Abstract
Plants have evolved complex regulatory mechanisms to control the defense response against microbial attack. Both temporal and spatial gene expression are tightly regulated in response to pathogen ingress, modulating both positive and negative control of defense. BLUFENSIN1 (BLN1), a small peptide belonging to a novel family of proteins in barley (Hordeum vulgare), is highly induced by attack from the obligate biotrophic fungus Blumeria graminis f. sp. hordei (Bgh), casual agent of powdery mildew disease. Computational interrogation of the Bln1 gene family determined that members reside solely in the BEP clade of the Poaceae family, specifically, barley, rice (Oryza sativa), and wheat (Triticum aestivum). Barley stripe mosaic virus-induced gene silencing of Bln1 enhanced plant resistance in compatible interactions, regardless of the presence or absence of functional Mla coiled-coil, nucleotide-binding site, Leu-rich repeat alleles, indicating that BLN1 can function in an R-gene-independent manner. Likewise, transient overexpression of Bln1 significantly increased accessibility toward virulent Bgh. Moreover, silencing in plants harboring the Mlo susceptibility factor decreased accessibility to Bgh, suggesting that BLN1 functions in parallel with or upstream of MLO to modulate penetration resistance. Collectively, these data suggest that the grass-specific Bln1 negatively impacts basal defense against Bgh.
The coevolution of plants and plant pathogens has generated a complex multilayered immune response (Jones and Dangl, 2006). Both temporal and spatial gene expression are regulated precisely in a system that balances both positive and negative control of defense. During the interaction of plants and plant pathogens, positive regulators potentiate defense by inducing genes involved in cell wall reinforcement, modification of the cytoskeleton, generation of toxic compounds (phytoalexins and peptides), formation of reactive oxygen species (ROS), and, potentially, programmed cell death in the form of the hypersensitive response (Schulze-Lefert and Panstruga, 2003; Brogden, 2005; Hückelhoven, 2005; Jones and Dangl, 2006; Wise et al., 2007b; Graham et al., 2008). Of these resistance mechanisms, antimicrobial peptides have been found to be one of the most fundamentally conserved among vertebrates, invertebrates, insects, and plants. These peptides have a broad range of toxic activities that inhibit the progression of pathogen invasion, such as membrane destabilization, interfering with transport, and inhibition of protein function (Ganz, 2003; Brogden, 2005). Many of the identified plant antimicrobial peptides fall into well-characterized families such as the γ-thionins, defensins, knottins, and protease inhibitors (Yount and Yeaman, 2004; Graham et al., 2008). Of these, both defensins and knottins can exceed 100 family members within a species. This abundance, resulting from family expansion, divergence, and unequal recombination events, reflects the selection process driven by an ongoing arms race between host and pathogen in developing new offensive weaponry (Silverstein et al., 2007; Graham et al., 2008).
With the generation of hundreds of secreted peptides during the defense response (Kwon et al., 2008), to what degree could these peptides have developed roles other than their known or predicted toxic effect? Several peptides have been characterized recently that have roles in wounding (systemin; Pearce et al., 1991), self-incompatibility (SCR; Schopfer et al., 1999), stomatal patterning (EPF1; Hara et al., 2007), cellular proliferation and expansion (PSY1; Amano et al., 2007), abscission (IDA; Butenko et al., 2003), pollen formation (TPD1; Yang et al., 2003), root development (RALF; Pearce et al., 2001), shoot meristem development (CLAVATA3; Fletcher et al., 1999), and innate immunity (AtPep1; Huffaker et al., 2006). In the case of systemin, PSY1, SCR, and CLAVATA3, the corresponding peptide receptors have been identified, suggesting a general model of hormone activity via ligand-receptor pairing (Matsubayashi et al., 2001). Although many of these peptides may have evolved independently of those associated with antimicrobial activity, they are all relatively small, probably secreted to the apoplast, and typically subjected to extensive posttranslational processing and/or modification (Lindsey et al., 2002).
Negative regulators of plant defense are essential components that temper the severity of the immune response (Lam, 2004). Several gain-of-function and loss-of-function mutants have revealed genes associated with basal defense and effector-triggered immunity (Büschges et al., 1997; Frye et al., 2001; Hückelhoven et al., 2003; Behn et al., 2004; Jones and Dangl, 2006; Wang et al., 2006; Shen et al., 2007). Many of these genes have been shown to have important roles in overlapping pathways, suggesting a complex interconnected network of regulation. Potentially, negative regulators may control the intensity of programmed cell death, preventing excessive responses from damaging more than just the intended target while not compromising defense. Of the negative regulators cloned in plants, there exist two distinct classes, based on mechanistic similarity to existing defense pathways. Edr1, AtWRKY58, HvWRKY1/2, and BAX inhibitor-1 (BI-1) are examples of well-characterized negative regulators in the known signal and transcriptional activation cascades. By contrast, genes such as the Mlo negative regulator of penetration resistance and several lesion-mimic mutants, such as lsd1, have only recently been characterized with regard to regulatory roles and importance during defense (Büschges et al., 1997; Frye et al., 2001; Hückelhoven et al., 2003; Wang et al., 2006; Shen et al., 2007). Although the former group of genes direct our attention to the complexity and redundancy of the regulatory network of plant defense, the latter set of genes are expanding our understanding of nonhost resistance, biotrophy, and the formation and/or progression of necrosis.
Over the past two decades, barley (Hordeum vulgare) powdery mildew, caused by Blumeria graminis f. sp. hordei (Bgh), has been developed as a model system to investigate host response to obligate fungal biotrophs (Bélanger et al., 2002; Panstruga, 2003, 2004; Schweizer, 2007). Pathogen recognition in barley-Bgh interactions is triggered in a pathogen race-specific manner by genes designated Ml (for Mildew resistance locus; Jørgensen, 1994). Approximately 30 distinct resistance specificities have been identified at the Mla locus; all cloned Mla alleles isolated so far encode coiled-coil, nucleotide-binding site, Leu-rich repeat (CC-NBS-LRR) resistance proteins (Wei et al., 2002; Shen et al., 2003; Halterman and Wise, 2004) that recognize, either directly or indirectly, corresponding fungal effector (AVR) proteins (Ridout et al., 2006). Programmed cell death mediated by MLA proteins occurs after fungal penetration, when primordial haustoria are presumed to secrete AVRa proteins. After recognition, MLA is translocated into the nucleus and binds the WRKY transcription factors HvWRKY1 and HvWRKY2, which instigate a signal cascade leading to the hypersensitive response (Shen et al., 2007). This MLA-AVRa race-specific mechanism of resistance contrasts with the nonspecific penetration resistance mediated by loss-of-function mutations in the seven-transmembrane-protein MLO (Büschges et al., 1997). mlo-mediated resistance during papillae formation is extremely effective, although in the rare case when penetration does occur, Bgh colonization of the leaf tissue proceeds normally (Jørgensen, 1994). Similarly, overexpression of the barley homolog of BI-1 in epidermal cells generates supersusceptibility (Hückelhoven et al., 2003). BI-1 was found to negatively regulate the penetration resistance mediated by mlo and almost restored the penetration efficiency (PE) of Bgh to wild-type levels (Hückelhoven et al., 2003). Thus, negative regulators play a direct role in modulating the defense response of barley to Bgh.
We have characterized a novel family of small peptides, designated blufensins, which are induced during Bgh infection and resemble Cys-rich peptides. We show that one of these, BLUFENSIN1 (BLN1), negatively impacts plant defense during Bgh infection. BLN1 is predicted to be secreted and contains both structural and sequence similarities to the family of knottins. Our results establish a previously unrecognized role for small peptides as negative regulators of plant defense.
