focus of this paper is the natural variation in VRN2.
In diploid wheat, the VRN2 locus includes two tandemly duplicated genes designated ZCCT1 and ZCCT2 (Yan et al., 2004b). These genes code for proteins that are 76% identical, each including a putative zinc finger and a CCT domain (for CONSTANS [CO], CONSTANS-LIKE [CO-like], and TIMING OF CAB EXPRESSION1 [TOC1]). The 43-amino acid CCT domain is present in proteins involved in photoperiod, light signaling, and circadian rhythms and is well conserved among different plant species (Griffiths et al., 2003; Yan et al., 2004b). Mutations within the CCT domain are known to alter the functions of proteins CO, TOC1, VRN2, and PPD-H1 (Wenkel et al., 2006). It has been shown recently that the CCT domain has similarities to a region of the yeast HEME ACTIVATOR PROTEIN2 (HAP2), a subunit of the HAP2/HAP3/HAP5 complex that binds to CCAAT boxes in the promoters of many eukaryotic genes and regulates their expression (Wenkel et al., 2006).
In wheat and barley accessions with winter growth habit, ZCCT transcripts show a progressive decrease during vernalization (under long days) that is not observed in control plants kept at nonvernalizing temperatures (Yan et al., 2004b, Trevaskis et al., 2006). Wheat and barley ZCCT genes are also down-regulated by short days (Dubcovsky et al., 2006; Trevaskis et al., 2006, 2007). GHD7 (=OsI), the closest homologous gene in rice (Oryza sativa; Xue et al., 2008), is also a long-day repressor of flowering down-regulated by short days.
All diploid wheat and barley accessions with winter growth habit studied so far have at least one functional ZCCT gene, whereas those with spring growth habit associated with recessive vrn2 alleles have deletions encompassing all ZCCT genes or carry mutations in conserved amino acids of the CCT domains (Yan et al., 2004b; Karsai et al., 2005; Cockram et al., 2007; Szücs et al., 2007). The presence of a single functional vrn2 allele in heterozygous plants is sufficient to confer some vernalization requirement, so only the homozygous recessive vrn2 allele results in spring growth habit (Takahashi and Yasuda, 1971; Tranquilli and Dubcovsky, 2000).
RNA interference of ZCCT1 in hexaploid winter wheat variety Jagger (T. aestivum; 2n = 42; genomes AABBDD) reduces ZCCT1 transcript levels and accelerates flowering, suggesting that the VRN2 locus also plays a significant role in the regulation of flowering in hexaploid wheat (Yan et al., 2004b) and likely in tetraploid wheat (Triticum turgidum; 2n = 28; genomes AABB), the donor of the A and B genomes to hexaploid wheat. Tetraploid wheats are divided into three subspecies: T. turgidum subsp. dicoccoides (wild accessions with disarticulating spikes), T. turgidum subsp. dicoccon (partially domesticated, with nondisarticulating spikes and non-free-threshing grains), and T. turgidum subsp. durum (modern free-threshing varieties). These species are characterized in this study for their natural allelic variation in the ZCCT1 and ZCCT2 genes. The homoeologous copies of the genes and loci from the A and B genomes are identified hereafter by including the genome designation before the gene or locus number, as is customary for wheat nomenclature (e.g. VRN-A2, VRN-B2, ZCCT-A1, and ZCCT-B1).
Allelic variation for VRN2 has not been described so far for tetraploid or hexaploid wheat, likely due to the fact that in polyploid wheat simultaneous loss-of-function mutations at all VRN2 homoeoloci are required to confer spring growth habit. In addition, allelic variation for VRN2 would be detected only when alleles for winter growth habit are present at all VRN1 loci, as alleles for spring growth habit are dominant and epistatic on VRN2.
RESULTS
ZCCT1 and ZCCT2 Sequences from Diploid and Tetraploid Wheat Species
The genomic region encompassing the VRN2 locus was previously sequenced from diploid wheat T. monococcum (Yan et al., 2004b; accession no. AY485644) and the A genome of tetraploid wheat T. turgidum subsp. durum var. Langdon (Dubcovsky and Dvorak, 2007; EF540321). A low-coverage sequencing of bacterial artificial chromosome (BAC) 738D05 including the VRN-B2 locus from T. turgidum subsp. durum revealed three ZCCT genes, designated ZCCT-B1 (FJ173819), ZCCT-B2a (FJ173823), and ZCCT-B2b (FJ173824), based on their sequence identity with previously reported ZCCT genes (Fig. 1). The coding regions of ZCCT-B2a and ZCCT-B2b are 100% identical, and their intron regions differ only by a 1-bp insertion/deletion (indel), which was used to develop a marker for the deletion. The genomic regions including the ZCCT-B2a and ZCCT-B2b genes (9.7 kb) are 99.7% identical, suggesting a recent duplication.
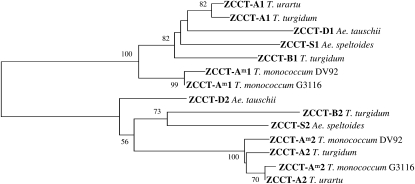
Figure 1.
Neighbor-joining phylogenetic analyses of wheat ZCCT proteins. Bootstrap values based on 1,000 replications are indicated above their respective nodes. Only values above 50 are presented.
Additional ZCCT genes were sequenced from winter accessions of diploid Triticum and Aegilops species with genomes similar to the A, B, and D genomes of hexaploid wheat. These species included Triticum urartu, the donor of the A genome (Dvorak et al., 1988); Aegilops speltoides (S genome), the closest extant diploid species to the B genome of tetraploid and hexaploid wheat (Dvorak and Zhang, 1990); and Aegilops tauschii, the donor of the D genome of hexaploid wheat (Kihara, 1944).
The sequences for ZCCT-D1 (FJ173818) and ZCCT-D2 (FJ173822) were obtained from Ae. tauschii BAC clones 2H24, 14E16, and 78I09 (Akhunov et al., 2005), whereas the ZCCT genes from the other diploid species were obtained directly by PCR from genomic DNA. Ae. tauschii ZCCT-D1 and -D2 sequences, together with T. urartu sequences for ZCCT-A1 (FJ173816) and ZCCT-A2 (FJ173820) and Ae. speltoides sequences for ZCCT-S1 (FJ173817) and ZCCT-S2 (FJ173821), were deposited in GenBank. The phylogenetic analysis grouped the predicted ZCCT protein sequences into two distinct clades (Fig. 1), one including the ZCCT1 proteins from all species and the other including the ZCCT2 proteins from all species. This suggests that the duplication that originated these two genes preceded the divergence of the diploid Triticum and Aegilops species.
RFLP Germplasm Screen
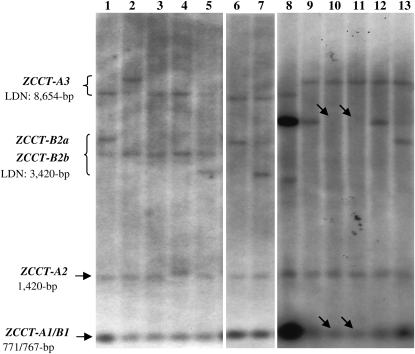
The hybridization of Southern blots including DraI-digested DNAs from wild and cultivated tetraploid and hexaploid Triticum accessions (see “Materials and Methods”) revealed contrasting patterns of allelic diversity for different ZCCT genes. The shortest overlapping DraI restriction fragments (767 bp from ZCCT-A1 and 771 bp from ZCCT-B1) showed limited variation among accessions. Limited variation was also found for the 1,420-bp fragment corresponding to the ZCCT-A2 gene (Fig. 2). On the contrary, the restriction fragments within the region corresponding to the ZCCT-B2a and ZCCT-B2b genes were variable in size (approximately 3–5 kb), generating multiple haplotypes (Fig. 2, lanes 1–5). Some cultivated durum lines showed two fragments in this region and others only one (Fig. 2).
Figure 2.
RFLP screening of polyploid Triticum accessions for variation in ZCCT genes. Lanes 1 to 7, T. turgidum subsp. durum accessions PI60727, PI60730, PI60731, PI60732, PI60733, PI7016, and PI10388. Lane 8, T. turgidum subsp. dicoccoides accession PI428107 (Rosh Pinna, Israel). Note the increased hybridization signal in fragments corresponding to ZCCT-B1 and ZCCT-B2 compared with the nonduplicated ZCCT-A2 1,420-bp band in the same accession. Lanes 9 to 13, T. turgidum subsp. dicoccon accessions PI470737, PI470739 (two lanes corresponding to two different plants), PI499973, and PI193881. Arrows indicate the deletion of the ZCCT-B2 bands and the reduced intensity of the ZCCT-A1/B1 overlapping bands. DNAs were digested with DraI and hybridized with the second exon of ZCCT-Am1.
The largest RFLP fragment corresponds to a third and more divergent ZCCT copy (ZCCT-A3). The ZCCT-A3 putative coding region is only 81% to 82% identical to the other two ZCCT genes and has a shorter first exon that does not include the predicted zinc finger characteristic of other ZCCT proteins. So far, ZCCT-A3 has been found only in the A genome, 16.2 kb upstream of ZCCT-A2 (Dubcovsky and Dvorak, 2007). It is not yet known if ZCCT-A3 is translated into a functional protein; therefore, it was not included in the allelic diversity study.
In addition to the variation in restriction fragment size, the ZCCT-B1 and ZCCT-B2 genes showed polymorphisms in copy number. Six T. turgidum subsp. dicoccoides accessions from Rosh Pinna, Israel, showed unusually strong hybridization signals at the restriction fragments corresponding to ZCCT-B1 and ZCCT-B2 (one accession is shown in Fig. 2, lane 8). The VRN-A2 fragments from the same accessions showed no increase in hybridization intensity, confirming equal loading of DNA on the Southern blots. Based on this result, we concluded that the copy number of the ZCCT-B1 and ZCCT-B2 genes was amplified in the Rosh Pinna accessions.
One spring accession of T. turgidum subsp. dicoccon (PI470739), collected in the mountains of Kars, Turkey (1,590 m above sea level), showed a deletion of the restriction fragments corresponding to the ZCCT-B2 gene(s) and reduced hybridization intensity of the 767/771-bp fragment corresponding to ZCCT-A1 and ZCCT-B1 overlapping fragments (Fig. 2, lanes 10 and 11). The deletion of both ZCCT-B1 and ZCCT-B2 genes in PI470739 was confirmed by PCR using primers specific for ZCCT-B1 (VRN2/B1/F3-R6) and ZCCT-B2 (VRN2/B2/F2-R5) genes (Supplemental Table S1). These primers failed to amplify any ZCCT fragment from PI470739.
Molecular Characterization of the 5A-5Am Translocation in BC3F2-521
The effect of the VRN2 loci on flowering time in tetraploid wheat was studied using a plant segregating simultaneously for the vrn-B2 deletion from PI470739 and the nonfunctional vrn-Am2 from T. monococcum accession DV92. The development of this plant is described in “Materials and Methods” and in Figure 3.
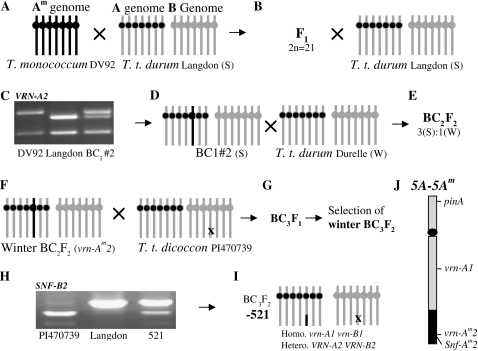
Figure 3.
Generation of a tetraploid line segregating for VRN-A2 and VRN-B2. A, Introgression of the nonfunctional vrn-Am2 allele from DV92 into tetraploid wheat. Black bars represent T. monococcum chromosomes, and gray bars represent T. turgidum (T. t.) chromosomes. B, First backcross to tetraploid wheat variety Langdon. C, Marker-assisted selection of line BC1#2 carrying the T. monococcum vrn-Am2 allele. D, Second backcross to tetraploid winter wheat variety Durelle. E, Selection of winter growth habit BC2F2 plants (3:1 segregation of winter to spring). F, Backcross of a selected winter BC2F2 plant carrying the vrn-Am2 allele to T. turgidum subsp. dicoccon accession PI470739 carrying the vrn-B2 deletion. G, Selection of winter BC3F2 plants (recessive vrn-A1 and vrn-B1 alleles). H, Codominant molecular marker for the vrn-B2 deletion from PI470739 based on tightly linked gene SNF-B2. I, Selected line BC3F2-521 heterozygous for VRN-A2 and VRN-B2 loci. J, Graphical representation of chromosome 5A from BC3F2-521. The T. monococcum chromosome 5Am segment carrying the vrn-Am2 allele was recombined with T. turgidum chromosome 5A between the VRN-A1 and VRN-Am2 genes. S, Spring growth habit; W, winter growth habit.
A total of 42 plants with winter growth habit were selected from the progeny of a BC3F1 line heterozygous for different vernalization genes (Fig. 3G). The winter growth habit indicates that these plants are homozygous for the recessive vrn-A1 and vrn-B1 alleles. Using molecular markers for VRN-A2 (Fig. 3C) and VRN-B2 (Fig. 3H), we selected plant 521 (BC3F2-521 hereafter), which was heterozygous for both VRN-A2 and VRN-B2 and homozygous for the recessive vrn-A1 and vrn-B1 alleles (Fig. 3). The progeny of this plant were used to test the effect of the different VRN2 alleles on flowering time.