RESULTS
Identification of Bln1 from Barley1 GeneChip Expression Profiles
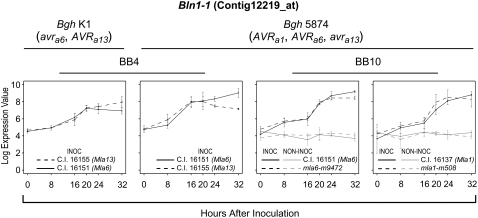
Bln1 was initially identified from a time-course microarray experiment designed to discover genes that had differential patterns of expression associated with either incompatibility or compatibility in barley-powdery mildew interactions (Caldo et al., 2004). Using the MIXED procedure in SAS, a contrast statement was developed to test the expression levels in incompatible pairings specified by Mla6-AVRa6 and Mla13-AVRa13 compared with compatible interactions determined by Mla6-avra6 and Mla13-avra13. Bln1 (represented by Barley1 GeneChip probe set Contig12219_at) was one of 22 genes found to be differentially expressed at a threshold P value of <0.0001 and a false discovery rate of <7% (Fig. 1, experiment BB4; Caldo et al., 2004). To further characterize Bln1 response in the scope of the barley transcriptome, we extended the analysis of Caldo et al. (2004) to another large expression-profiling experiment involving nearly isogenic lines C.I. 16151 (Mla6) and C.I. 16137 (Mla1) versus their respective loss-of-function mutants, mla6-m9472 and mla1-m508 (Fig. 1, experiment BB10). In addition to challenge with avirulent Bgh isolate 5874 (AVRa1, AVRa6), the BB10 experimental design included noninoculated samples, which allowed us to observe, conclusively, significant Bln1 induction over the 0- to 32-h time course in both incompatible and compatible pairings. The association with Bgh invasion (Caldo et al., 2004, 2006), in addition to strong induction by Fusarium graminearum (the causal agent of Fusarium head blight; Boddu et al., 2006, 2007), provided indirect evidence that Bln1 plays a role in mediating defense responses to fungal pathogens.
Figure 1.
Time-course expression profiles of Bln1-1 (Contig12219_at) in barley-Bgh interactions. The left two panels display data from BB4, an experiment described by Caldo et al. (2004), in which nearly isogenic barley lines harboring the contrasting Mla alleles, Mla6 and Mla13, were challenged in pairwise combinations with the alternately virulent and avirulent Bgh isolates 5874 (AVRa1, AVRa6, avra13) and K1 (AVRa1, avra6, AVRa13). A mixed linear model analysis (Wolfinger et al., 2001) using the SAS mixed procedure was conducted to identify genes whose average pattern of expression in one host-pathogen interaction category (e.g. compatibility) differed significantly from its average pattern of expression in its contrasting category (e.g. incompatibility). Time-specific differences between the average expressions (0, 8, 16, 20, 24, and 32 hai) were tested for equality using an F statistic (Caldo et al., 2004). The BB10 experiment (shown in the right two panels) compared wild-type (Mla) plants and derived loss-of-function deletion mutants inoculated with Bgh isolate 5874 (AVRa1, AVRa6). Identical noninoculated plants were included for each treatment. Normalized average signal intensities and se values were calculated based on three independent replications for both experiments. Derivations of se values are shown for illustration, as each contrast uses pooled variances when testing for significant differences between incompatible versus compatible interactions.
Bioinformatic Classification of the Blufensin Family of Small Peptides
A BLASTn search using Bln1 among the assembled ESTs used to create the Barley1 GeneChip (HarvEST:Barley assembly 21; http://138.23.191.142/hweb/; Altschul et al., 1990; Close et al., 2004) identified the family member Bln2, represented by Barley1 probe set Contig26496_at. Strong induction of Bln2 was observed after Bgh inoculation, but without a differential time-course expression pattern associated with incompatibility or compatibility, as was seen with Bln1. Bln2 was induced, however, upon challenge with Puccinia graminis f. sp. tritici, causal agent of stem rust (Zhang et al., 2008), whereas Bln1 was not. Conversely, it is possible that the observed noninduction of Bln1 in response to P. graminis f. sp. tritici was due to poor hybridization by allele-specific probes in Contig12219_at (see “Characterization of Bln1 Transcripts” below).
Proteins encoded by both genes were then examined using the suite of motif recognition software orchestrated via InterProScan (Quevillon et al., 2005). Two matches were found: a localization signal peptide and a transmembrane domain, both positioned in the N-terminal region of the predicted protein. TargetP 1.1 and WoLF PSORT II were used for signal peptide prediction, with both predicting secretion and a cleavage site between amino acid residues 29 and 30 (Nielsen et al., 1997; Horton et al., 2006; Emanuelsson et al., 2007). The best match to the signal peptide was Pa-AMP-1 (for Antimicrobial Protein-1) from Phytolacca americana (common pokeberry), a member of the knottin family of antimicrobial peptides (Liu et al., 2000).
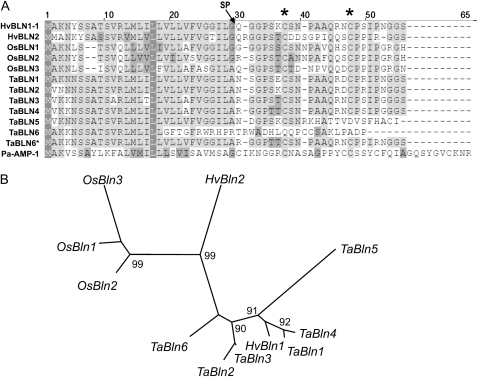
Next, we compared BLN1 and BLN2 to identify shared motifs or domains that may point to a known protein family. The use of InterProScan, BLAST, and the PANTHER database of motifs on all existing sequence information provided no information on the C-terminal regions of these two family members (Thomas et al., 2003; Mi et al., 2005). There were, however, several shared features between BLN1 and BLN2, including extensive amino acid conservation in the predicted signal peptide and cleaved peptide, the presence of only two Cys residues, and a single intron in a conserved position between the Cys residues. The last two features are hallmarks of small Cys-rich antimicrobial peptides, which generally have an even number of Cys residues (for the formation of disulfide bonds) and a conserved intron approximately 150 nucleotides in length positioned near the signal peptide/cleaved peptide border (Graham et al., 2008). Antimicrobial peptides are conserved across all living organisms; therefore, a literature search was performed on all small peptides found in vertebrates and invertebrates, whether or not they have been shown to have antimicrobial activity. We found that small peptides with only two Cys residues are uncommon; in fact, only six have been identified to date, from cow, insects, and frogs (Zasloff, 2002). Among these, the number of amino acids between the Cys residues range from five to seven, compared with eight and nine found in BLN1 and BLN2, respectively. The conservation of Cys positions and signal peptide among BLN1, BLN2, and Pa-AMP-1 suggests a possible evolutionary connection between the blufensin and knottin gene families (Fig. 2A).
Figure 2.
Multiple sequence alignment and phylogeny of the blufensin family. A, Alignment of protein sequences of blufensin family members in barley, rice, and wheat. TaBln6 has a single nucleotide insertion that generates a frameshift. TaBln6* represents the sequence that would result if the insertion were not present. SP designates the putative position of the signal peptide cleavage site, and asterisks at top designate the positions of the Cys residues. B, Unrooted phylogeny based on an alignment of the blufensin family coding sequences with 1,000 bootstraps. Support over 90% is shown at branch points in the phylogeny. The DNA alignment was used due to the poor bootstrap support generated from using the short protein sequence.
We next compared the blufensins with homologs in closely related species to determine the degree of residue conservation in this small peptide family. A tBLASTn search using BLN1 revealed three and six family members in rice (Oryza sativa) and wheat (Triticum aestivum), respectively (Fig. 2; Supplemental Table S1). No significant sequence similarity was found in available genomic sequences of species outside of the BEP clade of the Poaceae (grass) family, namely maize (Zea mays) and sorghum (Sorghum bicolor). Moreover, within the BEP clade, no significant similarity was found in the 4× Brachypodium sequence (as available on October 1, 2008), suggesting that preservation of blufensins within this clade may be incomplete. Multiple sequence alignment revealed high similarity in the signal peptide region and conservation of specific residues in the cleaved peptide region (Fig. 2). The identification of two genes in diploid barley (Triticeae H genome) correlates with the six found in hexaploid wheat (Triticeae A, B, and D genomes), based on available EST data. As illustrated in Figure 2B, phylogenetic analysis of this family grouped the rice blufensins distinct from the wheat. Curiously, HvBln1 grouped with its homologs in wheat, whereas HvBln2 occupied a branch distinct from both rice and wheat. Several of the wheat blufensins are clustered together with barley blufensins, indicating significant sequence conservation.