Several plants from the progeny of BC3F2-521 were analyzed for chromosome number and all showed 28 chromosomes, indicating that the vrn-Am2 gene was incorporated either as a complete chromosome substitution line or as a translocation line. To differentiate between these two possibilities, this line was analyzed with two molecular markers for the short and long arms of homoeologous group 5. The marker for the PINA gene (Bonafede et al., 2007), located in the distal region of the short arm of chromosome 5Am and deleted in tetraploid wheat, was not present in BC3F2-521, confirming that vrn-Am2 was transferred to a translocated chromosome. To determine the location of the 5A-5Am translocation, we developed a marker for the VRN-A1 gene, which is located on the middle of the long arm. The absence of the T. monococcum vrn-Am1 allele indicated that the recombination event occurred between the VRN-A1 (5AL) and VRN-Am2 (5AmL) loci. A marker for the deletion in the VRN-A1 intron (Fu et al., 2005) was used to confirm that the allele present in BC3F2-521 was the recessive vrn-A1 allele from Durelle, as expected from its winter growth habit. A representation of the recombined chromosome is shown in Figure 3J.
Effects of the Allelic Differences in VRN-A2 and VRN-B2 on Flowering Time
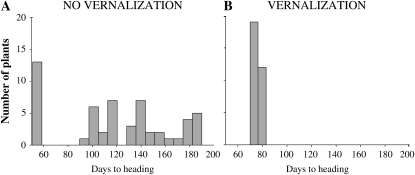
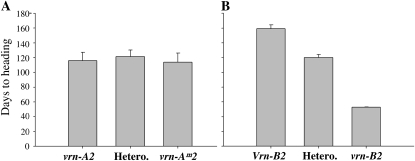
The nonvernalized progeny of line BC3F2-521 segregated into two nonoverlapping groups for flowering time. The first group included 13 early-flowering plants that headed in less than 60 d (average, 53.0 ± 0.4 d) and were classified as spring, whereas the second group included 41 late-flowering plants that took more than 90 d for heading (average, 139.1 ± 4.6 d) and were classified as winter (Fig. 4A). The group with spring growth habit was less variable than the group with winter growth habit, which showed two peaks, likely associated with the presence of homozygous and heterozygous lines (Fig. 4A). The observed ratio between spring and winter plants differed significantly from a 1:15 ratio segregation (two dominant genes; χ2 = 29.3, P < 0.0001) but not from a 1:3 ratio (one dominant gene; χ2 = 0.025, P = 0.88). The large differences in heading time observed among the nonvernalized plants disappeared when plants were vernalized (Fig. 4B).
Figure 4.
Segregation for heading time. Frequency distribution of days to heading for the progeny of winter plant BC3F2-521 (heterozygous for both VRN2 loci). A, Unvernalized plants (1:3 segregation of spring to winter growth habit). B, Vernalized plants (no segregation for flowering time).
The same plants were genotyped with VRN-A2 and VRN-B2 markers to determine which locus was responsible for the observed segregation in heading time (Fig. 5). No significant differences were detected among the VRN-A2 genotypic classes (Fig. 5A), indicating that both T. turgidum (PI470739) and T. monococcum DV92 have recessive vrn-A2 alleles. This indicates that none of the ZCCT genes present at the VRN-A2 locus is able to confer a vernalization requirement.
Figure 5.
Effects of VRN-A2 and VRN-B2 alleles on heading time in nonvernalized plants. A, No significant differences in heading time were observed between lines homozygous for the vrn-Am2 allele from T. monococcum DV92 and the one from T. turgidum subsp. dicoccon PI470739 (vrn-A2). B, Lines homozygous for the Langdon/Durelle VRN-B2 allele headed significantly later (P < 0.01) than the heterozygous or homozygous lines for the vrn-B2 deletion (PI470739). “Hetero.” indicates heterozygous plants, whereas the two other classes are homozygous for the indicated alleles.
Genotyping with the codominant SNF-B2 marker tightly linked to the VRN-B2 locus showed that all 13 spring plants were homozygous for the recessive vrn-B2 allele from PI470739 (Fig. 5B), a result that was confirmed using PCR primers specific for the ZCCT-B1 and ZCCT-B2 genes (Supplemental Table S1). These results indicate that variation at the VRN-B2 locus was responsible for the segregation in flowering time observed in the progeny from BC3F2-521.
The effect of the functional VRN-B2 allele on heading time was partially dominant. Plants homozygous for the functional VRN-B2 allele (Langdon/Durelle) flowered 159 ± 5.3 d after sowing, whereas those heterozygous for VRN-B2 flowered significantly (P < 0.001) earlier (120 ± 4.3 d after sowing). Heading time of the heterozygous plants was only 14 d later than the midpoint between the two homozygous classes (106 d). Using these numbers, the degree of dominance was calculated to be 0.26 (14 d/53 d, with 0 = completely additive and 1 = completely dominant). This value indicates a relatively small dominant effect.
The differences in heading time between the VRN-B2 allelic classes disappeared when the plants were vernalized. Vernalized plants carrying the functional VRN-B2 allele headed almost at the same time (73.7 ± 1.4 d) as the vernalized plants homozygous for the vrn-B2 deletion (74.1 ± 1.2 d; P = 0.26). Differences between the VRN-A2 allelic classes were also not significant (P = 0.13). A two-way factorial ANOVA including vernalization and VRN-B2 alleles as factors showed a highly significant interaction between VRN-B2 alleles and vernalization (P < 0.0001), confirming that the effect of this locus on heading time was the result of differences in vernalization requirement.
Sequence Diversity of the ZCCT1 and ZCCT2 Genes
The ZCCT1 and ZCCT2 genes present in the functional VRN-B2 and nonfunctional VRN-A2 loci from BC3F2-521 were sequenced to determine if the differences in functionality were associated with specific mutations in the conserved CCT domain. For comparison, a consensus CCT sequence was generated from different classes of CO-like proteins found in plants (Griffiths et al., 2003; Yan et al., 2004b) and was represented using WebLogo (Crooks et al., 2004). The consensus sequence was aligned with the CCT domain sequences from winter accessions of wild diploid progenitors of cultivated wheat (T. urartu, Ae. speltoides, and Ae. tauschii; Fig. 6). The main differences among species are summarized in Table I.
Figure 6.
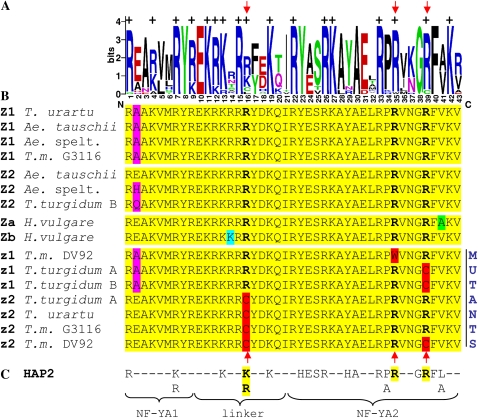
Alignment of the CCT domains from different plant species. A, WebLogo representation of consensus sequence from CCT domains from rice and Arabidopsis CO-like proteins (Griffiths et al., 2003; Yan et al., 2004b) and nonmutant ZCCT1 and ZCCT2 proteins. The sizes of the different letters are proportional to the frequency of the amino acid in the multiple sequence alignment. B, Alignment of the CCT domains from wheat and Aegilops ZCCT1 and ZCCT2 proteins. Arrows point to natural mutations discovered in conserved amino acids that may affect protein function. Ae. spelt., Ae. speltoides; T.m., T. monococcum. C, Conserved amino acids between CCT and HAP2. Subdomains NF-YA1 (interacts with HAP3 and HAP5) and NF-YA2 (interacts with CCAAT DNA sequences) are indicated below.