Characterization of Bln1 Transcripts
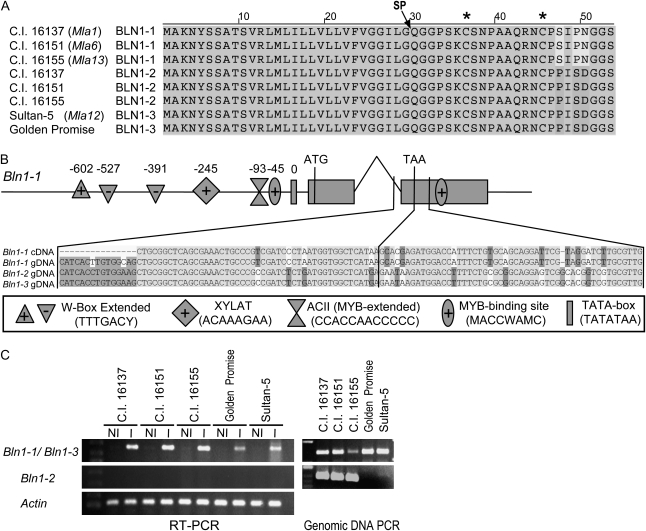
We also investigated Bln1 expression profiles in a third microarray data set (BB2) involving cv Sultan-5 (Mla12) as well as mla12 and rar1 loss-of function mutants derived from the Sultan-5 genotype (Torp and Jørgensen, 1986; Freialdenhoven et al., 1994; Caldo et al., 2006; retrieved from BarleyBase/PLEXdb; http://www.plexdb.org/). Rather surprisingly, induced transcript accumulation was not observed in Sultan-5 or its mutant derivatives. We suspected that this was due to the divergence of Bln1 sequences in Sultan-5, which would interfere with efficient hybridization to Barley1 GeneChip probe sets. Therefore, additional Bln1-homologous cDNA and genomic clones were isolated from C.I. 16137 (Mla1), C.I. 16151 (Mla6), C.I. 16155 (Mla13), Sultan-5 (Mla12), and Golden Promise by reverse transcription (RT)-PCR and inverse PCR, respectively. Genomic DNA sequence analysis revealed that there are two copies of Bln1 in C.I. 16137, C.I. 16151, and C.I. 16155, all of which are nearly isogenic derivatives of cv Manchuria (Moseman, 1972). These two copies were designated Bln1-1 and Bln1-2. As illustrated in Figure 3A, Bln1-1 is highly similar to Bln1-2, except that Bln1-2 has three single nucleotide polymorphisms within the open reading frame (ORF), which generate three nonsynonymous changes in the C-terminal end of the predicted protein. A single-copy chimera of Bln1-1 and Bln1-2 is contained within Sultan-5 and Golden Promise, which was designated Bln1-3 (Fig. 3A). Verification of copy number in each line was confirmed by Southern-blot analysis (data not shown). A conserved 124-nucleotide GT-AG-type intron was identified, with only two single nucleotide polymorphisms within the intron between Bln1-1 versus Bln1-2 and Bln1-3 (Fig. 3B). The 3′ untranslated regions (UTRs) of Bln1-2 and Bln1-3 were identical but highly dissimilar to Bln1-1 (data not shown). The coding region, intron, and 3′ UTR of Bln1-3 were identical to those of Bln1-2, yet the promoter of Bln1-3 was the same as that of Bln1-1 (Fig. 3B; data not shown).
Figure 3.
Comparison of Bln1 sequences and transcript accumulation. A, Amino acid alignment of Bln1-1, Bln1-2, and Bln1-3 alleles/paralogs. Gray shaded boxes indicate identity across all eight sequences. SP designates the putative position of the signal peptide cleavage site, and asterisks at top designate the positions of the Cys residues. B, Bln1-1 gene model with promoter analysis of a genomic DNA fragment cloned via inverse PCR. Gray boxes indicate exons, with a single intron between exon 1 and 2. Symbols shown in the box at bottom indicate the positions of several promoter elements associated with defense (W-box, WRKY, MYB, P-box) and xylem- and root-specific expression. Alignment of genomic DNA of Bln1-1 and Bln1-2 and cDNA of Bln1-1 shows the extensive nucleotide divergence between these paralogs, beginning near the end of the intron. C, Differential transcript accumulation of Bln1 paralogs upon inoculation with Bgh isolate 5874. RT-PCR was performed on RNA isolated from seedling leaves 24 h after Bgh inoculation (I) or from noninoculated controls (NI). Actin was used as the internal control in all samples. The genomic DNA PCR results shown demonstrate the existence of different paralogs in different genotypes.
As the Barley1 GeneChip could not measure transcript accumulation of Bln1-2 and Bln1-3, we designed primers (Supplemental Table S2) based on the newly discovered sequence polymorphisms to perform copy-specific RT-PCR of all three putative alleles or paralogs in response to Bgh. As illustrated in Figure 3C, Bln1-3-specific transcripts were amplified from RNA isolated from Bgh-inoculated leaves in all five cultivars, but no amplification product was detected from RNA isolated from noninoculated plants. However, when using primers specific for Bln1-2, no PCR product was detected from RNA isolated from either inoculated or noninoculated tissues. All sequenced ESTs in GenBank are identical to Bln1-1 (Supplemental Table S1); therefore, our working hypothesis is that Bln1-1/3 harbors a functional promoter, while Bln1-2 may have a nonfunctional promoter or one not associated with leaf or Bgh-induced expression.
Promoter Analysis of Bln1-1
The 5′ upstream regions of Bln1-1, Bln1-2, and Bln1-3 were isolated using inverse PCR from genomic DNA of cv C.I. 16151 and cv Golden Promise. As shown in Figure 3B and Table I, several common motifs associated with defense (W-box, WRKY, MYB, P-box), and xylem- and root-specific expression were identified in the upstream region of Bln1-1. Of those associated with defense, three WRKY transcription factor-binding sites or W-boxes (TTTGACY) were found, at −602, −526, and −391 bases from the TATA box (Rushton et al., 1996; Eulgem et al., 2000). Additionally, three MYB-binding sites (MACCWAMC) were found, at −92 and −45 nucleotides from the TATA box and within the 3′ UTR of Bln1-1 (Sablowski et al., 1994; Tamagnone et al., 1998). The xylem-specific expression elements ACII (CCACCAACCCCC) and XYLAT (ACAAAGAA) are located at −91 and −245 bases from the TATA box, respectively. ACII is an extended MYB-binding site motif with additional specific nucleotides that generate xylem-specific expression (Patzlaff et al., 2003; Gomez-Maldonado et al., 2004). Lastly, nine root-specific expression elements (ATATT) were found within −600 to −500 bases from the TATA box. The presence of these elements is consistent with EST evidence of expression in root tissue (Supplemental Table S1). All described motifs were highly oversampled with respect to prevalence in the rice genome (Table I). Specifically, the number of observed ACII and W-box motifs was only greater in less than 0.01% and 1.6% of all rice genes, respectively (quantile estimate based on rice V5).
Table I.
Analysis of sequence motifs in the Bln1-1 promoter compared with rice
| Sequence Motif | No. of Motifs
|
||
|---|---|---|---|
| Statistically Expecteda | Observed in Rice (Mean)b | Observed in Bln1-1 Promoter | |
| ACII (CCACCAACCCCC) | 0.00011 | 0.00067 | 1 (99.9%)c |
| MYB (MACCWAMC) | 0.22 | 0.35 | 2 (93.5%) |
| Root specific (ATATT) | 1.78 | 4.01 | 9 (89.5%) |
| W-box (TTTGACY) | 0.22 | 0.35 | 3 (98.4%) |
| XYLAT (ACAAAGAA) | 0.028 | 0.068 | 1 (93.5%) |
Expected number of motifs based on length of the Bln1-1 promoter.
Observed number of motifs in the rice genome per gene.
Percentage of promoters in the rice genome that have a lower number of motifs compared with Bln1-1.