Table I.
Summary of polymorphisms between ZCCT1 and ZCCT2 proteins from different species and genomes
Numbers below the protein names indicate the positions of the amino acids in the CCT domain. Del, Seven-amino acid deletion. Bold letters indicate mutated amino acids.
| Genotype | VRN2 Function | ZCCT1
|
ZCCT2
|
|||||
|---|---|---|---|---|---|---|---|---|
| Del | 16 | 35 | 39 | 16 | 35 | 39 | ||
| T. urartu | Functionala | Yes | R | R | R | C | R | R |
| Ae. speltoides | Functionala | No | R | R | R | R | R | R |
| Ae. tauschii | Functionala | No | R | R | R | R | R | R |
| T. monococcum 3116 | Functional | No | R | R | R | C | R | R |
| T. monococcum DV92 | Nonfunctional | No | R | W | R | C | R | C |
| T. turgidum 521 A genome | Nonfunctional | Yes | R | R | C | C | R | R |
| T. turgidum 521 B genome | Functional | No | R | R | C | R | R | R |
Based on winter growth habit but not confirmed by genetic studies.
ZCCT-A1
The predicted ZCCT-A1 protein corresponding to the nonfunctional VRN-A2 locus from BC3F2-521 has a mutation from R to C at position 39 of the CCT domain (designated R39C hereafter; Fig. 6). This position of the CCT domain is well conserved among CCT domains from other CO-like proteins and HAP2 proteins (Fig. 6).
The R39C mutation was detected in all 37 cultivated T. turgidum subsp. durum accessions analyzed in this study but was polymorphic in cultivated T. turgidum subsp. dicoccon (present in 11 of 22 accessions) and wild T. turgidum subsp. dicoccoides (present in 10 of 19 accessions; Table II). One accession from Asia Minor (PI355454) showed an additional R35Q mutation. The 11 accessions of T. turgidum subsp. dicoccon that lack the R39C mutation all have the R35W mutation. The R35W mutation was also found in eight of the nine accessions of T. turgidum subsp. dicoccoides that do not have the R39C mutation. T. turgidum subsp. dicoccoides accession 10-85 collected at Ammiad in Israel was the only one with no mutations in the CCT domain from ZCCT-A1 (Table II).
Table II.
Summary of different ZCCT-A1 and ZCCT-B1 alleles in a collection of wild T. turgidum subsp. dicoccoides, partially domesticated T. turgidum subsp. dicoccon, and modern cultivated T. turgidum subsp. durum
PI accessions are from the National Small Grain Collection (United States), and other numbers are population numbers from the University of Haifa. Numbers below the gene names indicate their positions within the CCT domain. All ZCCT-A1 and ZCCT-B1 proteins included here have an R at position 16. Bold letters indicate mutated amino acids.
| Germplasm | Country | Species | ZCCT-A
|
ZCCT-B
|
||||
|---|---|---|---|---|---|---|---|---|
| A1
|
A2
|
B1
|
B2
|
|||||
| 35 | 39 | 16a | 35 | 39 | No | |||
| 10-85 | Israel | T. turgidum subsp. dicoccoides | R | R | C | R | R | 1b |
| 1, 8, 17, 27, 41, PI428055 | Israel, Turkey | R | C | C | R | R | 1 | |
| PI428028, PI428036, PI428041, PI428066, PI428070, PI428072, PI428079, PI428082 | Turkey | W | R | C | R | R | 1 | |
| 5, 11, 30, 42 | Israel | R | C | C | R | C | 1 | |
| PI355498, CItr17676, PI606325, PI182743, PI352329, PI94627, PI352352, PI352357, PI352367 | Asia Minor, Israel, Syria, Turkey | T. turgidum subsp. dicoccon | W | R | C | R | R | 1 |
| PI319868, PI319869 | Turkey | W | R | C | R | C | 2 | |
| PI94640, PI254158, PI254180, PI347230, PI470737, PI470738, PI355496, CItr17675 | Iran, Israel, Turkey, Lebanon | R | C | C | R | R | 1 | |
| PI470739 | Turkey | R | C | C | Deletion
|
|||
| PI355454 | Asia Minor | Q | C | C | R | C | 2 | |
| PI352347 | Israel | R | C | C | R | C | 2 | |
| Adamello, Appio, Appulo, Capelli, Ciccio, Cirillo, Colosseo, Duilio, Karel, Latino, L35, Ofanto, Russello SG7, San Carlo, Saragolla, Trinakria, Valbelice, Valforte, Valnova, Varano, Vitron, WB881, Zenit, Inrat 69, Karim, Khiar, Exeldur, Durfort, Nefer, Neodur, Aconchi 89, Altar 84, Mexicali 75, Colorado, Kronos, Produra | Italy, Tunisia, France, Mexico, United States | T. turgidum subsp. durum | R | C | C | R | C | 2 |
| Messapia | Italy | R | C | C | R | C | 1 | |
Since the R16C mutation is fixed in ZCCT-A2 and ZCCT-Am2, we checked only one accession from each group for this mutation.
1 indicates that only one of the two polymorphic sites was detected but does not completely rule out the existence of the duplication.
In addition to the R39C mutation, the ZCCT-A1 protein has a deletion of seven amino acids relative to the ZCCT-Am1 protein from T. monococcum. These seven amino acids are located immediately downstream of the putative zinc finger domain from amino acids 49 to 55 (numbers are relative to the initial Met in ZCCT-Am1). A screening using primers VRN2/22F+R (Supplemental Table S1) showed that the same deletion was present in all 78 tetraploid wheats tested in this study (Table II) and in 103 of 107 T. urartu accessions. The ZCCT-A1 genes from 15 T. urartu accessions were fully sequenced, and none of them have mutations in the CCT domain.
ZCCT-A2
The predicted ZCCT-A2 protein corresponding to the nonfunctional VRN-A2 locus from BC3F2-521 has a mutation from R to C at position 16 of the CCT domain (R16C). This position is well conserved (R or K) among CCT domains from other CO-like proteins (except for CO-like group II) and HAP2 proteins (Fig. 6).
All 48 accessions of cultivated tetraploid wheat sequenced for this gene have the R16C mutation in the CCT domain. This mutation was also found in the 15 accessions of T. urartu and four accessions of T. monococcum (ZCCT-Am2) but was not detected in the predicted ZCCT2 proteins from Ae. tauschii or Ae. speltoides (Table I; Fig. 6). Three of the four T. monococcum accessions (including DV92) have an R39C mutation in addition to the R16C mutation in ZCCT-Am2.
ZCCT-B1
The predicted ZCCT-B1 protein corresponding to the functional VRN-B2 locus has the same R39C mutation as the ZCCT-A1 protein coded by the nonfunctional VRN-A2 locus (Table I; Fig. 6). The R39C mutation was conserved in the predicted ZCCT-B1 proteins from the 37 accessions of cultivated durum wheat (Table II) and was polymorphic in wild T. turgidum subsp. dicoccoides (present in four of 19 accessions) and cultivated T. turgidum subsp. dicoccon (present in four of 22 accessions). No additional changes in amino acids were detected in the CCT domain of ZCCT-B1 (Table II).