Functional Analysis of Bln1 via Barley Stripe Mosaic Virus-Induced Gene Silencing
A New DNA Bombardment-Based Silencing System for the Triticeae
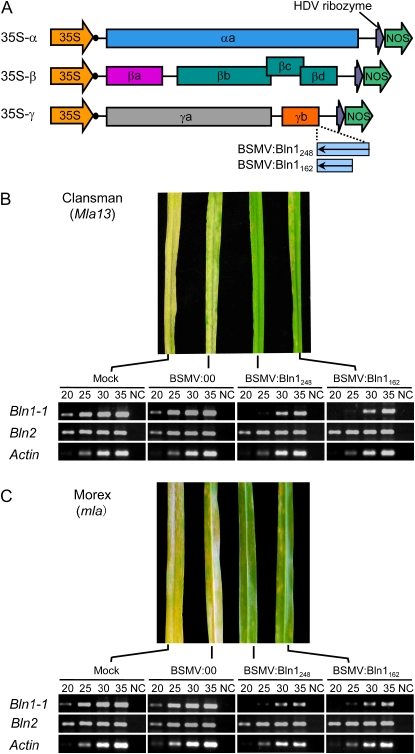
In recent years, virus-induced gene silencing (VIGS) has emerged as a powerful reverse genetics tool for the functional analysis of gene candidates in both model and crop plant species. In monocots, Brome mosaic virus has been utilized for functional genomics studies in rice and maize (Ding et al., 2006), whereas Barley stripe mosaic virus (BSMV) has been used for barley and wheat (Holzberg et al., 2002; Lacomme et al., 2003; Hein et al., 2005; Scofield et al., 2005). In these previous studies, BSMV vectors were under the control of the T7 promoter, which requires in vitro transcription to make infectious RNA transcripts for plant inoculation. We developed a modified BSMV-VIGS system using particle bombardment of DNA into barley seedlings, which eliminates the in vitro transcription step and is more amenable to high-throughput studies. As illustrated in Figure 4A, the new BSMV-VIGS DNA vector set consists of independent BSMV:α, BSMV:β, and BSMV:γ clones under the control of the cauliflower mosaic virus 35S promoter. Silencing of Phytoene desaturase (PDS; Holzberg et al., 2002) was used to quantify the efficacy of silencing with this approach, resulting in approximately 80% of the newly inoculated plants exhibiting a photobleaching phenotype.
Figure 4.
BSMV-VIGS of Bln1 in Clansman (Mla13) and Morex (mla). A, Schematic representing the DNA-based BSMV-VIGS constructs. The three subgenomes of BSMV are under the control of the cauliflower mosaic virus 35S promoter. Resulting transcripts are cleaved at the 3′ terminus by the HDV ribozyme. B, Clansman (Mla13) plants were subject to four treatments: mock (carborundum phosphate buffer), empty vector (BSMV:00), and test constructs (BSMV:Bln1248 and BSMV:Bln162). Plants were inoculated with Bgh at 12 d after treatment and photographed at 7 dai. Semiquantitative RT-PCR was performed with Bln1-1- and Bln2-specific primers to detect mRNA degradation of targeted transcripts. The lanes designated 20, 25, 30, and 35 indicate the number of amplification cycles performed for each sample. For each sample, lane NC shows the results of 35 cycles of PCR without RT, as a negative control. Actin transcripts served as a quantitative control for each sample. C, BSMV-VIGS of Bln1 in Morex. Protocols were as described for B.
Silencing of Bln1 Enhances Plant Resistance in Compatible Interactions
To examine the role of Bln1 in the barley defense response to Bgh, we used the bombardment-based BSMV-VIGS approach to down-regulate Bln1 gene expression. Figure 4A illustrates two Bln1 cDNA fragments of different lengths inserted downstream of the stop codon of γb, designated BSMV:Bln1248 and BSMV:Bln1162. Wild-type γ-BSMV:00 was used as a negative control. After a survey of BSMV-bombarded cultivars, Clansman (Mla13) and C.I. 16151 (Mla6) were chosen for VIGS assays, since silencing of PDS in these genotypes resulted in fewer virus infection symptoms but significant photobleaching. Plants were inoculated with Bgh 5874 (avra13, AVRa6) 12 d after BSMV treatment, and third leaves were scored for Bgh infection type 7 d later.
Three independent experiments with Clansman infected with Bgh 5874 demonstrated that silencing Bln1 visibly enhanced resistance. Microscopic inspection was carried out to determine PE, as calculated by the percentage of total conidiospores that produced haustoria and secondary hyphae. As shown in Figure 4B, BSMV:Bln1248- and BSMV:Bln1162-inoculated plants were significantly less susceptible at 7 d after inoculation (dai) than the inoculated BSMV:00 and non-BSMV-inoculated control plants (mock), resulting in a PE of 21% for BSMV:00, compared with 11% in BSMV:Bln1248-silenced plants and 12% in BSMV:Bln1162-silenced plants, respectively. In incompatible interactions, C.I. 16151 plants were fully resistant in BSMV:Bln1248- and BSMV:Bln1162-infected plants, with no significant difference observed between inoculated BSMV:00 and mock control plants. When C.I. 16151 test plants were inspected microscopically, no Bgh secondary hyphae were detected up to 7 dai in either silenced or control plants. Thus, the significant reduction in susceptibility in compatible interactions suggests that Bln1 could function as a negative regulator of barley defense response to Bgh infection.
Semiquantitative RT-PCR of Bln1 and Bln2 mRNA from VIGS-Treated Plants
Transcript accumulation of Bln1-1 was assayed to monitor the level of Bln1 gene silencing. The third leaves of BSMV-treated plants were used for RT-PCR assays 24 h after Bgh inoculation. Barley Actin mRNA was used as an internal quantitative control for all samples (Halterman et al., 2003). Using Bln1-1-specific primers (Supplemental Table S2), semiquantitative RT-PCR revealed the reduction of Bln1-1 transcripts in both BSMV:Bln1248- and BSMV:Bln1162-infected leaves compared with inoculated BSMV:00 and mock-inoculated plants (Fig. 4, B and C). There were no detectable amplicons at 20 cycles in Bln1-1-silenced plants. However, amplicons could be observed when using 25 cycles or more, indicating that silencing efficiency is not 100%. This is consistent with the observed heterogeneous silencing pattern observed in BSMV:PDS plants (Scofield et al., 2005). To check for off-target silencing of associated blufensin family members, Bln2-specific primers (Supplemental Table S2) were used for semiquantitative RT-PCR on the same RNA samples. Bln2 mRNA levels were equivalent in BSMV:Bln1248-, BSMV:Bln1162-, BSMV:00-, and mock-inoculated control plants, indicating a low probability of cross-silencing with Bln1-1 (Fig. 4, B and C). These results imply that the reduced susceptibility to Bgh in BSMV-VIGS-treated plants is due to the suppression of Bln1 and not to silencing of its family member, Bln2.
Bln1-1 Is Highly Inducible in All Barley Genotypes Tested, But Silencing Consequences Differ
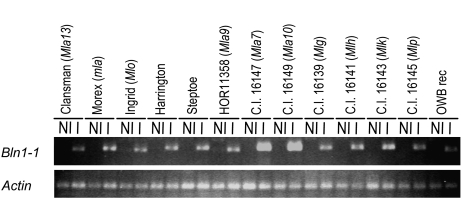
To further understand the role of Bln1 in barley defense response to Bgh, we silenced Bln1 in 12 additional barley genotypes and recorded the resulting infection types in compatible interactions with Bgh 5874 (Table II). Barley cv Black Hull-less seedlings were bombarded with BSMV:Bln1248, BSMV:Bln1162, and BSMV:00 constructs. Seven days after bombardment, BSMV-infected leaves that showed a visible stripe mosaic phenotype were utilized to recover recombinant virions, which, in turn, were uniformly applied to test plants by mechanical inoculation. After 12 d of silencing, plants were inoculated with the Bgh 5874 isolate. Seven days after Bgh inoculation, six genotypes, including Morex (mla; Fig. 4C), exhibited a significant reduction in susceptibility in BSMV:Bln1248- and BSMV:Bln1162-transformed plants compared with BSMV:00 and mock plants, whereas the other seven were not significantly different from the BSMV:00 control (Table II). Moreover, silencing generated significantly reduced susceptibility in Bln1-silenced Ingrid (Mlo) plants at 7 dai with Bgh (Table II), while silencing in mlo-5 BC7 Ingrid had no effect. When plants were inspected microscopically, we observed a reduction in PE from 38% in BSMV:00 plants to 22% in BSMV:Bln1248-silenced and 28% in BSMV:Bln1162-silenced Mlo plants. Therefore, silencing of Bln1 generates reduced susceptibility in the presence of wild-type Mlo.
Table II.