ZCCT-B2
The predicted ZCCT-B2a and ZCCT-B2b proteins corresponding to the functional VRN-B2 locus from tetraploid variety Langdon have no mutations in any of the conserved amino acids of the CCT domain (Table I; Fig. 6). This was also the case for the other 37 cultivated durum accessions (Fig. 6).
A screening for the 1-bp indel characteristic of the ZCCT-B2 gene duplication using PCR primers VRN2/B2/F2+R5 (Supplemental Table S1) failed to detect the duplication in T. turgidum subsp. dicoccoides. Although the detection of the two ZCCT-B2 forms with this PCR marker is sufficient to confirm the presence of the duplication, the detection of a single sequence needs to be interpreted with caution because it can indicate either the absence of the duplication or the absence of the 1-bp indel polymorphism. The fact that all of the T. turgidum subsp. dicoccoides accessions included in the RFLP screen show a single ZCCT-B2 fragment (with the exception of the Rosh Pinna accessions) provides additional indirect evidence for the absence of the duplication in most wild accessions.
In T. turgidum subsp. dicoccon, the presence of the ZCCT-B2 duplication was confirmed in the four accessions that carry the R39C mutation at the ZCCT-B1 gene (PI319868, PI319869, PI355454, and PI352347; Table II). The presence of the ZCCT-B2 duplication was also confirmed among most of the modern T. turgidum subsp. durum varieties (36 of 37), with Messapia as the only exception (Table II). Many of the cultivated durum varieties showed two fragments in the RFLP screening (Fig. 2).
Expression of ZCCT1 and ZCCT2 Genes in Tetraploid Wheat
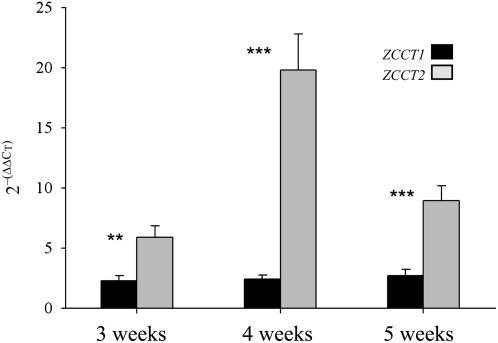
Quantification of transcript levels of ZCCT1 and ZCCT2 in tetraploid wheat leaves collected from 3-, 4-, and 5-week-old plants showed that the average transcript levels of ZCCT2 were significantly higher than those of ZCCT1 for all three time points (Fig. 7). Since quantitative reverse transcription-PCR primers (Supplemental Table S1) were designed to differentiate ZCCT1 from ZCCT2 but not A from B genome copies of the same gene, the transcript levels presented in Figure 7 include both A and B homoeologues for each gene.
Figure 7.
Expression of ZCCT1 and ZCCT2 in tetraploid wheat. Transcript levels of ZCCT1 and ZCCT2 were compared in samples from leaves of a tetraploid winter line derived from BC3F2-521 (homozygous for the functional alleles VRN-A2 from T. turgidum subsp. dicoccon accession PI470739 and VRN-B2 from varieties Langdon or Durelle). Eight plants were sampled from 3-, 4-, and 5-week-old plants. Gray bars represent ZCCT2 transcripts, and black bars represent ZCCT1 transcripts. Amplification primers are conserved for the A and B homoeologous copies of each gene. Asterisks indicate significant differences (t test): ** P ≤ 0.01, *** P ≤ 0.001.
DISCUSSION
The results presented here indicate that the differences among ZCCT proteins coded by genes corresponding to functional and nonfunctional VRN2 alleles are concentrated in the CCT domain. This 43-amino acid domain is well conserved in CO and CO-like proteins (defined as being more similar to CO than to other Arabidopsis proteins like TOC1) from mosses, gymnosperms, and angiosperms, indicating an ancient origin (Griffiths et al., 2003). The CCT domains are involved in the nuclear localization of CO and CO-like proteins but also have additional roles. In Arabidopsis, the co-7 CCT mutation does not alter the nuclear localization of the CO protein but delays flowering significantly (Robson et al., 2001). It is possible that some mutations in the CCT domain may limit its ability to interact with other proteins (Kurup et al., 2000).
It was shown recently that the CCT domains from Arabidopsis CO and COL15 can interact with several AtHAP3 and AtHAP5 proteins in yeast, and this interaction was confirmed in plant cells and in vitro (Ben-Naim et al., 2006; Wenkel et al., 2006). Wenkel et al. (2006) proposed that CCT proteins act by replacing the HAP2 subunit of the HAP2/HAP3/HAP5 complex, altering the ability of this complex to bind to the CCAAT boxes in the promoters of target genes. Overexpression of AtHAP3b was shown to promote early flowering, probably through an interaction with CO or COL proteins, while hap3b, a null mutant of HAP3b, delayed flowering under long days but not under short days (Cai et al., 2007).
CCT domains and HAP2 proteins have similar amino acids at 18 positions, which are also well conserved within each group of proteins from mosses to vascular plants (Wenkel et al., 2006). Conservation of these amino acids for more than 400 million years suggests that they play critical roles in the proper function of these proteins. Six independent mutations at four of these conserved positions have been shown to disrupt the function of CO (Putterill et al., 1995; Robson et al., 2001), VRN2 (Yan et al., 2004b), TOC1 (Strayer et al., 2000), and PPD-H1 (Turner et al., 2005) proteins. These six mutations are all located within the CCT domain region that corresponds to the NF-YA2 subdomain of the HAP2 protein (Fig. 6). This HAP2 subdomain has been modeled and predicted to interact with the DNA of the CCAAT box (Romier et al., 2003). In addition to the NF-YA2 subdomain, the HAP2 protein has another NF-YA1 subdomain proposed to interact with the HAP3/HAP5 dimer and a linker region between these two subdomains (Romier et al., 2003). It is tempting to speculate that the ZCCT proteins may also regulate flowering time through interactions with HAP proteins and that mutations in the CCT domain may affect the ability of the ZCCT proteins to interact with DNA or with HAP3/HAP5 dimers. We recently confirmed that ZCCT proteins can interact with several wheat HAP3 and HAP5 proteins in yeast two-hybrid systems (C. Li, A. Distelfeld, and J. Dubcovsky, unpublished data), providing additional support for this hypothesis.
Interestingly, the three CCT mutations identified here in ZCCT proteins coded by genes located in nonfunctional VRN2 loci are located at positions 16, 35, and 39, which are conserved both between and within the CCT domains and HAP2 proteins (Fig. 6). These three positions of the CCT domain are conserved in many plants, including mosses. Position 16 is either K or R (both positively charged amino acids) in Arabidopsis CO, CO-like (except those of class II), and all HAP2 proteins, whereas Arg residues at positions 35 and 39 are invariant among the same proteins (Fig. 6). All three mutations (R16C, R35W, and R39C) are associated with high negative BLOSUM62 scores (−3), which are indicative of changes involving amino acids with very different biochemical properties.