Bgh 5874 infection types after BSMV-VIGS of Bln1 in 13 compatible barley-Bgh interactions
| No. | Genotypea | Mock | BSMV:00 | BSMV:Bln1248 | BSMV:Bln1162 |
|---|---|---|---|---|---|
| 1 | Clansman (Mla13) | 3–4b | 3 | 2 | 2 |
| 2 | Morex (mla) | 3–4 | 3–4 | 2 | 2 |
| 3 | Ingrid (Mlo) | 4 | 4 | 2 | 2 |
| 4 | Harrington | 3–4 | 3–4 | 1–2 | 1–2 |
| 5 | Steptoe | 3–4 | 3–4 | 2 | 2 |
| 6 | HOR11358 (Mla9) | 4 | 3–4 | 1–2 | 1–2 |
| 7 | C.I. 16147 (Mla7) | 3 | 3 | 3 | 3 |
| 8 | C.I. 16149 (Mla10) | 3 | 3 | 3 | 3 |
| 9 | C.I. 16139 (Mlg) | 4 | 4 | 4 | 4 |
| 10 | C.I. 16141 (Mlh) | 4 | 4 | 4 | 4 |
| 11 | C.I. 16143 (Mlk) | 3 | 3 | 3 | 3 |
| 12 | C.I. 16145 (Mlp) | 3 | 3 | 3 | 3 |
| 13 | OWB rec | 4 | 4 | 4 | 4 |
Genotypes in rows 1 to 6 exhibited significantly reduced susceptibility, while genotypes in rows 7 to 13 were not significantly different after silencing. Order of genotypes is identical to that in Figure 5.
Plants were inoculated with Bgh isolate 5874 (AVRa1, AVRa6). The rates of severity of Bgh infection are presented as 0 to 4, indicating levels of sporulation from completely resistant (0) to completely susceptible (4).
The above results contrast with the level of Bln1-1 transcript accumulation at 24 h after inoculation (hai) in all 13 genotypes, as demonstrated by RT-PCR analysis (Fig. 5). It is possible that the ability of the host plant to tolerate virus accumulation interfered with the efficiency of BSMV-induced gene silencing, since the seven genotypes with no significant reduction in susceptibility to Bgh exhibited significant necrosis along the mid and lateral veins in the upper half of the second leaf, a strong BSMV infection symptom. Specific cultivars must be utilized that provide a suitable genetic background to tolerate the substantial levels of BSMV accumulation that are required to elicit a significant VIGS response (Hein et al., 2005). Thus, although Bln1-1 is highly expressed in all genotypes upon inoculation of Bgh, there were diverse phenotypic effects of attempted Bln1 silencing in different genotypes.
Figure 5.
RT-PCR to detect Bln1 transcript accumulation in 13 barley genotypes. RNA was isolated from seedling leaves at 24 h after Bgh inoculation (I) or from noninoculated controls (NI). Actin transcripts served as a quantitative control for each sample. Order of genotypes is identical to that in Table II.
Overexpression of Bln1 Results in Hypersusceptibility to Bgh
In light of the enhanced resistance to Bgh in compatible interactions due to Bln1 silencing, we hypothesized that overexpression of Bln1 should render comparable epidermal cells supersusceptible. To test this, we utilized single-cell-transient overexpression of Bln1 in barley epidermal cells (Shirasu et al., 1999). The full-length Bln1-1 ORF was cloned into the vector pUbi:Nos to create the expression construct pUbi:Bln1. The pUbi:Bln1 plasmid was then cobombarded with the pUGN GUS expression vector (Nielsen et al., 1999) into Clansman (Mla13) epidermal cells and subsequently challenged with the virulent Bgh isolate 5874 (avra13). Control bombardments were performed with the pUGN reporter construct alone. Fungal PE was calculated as the ratio of GUS-marked cells exhibiting elongating secondary hyphae to the total transformed cells attacked by Bgh.
As shown in Table III, generalized linear mixed-model analyses for three independent experiments revealed that the formation of elongating secondary hyphae in compatible interactions (an indicator of PE) was significantly more likely for constructs pUGN + pUBI:Bln1 than for construct pUGN alone (P = 0.0028). Overexpression of Bln1 in C.I. 16151 (Mla6) cells did not compromise resistance in incompatible interactions with Bgh isolate 5874 (AVRa6). Combined with the BSMV-VIGS experiments above, results from the overexpression experiments provide additional support for the hypothesis that Bln1 negatively regulates basal defense but does not compromise effector-triggered Mla6-mediated race-specific resistance.
Table III.
Results of overexpression of Bln1 in cultivars Clansman (compatible) and C.I. 16151 (incompatible) after inoculation with Bgh 5874
| Cultivar (R Gene) | Construct |
Bgh 5874 (AVRa6, avra13)
|
|||
|---|---|---|---|---|---|
| Total No. of GUS Cells with Conidiosporea | Total No. of GUS Cells with Elongating Secondary Hyphaea | PEb | P (Control Versus Bln1)c | ||
| % | |||||
| Clansman (Mla13) | Ubi:GUS | 281 | 84 | 29.9 | |
| Clansman (Mla13) | Ubi:GUS + pUBI:Bln1 | 505 | 273 | 54.1 | 0.0028 |
| C.I. 16151 (Mla6) | Ubi:GUS | 263 | 0 | 0 | |
| C.I. 16151 (Mla6) | Ubi:GUS + pUBI:Bln1 | 372 | 1 | 0.3 | n.s. |
Raw numbers indicate the combined results of three independent experiments.
PE represents GUS-stained cells with secondary hyphae among the total number of GUS-stained cells with spores attached.
P values were obtained using a generalized linear mixed model to test for significant differences in secondary hyphae formation by comparing the test constructs versus the empty vector negative control. n.s., Not significant.
DISCUSSION
BLN1 Plays a Key Role for Powdery Mildew Susceptibility in Barley
We have shown here that BLN1, a small peptide of the novel blufensin gene family, negatively impacts the defense response to barley powdery mildew. Based on the expression profiling results of Caldo and colleagues (2004), Bln1 was one of several genes that exhibited an equivalent pattern of transcript accumulation in both incompatible and compatible interactions during germination of Bgh conidiospores and formation of appressoria (Fig. 1). However, during establishment of the perihaustorial interface between penetrating Bgh and host epidermal cells, divergent expression of these transcripts occurred, in which compatible interactions led to lower accumulation of transcripts compared with paired incompatible interactions.
In gene-for-gene-mediated incompatible interactions, the increase in Bln1 transcript accumulation could be interpreted to imply that Bln1 transcript accumulation is intimately associated with Bgh defense. However, lower Bln1 transcript accumulation in compatible interactions would suggest that its expression was influenced by Bgh invasion and that its reduction is correlated with increased susceptibility (Caldo et al., 2004). In fact, we observed the opposite. Decreased susceptibility in compatible interactions was observed via BSMV-VIGS-mediated suppression of Bln1, whereas susceptibility was enhanced after Bln1 overexpression. Bgh-induced Bln1 transcript accumulation was evident in all 18 barley genotypes tested, implicating a conserved mechanism of regulatory control. Bln1 silencing enhanced plant resistance in compatible interactions, regardless of the presence or absence of Mla CC-NBS-LRR alleles, indicating that BLN1 can function in a R-gene-independent manner. Based on the phenotypic observations described above, we propose two hypotheses that model the function of BLN1: (1) BLN1 is a negative regulator of penetration defense, resulting in the attenuation of host defenses that retard fungal infection, similar to mlo-mediated resistance; and (2) BLN1 is a susceptibility factor that is required for promoting fungal establishment, penetration, and/or colonization. Indeed, these hypotheses are not mutually exclusive, because the difference could be considered semantic in that a negative regulator of defense could be considered one class of susceptibility factors.
Host Accessibility and Susceptibility Factors in Plant Defense
The possibility that BLN1 has been recruited by Bgh to take advantage of host factors normally utilized for basic metabolism and defense is not without reason. To acquire nutrients from host cells, obligate biotrophic fungi have evolved mechanisms to secrete effectors to suppress host defenses (Dodds et al., 2004; Catanzariti et al., 2006; Wang et al., 2007) and to induce host susceptibility genes (Schulze-Lefert and Panstruga, 2003; Hückelhoven, 2005). In Arabidopsis (Arabidopsis thaliana) and barley, several host susceptibility factors have been identified for powdery mildew, but how pathogens utilize these host genes remains unclear (Schultheiss et al., 2002, 2003; Hückelhoven et al., 2003; Hückelhoven, 2005). The observation of proteins similar to the plant blufensin family within many Ascomycota species may indicate functional mimicry (Abramovitch et al., 2006). To determine if convergent coevolution of the blufensin family with host pathogens might exist, both BLAST and pattern matching using regular expression identified conserved family members (M. Moscou and R. Wise, unpublished data). Interestingly, matches were found in the genomic sequences of the two grass fungal pathogens, Magnaporthe grisea and Blumeria graminis f. sp. hordei, but no significant homology was found outside of the Ascomycota phylum. If these fungal proteins are expressed in hyphae and at the perihaustorial interface, they may suggest a role in plant susceptibility, the establishment of feeding structures, and/or biotrophic interactions between the plant and the pathogen (Dodds et al., 2004). An example of functional mimicry is provided by a root-knot nematode-secreted protein found to have high similarity to the peptide hormone CLAVATA3, which binds to CLAVATA1 to stimulate root formation (Huang et al., 2006). Alternatively, the possibility exists that Bgh induces a gene in barley that acts as a stimulant to fungal growth. This notion of the induction of host susceptibility factors and/or functional mimicry of plant and pathogen signaling peptides presents a coevolutionary model of selection for and against factors that mediate this biotrophic interaction.