Taken together, the high negative BLOSUM62 scores and the conserved CCT/HAP2 positions where these mutations occurred suggest that these three mutations have a high probability of disrupting or altering the function of the mutant ZCCT proteins. The importance of CCT position 35 has been confirmed independently in Arabidopsis CO, as an induced ethylmethane sulfonate mutation at this position (co-7) produces a severe effect on flowering time (Robson et al., 2001).
The Two-ZCCT Hypothesis
Assuming that the mutations at CCT positions 16, 35, and 39 can disrupt the function of the ZCCT proteins, the following model can explain the complex results presented here. We propose that both ZCCT1 and ZCCT2 have the ability to delay flowering and confer a vernalization requirement. We will refer to this model hereafter as the “two-ZCCT” hypothesis to facilitate the discussion. The first corollary of this hypothesis is that the presence of a functional copy of at least one of these two genes would be sufficient to confer a vernalization requirement. The second corollary of this hypothesis is that mutations in both genes are required to completely disrupt the function of a particular VRN2 locus. The following arguments are presented to support this hypothesis.
Similarity of ZCCT1 and ZCCT2 CCT Domains
The CCT domains from ZCCT1 and ZCCT2 are almost identical among functional alleles from different species (Fig. 6). The only difference between them is found at the second amino acid, which is fixed for A in the ZCCT1 proteins and varies between E, H, and Q in the wheat ZCCT2 proteins (and the barley ZCCT proteins). The second amino acid of the CCT domain is also variable among CO-like proteins and is not conserved with the HAP2 protein (Fig. 6), suggesting that it may not be a critical position for the function of the CCT domain. Therefore, it is reasonable to assume that ZCCT1 and ZCCT2 may have the ability to perform similar functions.
Nonfunctional vrn2 Alleles
The two-ZCCT hypothesis predicts that all recessive vrn2 alleles would have nonfunctional mutations at both ZCCT1 and ZCCT2 proteins. In agreement with this prediction, the recessive vrn-A2 allele from BC3F2-521 has the R39C mutation in ZCCT-A1 and the R16C mutation in ZCCT-A2 (Table I). The deletion of seven amino acids found downstream of the putative zinc finger in the ZCCT-A1 protein in tetraploid variety Langdon (Dubcovsky and Dvorak, 2007) and BC3F2-521 (from PI470739) does not seem to be critical for the function of the ZCCT-A1 protein, since a similar deletion was observed in T. urartu accession PI428180, which has a winter growth habit (functional VRN-A2 allele) and a likely nonfunctional ZCCT-A2 protein (Table I).
The available information from T. monococcum also supports the two-ZCCT hypothesis. Cultivated T. monococcum accession DV92 has a recessive vrn-Am2 allele that is associated with the R35W mutation in ZCCT-Am1 and both R16C and R39C mutations in ZCCT-Am2 (Yan et al., 2004b). An additional survey of 39 spring accessions of cultivated T. monococcum carrying the recessive vrn-Am2 allele showed that all have either deletions encompassing both ZCCT-Am1 and ZCCT-Am2 genes (17 accessions) or R35W mutations in ZCCT-Am1 (22 accessions; Yan et al., 2004b). Although ZCCT-Am2 was not analyzed in detail, all four accessions of T. monococcum for which the sequence of this gene is available have the R16C mutation in ZCCT-Am2. Since the R16C mutation has been detected in all of the ZCCT-A2 (T. urartu and T. turgidum) and ZCCT-Am2 genes sequenced so far, it is reasonable to assume that this mutation occurred before the divergence of the A and Am genomes and, therefore, that it is likely fixed in the polyploid wheat species.
Based on the limited information available at the time of cloning VRN-Am2, Yan et al. (2004b) concluded that ZCCT-Am1 was VRN-2 and that ZCCT-Am2 was not important for the determination of the winter growth habit. At that point, it was not clear that the ZCCT-Am2 gene in T. monococcum was fixed for a nonfunctional allele, simplifying the detection of the segregation for the R35W mutation in ZCCT-Am1. Although the published conclusion is valid for T. monococcum, our results indicate that it cannot be generalized to all Triticeae species.
Functional VRN2 Alleles
None of the 16 winter accessions of cultivated T. monococcum screened so far for ZCCT-Am1 has the R35W mutation (Yan et al., 2004b). Available sequences for ZCCT-Am2 for two of these winter accessions (AY485976 and AY485975) showed that they both have the R16C and R39C mutations, suggesting that winter growth habit in T. monococcum is conferred only by the ZCCT-Am1 protein.
The same is true for the winter accessions of T. urartu. The 15 accessions of T. urartu sequenced so far all have the R16C mutation in the CCT domain of ZCCT-A2 and no mutations in ZCCT-A1 (Table I). This suggests that the winter growth habit in T. urartu is also conferred by ZCCT-A1.
The molecular characterization of the functional VRN-B2 allele provided the strongest support to the two-ZCCT hypothesis. The ZCCT-B1 protein found in the parental lines of BC3F2-521 (Langdon/Durelle) has an R39C mutation identical to the one found in the ZCCT-A1 protein from the nonfunctional VRN-A2 allele (Table I). The low BLOSUM62 score (−3) and the fact that this mutation alters a conserved position across HAP2 proteins and CCT domains (Fig. 6) suggest that this ZCCT-B1 protein is nonfunctional. In contrast, the ZCCT-B2 protein has no mutations in the conserved amino acids of the CCT domain. The Q mutation found in ZCCT-B2 is associated with a positive BLOSUM62 score (+2), indicative of similar biochemical properties. In addition, CCT position 2 is variable among the CCT domains of ZCCT2 and CO-like proteins and is not conserved with the HAP2 proteins (Fig. 6). These observations suggest that this mutation may not have a negative impact on the structure or function of ZCCT-B2 and that this protein rather than ZCCT-B1 is the one conferring the strong vernalization requirement observed in the late-flowering lines from the BC3F2-521 progeny.
Functional VRN-S2 (Ae. speltoides) and VRN-D2 (Ae. tauschii) Alleles
Most of the Ae. speltoides and Ae. tauschii accessions have a winter growth habit, which suggests that they have functional VRN2 alleles. The ZCCT1 and ZCCT2 proteins from both species showed no mutations in the conserved amino acids of the CCT domains. The Ae. speltoides ZCCT2 protein has an H mutation at the second amino acid of the CCT domain. However, since this position is not conserved, this mutation has a small probability of disrupting the function of the Ae. speltoides ZCCT2 protein.
The lack of mutations in ZCCT1 and ZCCT2 in the functional VRN2 alleles from these two diploid species is consistent with the two-ZCCT hypothesis, but it does not provide new information about the relative importance of these genes for the establishment of the vernalization requirement. The absence of mutations in the CCT domain of the ZCCT-D1 and ZCCT-D2 genes in diploid Ae. tauschii (Table I) suggests that the D genome has the potential to contribute two functional ZCCT copies to common wheat.
In summary, the hypothesis that both ZCCT1 and ZCCT2 genes can confer vernalization requirement explains well the different results on VRN2 allelic variation described in this and previous studies.