Negative Regulators in Plant Defense
Our early understanding of disease defense came via studies involving R gene-mediated resistance, also known as effector-triggered immunity, in which a rapid and evolutionarily adapted response is generated after recognition of an invading pathogen. This is in contrast to pathogen-associated molecular patterns-triggered immunity or basal defense, which expresses a nonspecific and broader type of resistance response. Negative regulation of the basal defense pathway prevents unchecked potentiation of the response and deleterious effects on normal cell functions (Alexander and Hilton, 2004; Ge et al., 2007). As we demonstrated that Bln1 did not require a functional effector-triggered resistance, we surmise that Mla-mediated postpenetration resistance is epistatic to the negative regulation of Bln1-mediated suppression.
MLO, as a negative regulator of penetration resistance, but not Mla-mediated hypersensitive response, is essential for compatibility to all known Bgh isolates (Büschges et al., 1997; Piffanelli et al., 2002). Hypothesized to be a host susceptibility factor, it is believed that MLO is recruited by Bgh to diminish the plant defense response (Büschges et al., 1997; Devoto et al., 1999). A small GTP-binding protein of the barley RAC family is associated with MLO-mediated suppression of Bgh defense (Schultheiss et al., 2002), and RACs can regulate subcellular gradients of Ca2+ (Schultheiss et al., 2003). A domain that mediates a Ca2+-dependent interaction with calmodulin has been identified in MLO, and loss of calmodulin binding inhibits the capacity for MLO to negatively regulate Bgh defense (Kim et al., 2002). Like HvCaM3, silencing of Bln1 also enhanced resistance to Bgh in plants harboring wild-type Mlo but not in mlo-5 mutants, suggesting that BLN1 functions in parallel with or upstream of MLO to modulate penetration resistance. Preliminary experiments using a BLN1-GFP fusion construct bombarded into barley epidermal cells indicated that BLN1 was undetectable in the nucleus and located mainly in the cytoplasm and the cell periphery. By contrast, the GFP control was mainly localized in the nucleus and the cytoplasm (Y. Meng and R. Wise, unpublished data). Computational analysis of the BLN1 signal peptide predicts that BLN1 is secreted into the apoplast, which is consistent with these early fusion assays. MLO is localized to the plasma membrane (Devoto et al., 1999). Indeed, if Bln1 is secreted into the apoplast, it may act as a ligand to generate a signal transduction cascade, influencing Bgh accessibility.
Alternative Modes of Action of Bln1 Function
Several functional models can account for the process by which Bln1 mediates the balance between susceptibility and resistance. These are based on a specific tissue or compartment in which BLN1 functions. If BLN1 were localized to the cytoplasm, it may act as an oxidation sensor (Cumming et al., 2004). Normally, the reductive environment of the cytoplasm does not permit stable disulfide bonds. But with the formation of ROS, disulfide binding is known to alter the structures of proteins involved in several pathways, including the master regulator of defense, NPR1, which loses intermolecular disulfide bonds after being catalyzed by thioredoxins (Cumming et al., 2004; Tada et al., 2008). In this scenario, Bln1 could activate a negative regulatory response due to the formation of a disulfide bond after exposure to ROS formed at any of several stages of the defense response.
High-Throughput DNA-Based BSMV-VIGS Promotes Functional Analysis of Genes Associated with Defense
Recently, a DNA-based Bean pod mottle virus (genus Comovirus) was developed as an efficient tool for a wide range of applications in soybean (Glycine max) functional genomics (Zhang et al., 2009). Accordingly, functional analysis of barley genes associated with resistance to Bgh was facilitated by the development of a similar, DNA-based BSMV-VIGS system. Relative to the commonly used RNA-based BSMV-VIGS, which uses mechanical inoculation of in vitro-generated transcripts (Hein et al., 2005; Scofield et al., 2005), the biolistic-based delivery system is easier to handle and cost efficient. Since the experimental substrate is DNA, as opposed to RNA, constructs are more stable and increase the probability of obtaining silenced plants. Using biolistic transfer of wild-type BSMV constructs to barley cv Black Hull-less, 80% to 100% of the plants normally display a BSMV infection phenotype at 7 d after bombardment.
BSMV-based VIGS constructs can be passaged through the barley host, thus inexpensively amplifying recombinant virions. Since we usually test the effect of silencing on multiple plants from one cultivar or multiple cultivars, utilization of this traditional “plant pathology” step makes this system more amenable to high-throughput applications. An intermediate mechanical infection step has also been adopted to infect Arabidopsis using the sap of Nicotiana benthamiana infected with Tobacco rattle virus (Lu et al., 2003) and to infect rice using sap from barley infected with Brome mosaic virus (Ding et al., 2006). In our hands, recombinant virions from one infected Black Hull-less plant could be used to test approximately 30 additional plants of different cultivars. The one drawback is that the additional 7 d required for the secondary BSMV infection can result in instability of the recombinant inserts during viral replication (Bruun-Rasmussen et al., 2007). To verify the stability of our constructs, RT-PCR was conducted on RNA isolated from BSMV-VIGS-treated leaf tissue at 24 hai with Bgh. About 50% of BSMV:Bln1248- and BSMV:Bln1162-derived transcripts contained the Bln1 inserts (data not shown). Even so, these plants displayed significant reduction in susceptibility (Fig. 4).
The BSMV vector was useful in many different cultivars. Since BSMV has a broad host range among the grasses (e.g. oat [Avena sativa], maize, and wheat), we anticipate that this system could be used as a powerful tool for functional studies in a wide range of economically important plant species. In this study, this effective reverse genomics tool was used to characterize a novel Blufensin family member, Bln1, which negatively impacts barley basal defense response to Bgh. Other Bln family members in barley, rice, and wheat may also have associated functions in crop defense response to biotrophs. Functional identification of this novel gene family may shed light on mechanisms that are required for the regulation of grass disease resistance.
CONCLUSION
The development of new technology that translates primary research in model systems to agronomic traits of interest in crop species is now feasible. The high-throughput silencing assay permitted our investigation of the negative regulatory role of BLN1 during disease defense, implicating another protein in addition to MLO, BI-1, and RACB. As these and new regulators are identified, our understanding of the delicate balance between resistance and susceptibility will broaden to a spectrum of quantitative regulatory network responses.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Fungal Isolates
For functional analysis, seedlings of barley (Hordeum vulgare) lines C.I. 16151 (Mla6), C.I. 16137 (Mla1), C.I. 16155 (Mla13), Clansman (Mla13), Sultan-5 (Mla12), Golden Promise, C.I. 16147 (Mla7), C.I. 16149 (Mla10), HOR11358 (Mla9), C.I. 16143 (Mlk), C.I. 15229 (Steptoe), Ingrid (Mlo), Harrington, C.I. 16139 (Mlg), OWB rec, C.I. 16145 (Mlp), C.I. 16141 (Mlh), mlo-5 BC7 Ingrid, and C.I. 15773 (Morex) were grown in a temperature-controlled greenhouse with supplemental lighting. Following BSMV-VIGS bombardment/mechanical inoculation, plants were transferred to a temperature-controlled growth chamber with a 16-h photoperiod with light intensity ranging from 400 to 1,000 μmol m−2 s−1, a daytime temperature of 24°C, and a dark temperature of 20°C. Subsequent to Blumeria graminis f. sp. hordei (Bgh) inoculation, plants were kept in the Bgh chamber. Bgh isolates 5874 (AVRa1, AVRa6, avra7, avra9, avra10, AVRa12, avra13, avrg, avrh, avrk, avrp), K1 (AVRa1, avra6, AVRa13), and CC148 (AVRa1, avra6, AVRa13) were propagated on Manchuria barley (C.I. 2330) in separate controlled-growth chambers at 18°C (16 h of light/8 h of darkness).
Isolation of Fast-Neutron-Derived, mla6 Loss-of-Function Mutants
The C.I. 16151 line was obtained by introgression of the Mla6 gene into the universal susceptible cv Manchuria (Moseman, 1972). Seeds of C.I. 16151 were treated with fast neutrons at 4 Gy Nf at the International Atomic Energy Agency. M1 seeds were space planted at the U.S. Department of Agriculture-Agricultural Research Service Small Grains Laboratory in Aberdeen, Idaho. Single spikes from each individual M1 plant were harvested to represent the M2 family, which was screened for mutant segregants by sowing intact spikes consisting of 25 to 40 seeds in potting soil following the method of Wise and Ellingboe (1985). Each of 40 M2 families as well as the susceptible control (cv Manchuria, C.I. 2330) were sown per flat. When the first leaves were completely unfolded (approximately 10 cm high), plants were inoculated with Bgh isolate 5874 (AVRa6), and families were scored for infection type 7 d after inoculation. Seedlings that produced cell death symptoms or sporulating Bgh colonies were selected for rescue. Putative mutants deemed as homozygous by a 1:3 mutant:wild-type segregation ratio were advanced to the M3 generation and then retested with Bgh 5874. Selected mutants that displayed sporulating Bgh colonies were crossed pairwise among each other as well as to mla1-m508, mla1-m600 (Zhou et al., 2001), and rar1-1 (Torp and Jørgensen, 1986; Freialdenhoven et al., 1994; Jørgensen, 1996). mla6-m9472 was confirmed by genetic complementation, Southern-blot (Halterman et al., 2001), and Barley1 GeneChip (Caldo et al., 2004) analyses.
Expression Profiling and Analysis
The Barley1 GeneChip probe array (part no. 900515) is distributed by Affymetrix. The array includes 22,792 probe sets derived from 350,000 ESTs clustered from 84 cDNA libraries, in addition to 1,145 barley gene sequences from the National Center for Biotechnology Information (NCBI) nonredundant database (Close et al., 2004). Total RNA was isolated using a hot (60°C) phenol/guanidine thiocyanate method as described by Caldo et al. (2004). Probe synthesis and labeling were performed at the Iowa State University GeneChip Core facility (http://www.biotech.iastate.edu/facilities/genechip/Genechip.htm). All detailed protocols can be accessed online within the BarleyBase/PLEXdb parallel expression database for plants and plant pathogens (http://barleybase.org/; http://plexdb.org/; Shen et al., 2005; Wise et al., 2007a).
Plants harboring Mla6 (Rar1 dependent) and Mla1 (Rar1 independent) both exhibit rapid and absolute resistance responses when challenged by Bgh isolates that carry cognate AVRa6 and AVRa1 genes, respectively (Wise and Ellingboe, 1983; Boyd et al., 1995). mla6-m9472 is a fast-neutron-derived, Mla6 deletion mutant derived from C.I. 16151 (see above). mla1-m508 is a γ-radiation-derived, Mla1 deletion mutant derived from C.I. 16137 (Zhou et al., 2001). Time-course GeneChip expression profiling was used to compare barley lines that harbor Mla6 and those with mla6-m9472 as well as the nearly isogenic line harboring Mla1 and those with mla1-m508. The experiment (designated BB10) was based on a split-split plot design described for BB2 by Caldo et al. (2006), with barley first leaves harvested at 0, 8, 16, 20, 24, and 32 h after inoculation with Bgh isolate 5874 (AVRa1, AVRa6). Identical noninoculated plants were included for each treatment. The BB10 study consisted of 144 Barley1 GeneChip hybridizations (4 genotypes × 6 time points × 2 inoculation treatments × 3 biological replications) and the BB4 (Caldo et al., 2004) study consisted of 108 hybridizations (3 genotypes × 6 time points × 2 isolates × 3 biological replications), resulting in a total of 84 treatment combinations for the two experiments. Both studies were conducted under identical conditions, except that inoculations for BB4 were performed in 2002 and inoculations for BB10 were conducted in 2004. Interpretation of results was based on gene expression data within each experiment (Stevens and Doerge, 2005).
Normalization and Data Analysis
Normalization, data transformation, and mixed linear model analysis (Wolfinger et al., 2001) for the BB10-derived microarray data were patterned after the methods used by Caldo et al. (2004). The mixed linear model analysis was performed using the SAS MIXED procedure. Contrast statements in SAS were made to compare mRNA expression over time in noninoculated and inoculated plants for the individual genotypes.
Microarray Data Access
All detailed data and data from expression profiling have been deposited in BarleyBase/PLEXdb (http://barleybase.org; http://plexdb.org/), a MIAME-compliant expression database for plant GeneChips (Shen et al., 2005). Files are categorized under accession numbers BB4 for the 108 GeneChip Caldo et al. (2004) study and BB10 for the 144 GeneChip, Mla genotypes and derived mutant experiment. Data files have also been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) with accession numbers E-MEXP-142 for the Caldo et al. (2004) study and E-TABM-142 for the BB10 investigation. Barley1 GeneChip data files for the Fusarium graminearum (Boddu et al., 2006) and Puccinia graminis f. sp. tritici (Zhang et al., 2008) experiments are categorized under PLEXdb accession numbers BB9 and BB49, respectively.
Identification of the Blufensin Family
Unigene numbers used refer to those originally assigned in assembly 21 from Close et al. (2004), which was the template used for designing the Affymetrix Barley1 GeneChip. BLAST version 2.2.13 from the NCBI (www.ncbi.nlm.nih.gov) was used for all sequence database queries. The Web site interface of InterProScan (www.ebi.ac.uk/interpro/; July 2008) was utilized for domain and structure prediction. Databases used for identifying family members were Gramene (www.gramene.org) for rice (Oryza sativa), PlantGDB (www.plantgdb.org) for all other plant species, and both NCBI (www.ncbi.nlm.nih.gov) and UniProt (www.uniprot.org) for targeting all other organisms.
Multiple Sequence Alignment and Phylogeny of the Blufensin Family
The VectorNTI program AlignX was used to align the unigenes, ORFs, and peptides of the blufensin family. As the sequences are short, visual inspection of the alignment was used to correct any misalignments. The phylogeny was generated using the software package Phylip, using dnapars and protpars for DNA and protein sequence, respectively. Bootstrap support was performed with 1,000 replications, with only support values above 90% shown.
Promoter Analysis of Barley and Rice Blufensins
Promoters were subjected to motif search using the Plant Cis-Acting Regulatory DNA Elements database with the release version of February 2007 (Higo et al., 1999). As an extensive barley promoter sequence is currently unavailable, rice promoter sequence was used to determine if predicted occurrences of motifs were similar to those observed biologically. A Python script was developed to parse the promoter elements of the rice genome (version 5) using regular expressions to determine the occurrence of different motifs in both the forward and reverse strands of gene promoters using the same amount of sequence available for Bln1-1.
Biolistic-Based BSMV Vector Construction
The DNA-based BSMV constructs used in this study were modified from in vitro transcription-based BSMV clones (Scofield et al., 2005). Full-length cDNAs of BSMV α-, β-, and γ-subunits were amplified using high-fidelity Platinum Taq DNA polymerase (Invitrogen); primers are listed in Supplemental Table S2. The BSMV-Rev universal reverse primer was used in combination with each of the specific forward primers to amplify cDNA of BSMV α-, β-, and γ-subunits. PCR products were then inserted into 35S expression vector SMVNVEC (provided by Dr. Alan Eggenberger, Iowa State University) between StuI and SmaI sites. The 3′ hepatitis delta virus (HDV) ribozyme will self-cleave to generate an authentic 3′-end BSMV genome RNA.
Silencing Constructs
Total RNA was extracted from C.I. 16151 (Mla6) plants at 20 hai with Bgh isolate 5874 (AVRa6) according to the method of Caldo et al. (2004). First-strand cDNA was synthesized using 2 μg of total RNA, oligo(dT)20 primer, and SuperScript III reverse transcriptase (Invitrogen). Subsequently, first-strand cDNA was used as the template to amplify two independent fragments with lengths of 248 and 162 bp, respectively. Primers were designed according to the Bln1 EST sequence (GenBank accession no. BE216690) and are listed in Supplemental Table S2. Positions of the two fragments on the EST sequence were from 28 to 275 bp and from 39 to 200 bp. Amplified PCR fragments each contained introduced PacI and NotI recognition sites at the 5′ and 3′ ends, respectively, and were inserted into the PacI and NotI sites of BSMV:γ. The resulting vectors were designated BSMV:Bln1248 and BSMV:Bln1162, respectively.