Allelic Diversity in VRN2 Alleles in Tetraploid Wheat
The R16C mutation in the ZCCT-A2 protein seems to be fixed in the A genome of tetraploid wheat, since it is present in all A and Am diploid species sequences determined so far. However, the R39C mutation in the ZCCT-A1 protein is still polymorphic among the wild and cultivated T. turgidum subsp. dicoccoides. Approximately half of the accessions of these two subspecies have the R39C mutation, whereas the others do not. The R39C mutation was present in all 37 T. turgidum subsp. durum varieties analyzed in this study (Table II), suggesting that this mutation was fixed during the domestication of the modern free-threshing tetraploid wheats.
Eight of the nine T. turgidum subsp. dicoccoides accessions and all of the T. turgidum subsp. dicoccon accessions that lack the R39C mutation in ZCCT-A1 carry a R35W mutation identical to the one detected in T. monococcum accession DV92 (Table II). Since there is strong evidence indicating that mutations at CCT position 35 result in nonfunctional proteins (Robson et al., 2001; Yan et al., 2004b) and that the ZCCT-A2 protein from tetraploid wheat is fixed for the R16C mutation, it is very likely that most of the wild and cultivated tetraploid accessions have no functional VRN-A2 alleles. The only possible exception was T. turgidum subsp. dicoccoides accession 10-85 from Israel (Amiad population), which showed no mutations in the CCT domain of ZCCT-A1 (Table II). We plan to cross T. turgidum subsp. dicoccoides accession 10-85 with a line homozygous for recessive vrn-A2 and vrn-B2 alleles to test the effect of the 10-85 VRN-A2 allele on flowering time.
The R39C mutation in the ZCCT-B1 gene was also polymorphic among the T. turgidum subsp. dicoccon and T. turgidum subsp. dicoccoides accessions but was fixed in all of the T. turgidum subsp. durum varieties analyzed here (Table II). On the contrary, none of the ZCCT-B2 proteins from these 37 accessions of cultivated durum wheat has mutations in the CCT domain. This result suggests that winter growth habit in cultivated tetraploid wheat is conferred mainly by the ZCCT-B2 gene(s) and that in some T. turgidum subsp. dicoccon and T. turgidum subsp. dicoccoides accessions both the ZCCT-B1 and ZCCT-B2 genes can delay flowering under long days.
VRN-B2 Dosage Effect
The analysis of the progeny of BC3F2-521 showed that the effect of the functional VRN-B2 locus on heading time was partially dominant (degree of dominance = 0.26), which agrees with previous results reported in barley (Szücs et al., 2007). This partial dominant effect indicates that allelic variation in the number of functional copies of ZCCT1 and ZCCT2 can affect heading time in tetraploid wheat. Therefore, the duplication of the functional ZCCT-B2 gene found in most cultivated durum wheat and in some T. turgidum subsp. dicoccon accessions may have contributed to the variation in heading time in tetraploid wheat. The high sequence identity between the two copies (99.7% identity) suggests that this duplication originated recently.
The duplication of the functional ZCCT-B2 locus provides a simple explanation for the higher transcript levels of ZCCT2 relative to ZCCT1 in tetraploid wheat (Fig. 7, A and B copies combined). The opposite result was observed before in T. monococcum, where ZCCT-Am1 transcripts were more abundant than those of ZCCT-Am2 (Yan et al., 2004b). It is interesting that in both cases the most abundant transcripts were those including the functional alleles (ZCCT-Am1 in T. monococcum accession G3116 and ZCCT-B2 in tetraploid wheat). A possible explanation for this observation could be the progressive degradation of regulatory elements of genes that are no longer functional. Deletions or mutations in binding sites for regulatory elements in the promoters of nonfunctional alleles would have no effect on flowering time and therefore would not be affected by purifying selection.
In addition to the internal duplication of the ZCCT-B2 gene in cultivated wheat, other deletion and duplication events affected the complete VRN-B2 locus. The deletion of all ZCCT genes from the B genome found in T. turgidum subsp. dicoccon accession PI470739 was instrumental in demonstrating the dosage effect of functional ZCCT genes in polyploid wheat. The RFLP screening also revealed the existence of a duplication of the complete VRN-B2 locus affecting both ZCCT-B1 and ZCCT-B2 genes (T. turgidum subsp. dicoccoides from Rosh Pinna). The copy number of ZCCT-B1 and ZCCT-B2 in these accessions is currently unknown, but the intensity of the hybridization signal suggests the presence of several copies (Fig. 2). We have initiated the crosses required to study the effect of this duplication on flowering time.
CONCLUSION
Accessions with a spring growth habit determined only by deletions or mutations in the VRN2 locus are frequent in cultivated barley (Dubcovsky et al., 2005; Szücs et al., 2007) and diploid wheat (Yan et al., 2004b). These VRN2 mutations are also found in combination with dominant VRN1 alleles. These results suggest that vrn2 mutations alone or in combination with some dominant VRN1 alleles might confer different responses to environmental cues from those conferred by those VRN1 mutations alone.
The discovery that durum wheat varieties have nonfunctional vrn-A2 alleles and the development of a codominant marker tightly linked to the vrn-B2 deletion (PI470739) will facilitate the development of spring durum wheat varieties with no functional VRN2 loci. These nonfunctional VRN2 alleles can then be used alone or in combination with different dominant VRN1 alleles to develop spring durum wheat varieties with new allelic diversity in heading time.
Allelic variation for VRN2 can be widened also in the opposite direction by adding more copies of functional ZCCT genes to cultivated durum wheat. This is expected to increase vernalization requirement and/or delay flowering, although its final effect will depend on other vernalization genes present in the genetic background. The ZCCT-A1 allele with no mutations in the CCT domain (T. turgidum subsp. dicoccoides accession 10-85) can be used to replace the nonfunctional ZCCT-A1 gene in cultivated durum wheat. In addition, the duplicated VRN-B2 allele present in the T. turgidum subsp. dicoccoides accessions from Rosh Pinna may be deployed in cultivated durum wheat.
Allelic variation in the ZCCT closest homolog in rice (Ghd7) has shown significant contributions of this locus to both the productivity and adaptability of cultivated rice on a global scale (Xue et al., 2008). Hopefully, the ZCCT allelic diversity described here would be useful to fine-tune heading time and improve or expand the adaptability of tetraploid and hexaploid wheat varieties to different environments.
MATERIALS AND METHODS
Plant Materials and Growing Conditions
Triticum monococcum accession DV92 was the source of the nonfunctional vrn-Am2 allele (Yan et al., 2004b), and Triticum turgidum subsp. dicoccon accession PI470739 was the source of the recessive vrn-B2 allele. The winter tetraploid durum variety Durelle was used as the source of the recessive vrn-A1 and vrn-B1 alleles. The RFLP screening included a previously described collection (Dvorak et al., 2006) including 614 wild and cultivated tetraploid accessions, 445 hexaploid accessions, and 443 diploid wheat accessions.
Seeds were imbibed for 24 h at 4°C to promote synchronized germination. Seedlings were transferred to pots and watered with nutrition solution. Unvernalized plants were grown in a greenhouse at room temperature (20°C−25°C) and long-day photoperiod (8 h of dark/16 h of light). For the vernalization experiments, plants were first grown for 3 weeks at the same conditions described above, transferred to a cold room at 4°C and a long-day photoperiod for 4 weeks, and then transferred back to the greenhouse to score heading date. Heading date was recorded at complete spike emergence.
Methods used for sequencing BAC clone 738D05 (VRN-B2 locus), hybridization, PCR, and quantitative reverse transcription PCR, together with the markers for VRN-Am2, VRN-B2, VRN-A1, and PINA loci, are described in the Supplemental Data. Primers for all of the experiments are described in Supplemental Table S1.
Development of a Tetraploid Wheat Line Segregating for VRN-A2 and VRN-B2
The following crosses and selections were performed to introduce the nonfunctional vrn-Am2 allele (R35W) from T. monococcum accession DV92 and the null vrn-B2 allele from T. turgidum subsp. dicoccon PI470739 into tetraploid wheat. T. monococcum accession DV92 was crossed with cultivated tetraploid wheat Langdon (Fig. 3A), which carries dominant VRN-A1 and recessive vrn-B1 alleles (Fu et al., 2005). The hybrid from this cross was backcrossed to Langdon (Fig. 3B), and two BC1 plants were obtained. Plant BC1#2 was confirmed to carry the recessive vrn-Am2 allele using a cleaved amplified polymorphic sequence (CAPS) marker (see below and Fig. 3C).
Plant BC1#2 was crossed with the tetraploid winter wheat Durelle to incorporate the recessive vrn-Am2 allele into a winter background (Fig. 3D). The BC2F1 plant from this cross was self-pollinated, and a population of 80 BC2F2 plants was generated and grown in a greenhouse without vernalization (Fig. 3E). This population showed a 3:1 (62:18) segregation between winter and spring growth habit, as expected for a population segregating only for VRN-A1. Winter BC2F2 lines (homozygous for recessive vrn-A1 and vrn-B1 alleles) were screened with the VRN-Am2 CAPS marker, and three lines homozygous for the recessive vrn-Am2 allele were selected (Fig. 3F).
The selected BC2F2 lines were crossed with T. turgidum subsp. dicoccon accession PI470739 (Fig. 3F), which is homozygous for a deletion encompassing both ZCCT-B1 and ZCCT-B2 genes (recessive vrn-B2 allele). Three BC3F1 plants were self-pollinated, and the resulting BC3F2 seeds were grown in a greenhouse without vernalization to select winter BC3F2 plants (Fig. 3G). The winter lines (homozygous vrn-A1 and vrn-B1) were then screened with the VRN-Am2 CAPS marker and with a codominant marker for SNF-B2 (Fig. 3H), a gene tightly linked to VRN2 (Yan et al., 2004b), to select plants heterozygous for both VRN2 loci. Plant BC3F2-521 was selected, and its progeny were used for the genetic analysis presented in this study (Fig. 3I).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ173816 to FJ173824 and FJ427399.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers and genetic markers.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Dr. J. Dvorak and M.C. Luo for supplying seeds for several wheat accessions and the Southern blots used in Figure 2, the National Small Grains Collection for supplying germplasm, and X. Zhang, C. Miguita, and M. Lau for excellent technical assistance.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2007–35301–17737) and by a Vaadia-Binational Agricultural Research and Development Fund Postdoctoral Fellowship Award (grant no. FI–386–06 to A.D.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Assaf Distelfeld (adistel@ucdavis.edu) and Jorge Dubcovsky (jdubcovsky@ucdavis.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akhunov ED, Akhunova AR, Dvorak J (2005) BAC libraries of Triticum urartu, Aegilops speltoides and Ae. tauschii, the diploid ancestors of polyploid wheat. Theor Appl Genet 111 1617–1622 [DOI] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46 462–476 [DOI] [PubMed] [Google Scholar]
- Bonafede M, Kong L, Tranquilli G, Ohm H, Dubcovsky J (2007) Reduction of a Triticum monococcum chromosome segment carrying the softness genes Pina and Pinb translocated to bread wheat. Crop Sci 47 821–826 [Google Scholar]
- Cai XN, Ballif J, Endo S, Davis E, Liang MX, Chen D, DeWald D, Kreps J, Zhu T, Wu YJ (2007) A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol 145 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Chiapparino E, Taylor SA, Stamati K, Donini P, Laurie DA, O'Sullivan DM (2007) Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor Appl Genet 115 993–1001 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol (in press) [DOI] [PubMed]
- Dubcovsky J, Chen C, Yan L (2005) Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed 15 395–407 [Google Scholar]
- Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L (2006) Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol 60 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Akhunov ED, Akhunov AR, Deal KR, Luo MC (2006) Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol Biol Evol 23 1386–1396 [DOI] [PubMed] [Google Scholar]
- Dvorak J, McGuire PE, Cassidy B (1988) Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 30 680–689 [Google Scholar]
- Dvorak J, Zhang HB (1990) Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc Natl Acad Sci USA 87 9640–9644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Szücs P, Yan L, Helguera M, Skinner J, Hayes P, Dubcovsky J (2005) Large deletions in the first intron of the VRN-1 vernalization gene are associated with spring growth habit in barley and polyploid wheat. Mol Genet Genomics 273 54–65 [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B (2008) Low temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol 147 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai I, Szücs P, Meszaros K, Filichkina T, Hayes PM, Skinner JS, Lang L, Bedo Z (2005) The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110 1458–1466 [DOI] [PubMed] [Google Scholar]
- Kihara H (1944) Discovery of the DD-analyser, one of the ancestors of Triticum vulgare (Japanese). Agric Hortic (Tokyo) 19 13–14 [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21 143–155 [DOI] [PubMed] [Google Scholar]
- Lantican MA, Dubin HJ, Morris ML (2005) Impacts of International Wheat Breeding Research in the Developing World, 1988-2002. International Maize and Wheat Improvement Center, Mexico, D.F.
- Li C, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857 [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MMR, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28 619–631 [DOI] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D (2003) The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem 278 1336–1345 [DOI] [PubMed] [Google Scholar]
- Shitsukawa N, Ikari C, Shimada S, Kitagawa S, Sakamoto K, Saito H, Ryuto H, Fukunishi N, Abe T, Takumi S, et al (2007) The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet Syst 82 167–170 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771 [DOI] [PubMed] [Google Scholar]
- Szücs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM (2007) Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genomics 277 249–261 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Yasuda S (1971) Genetics of earliness and growth habit in barley. In RA Nilan, ed, Proceedings of the 2nd International Barley Genetics Symposium. Washington State University Press, Pullman, WA, pp 388–408
- Tranquilli GE, Dubcovsky J (2000) Epistatic interactions between vernalization genes Vrn-Am1 and Vrn-Am2 in diploid wheat. J Hered 91 304–306 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ (2007) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12 352–357 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034 [DOI] [PubMed] [Google Scholar]
- vonZitzewitz J, Szücs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59 449–467 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059 [DOI] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, et al (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40 761–767 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004. a) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109 1677–1686 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004. b) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.