Microprojectile Bombardment
All constructs were screened in at least three independent experiments. Biolistic bombardment of barley plants was carried out according to Halterman and Wise (2004) using a biolistic PDS-1000/He system (Bio-Rad) with minor modifications. Gold particles (Bio-Rad) were coated with plasmid BSMV:α, BSMV:β, and BSMV:γ (or the recombinant BSMV:Bln1248 or BSMV:Bln1162) at a molar ratio of 1:1:1, and the mixture was delivered to leaves using 900-psi rupture discs using a Hepta adaptor microcarrier. Eight 7-d-old Black Hull-less barley seedlings (susceptible to BSMV) were used per bombardment. Subsequently, plants were transferred to 7.5- × 7.5-cm pots for 7 to 10 d for viral replication and systemic infection. Virus-infected barley was maintained in a growth chamber (Percival Scientific) at 24°C with 16 h of light (550 μmol m−2 s−1) and 8 h darkness at 20°C.
Mechanical Infection of BSMV and Powdery Mildew Inoculation
Seven to 10 d after bombardment, plants displaying a BSMV infection phenotype (brown streak on the first leaf and chlorotic mosaics on the second leaf) were selected. Leaves from the infected plant were ground with 2 to 5 volumes of 0.05 m phosphate buffer (pH 7.2) in an ice-cold mortar. Carborundum (0.05 g; Sigma-Aldrich) was added to the buffer for optimal grinding. Seven-day-old healthy barley seedlings were then infected with the appropriate recombinant virions by rubbing the first leaf with crude virus extract four to six times between thumb and index finger, with new gloves used for each construct to prevent contamination. Twelve days after mechanical infection, plants displaying a BSMV infection phenotype (brown stripe on the first leaf) were inoculated with fresh Bgh conidiospores and placed in an 18°C growth chamber (16 h of light/8 h of darkness). Bgh infection types were scored at 7 dai.
Staining and Microscopy
The staining process was performed according to Hein et al. (2005) with minor modifications. Leaves were fixed for 24 h on filter paper soaked with 1:1 (v/v) ethanol:acetic acid and for 48 h on filter paper soaked with lactoglycerol (1:1:1 [v/v] lactic acid:glycerol:water) and stained with Coomassie Brilliant Blue R-250 stain (0.05% [w/v] Coomassie Brilliant Blue in 50% methanol and 10% acetic acid). A Zeiss Axio Imager M.1 microscope was used for observation.
Semiquantitative RT-PCR
Primers for semiquantitative RT-PCR are listed in Supplemental Table S2. Third leaves from BSMV-VIGS-treated plants that displayed a typical mosaic virus infection symptom were sampled for RT-PCR. Barley total RNA was isolated using a hot (60°C) phenol/guanidine thiocyanate method as described previously (Caldo et al., 2004) and treated with DNase I (Ambion). Two micrograms of RNA was transcribed into cDNA with an oligo(dT)20 primer by SuperScript III reverse transcriptase (Invitrogen). First-strand cDNAs were used as templates for amplifying target gene fragments at cycling conditions of 92°C for 20 s, 58°C for 20 s, and 68°C for 15 s for 20, 25, 30, 35, and 40 cycles. Actin was used as an internal constitutive expression control for cDNA quantitative normalization. The intensities of PCR-generated fragments were analyzed and quantified using Gel Doc 2000 and Quantity One version 4.2.1 (Bio-Rad).
Bln1 Transient Overexpression
The full-length ORF of Bln1 was amplified from vector BSMV:Bln1248 using with both sense (5′-TCAAAGCTTACGAGGATATGGCAAAGAACTAC-3′) and antisense (5′-AGTGATATCTTATGAGCCACCATTAGGGATCG-3′) primers. EcoRV and HindIII were used to double digest the PCR product, which was inserted into the expression vector pUbi:Nos, which was also digested with the same enzymes. The newly constructed vector, pUbi:Bln1, was cobombarded with pUGN (Nielsen et al., 1999) into barley epidermal cells in three independent experiments. A generalized linear mixed model was fit to the data from the three experiments. The model assumed a binomial response for each leaf. The logit of the binomial success probability (probability of hyphae formation) was modeled as a linear function of an overall mean, fixed construct effects, random experiment effects, and random effects for leaves within experiments and constructs.
Inverse PCR
Inverse PCR was performed according to Meng et al. (2007) with minor modifications. One microgram of genomic DNA sample was subjected to overnight digestion with 5 units of AflIII, MspI, and NcoI. The primers used are listed in Supplemental Table S2. The conditions used for PCR were as follows: 94°C for 2 min, followed by 40 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 3 min. A final extension step was performed at 72°C for 10 min.
Accession Numbers
Accession numbers are FJ156737 (C.I. 16151), FJ156738 (C.I. 16155), and FJ156739 (C.I. 16137) for Bln1-1 genomic sequences; FJ156740 (C.I. 16151), FJ156741 (C.I. 16155), and FJ156742 (C.I. 16137) for Bln1-2 genomic sequences; and FJ156743 (Sultan-5) and FJ156744 (Golden Promise) for Bln1-3 genomic sequences. Accession numbers for Bln2 genomic sequences are FJ156745 (C.I. 16151), FJ156746 (C.I. 16155), FJ156747 (C.I. 16137), FJ156748 (Sultan-5), and FJ156749 (Golden Promise).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Annotations of Blufensin family members in barley, rice, and wheat.
Supplemental Table S2. Primers used for inverse PCR, Bln1 transcript detection, BSMV-VIGS vector construction, Bln1 silencing constructs, and semiquantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Steve Scofield for the gift of in vitro transcription-based BSMV clones, Alan Eggenberger for the SMVNVEC 35S expression vector, Liz Miller for technical assistance during isolation of the C.I. 16151-derived fast-neutron mutants, Shauna Somerville for the mla1-m508 mutant, Rico Caldo for initial analysis of the BB10 data set, Dan Nettleton and Tim Bancroft for statistical analysis, and Adam Bogdanove for critical review of the manuscript.
This work was supported by the National Science Foundation (Plant Genome grant no. 05–00461) and the U.S. Department of Agriculture-Agricultural Research Service (CRIS project no. 3625–21000–049–00D). This is a joint contribution of the Iowa Agriculture and Home Economics Experiment Station and the Corn Insects and Crop Genetics Research Unit, U.S. Department of Agriculture-Agricultural Research Service.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Roger P. Wise (rpwise@iastate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander WS, Hilton DJ (2004) The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol 22 503–529 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behn A, Hartl L, Schweizer G, Wenzel G, Baumer M (2004) QTL mapping for resistance against non-parasitic leaf spots in a spring barley doubled haploid population. Theor Appl Genet 108 1229–1235 [DOI] [PubMed] [Google Scholar]
- Bélanger RR, Bushnell WR, Dik AJ, Carver TLW (2002) The Powdery Mildews: A Comprehensive Treatise. APS Press, St. Paul
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ (2006) Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol Plant Microbe Interact 19 407–417 [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Muehlbauer GJ (2007) Transcriptome analysis of trichothecene-induced gene expression in barley. Mol Plant Microbe Interact 20 1364–1375 [DOI] [PubMed] [Google Scholar]
- Boyd LA, Smith PH, Foster EM, Brown JKM (1995) The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J 7 959–968 [Google Scholar]
- Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3 238–250 [DOI] [PubMed] [Google Scholar]
- Bruun-Rasmussen M, Madsen CT, Jessing S, Albrechtsen M (2007) Stability of Barley stripe mosaic virus-induced gene silencing in barley. Mol Plant Microbe Interact 20 1323–1331 [DOI] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88 695–705 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldo RA, Nettleton D, Peng J, Wise RP (2006) Stage-specific suppression of basal defense discriminates barley plants containing fast- and delayed-acting Mla powdery mildew resistance alleles. Mol Plant Microbe Interact 19 939–947 [DOI] [PubMed] [Google Scholar]
- Caldo RA, Nettleton D, Wise RP (2004) Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D (2004) Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem 279 21749–21758 [DOI] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem 274 34993–35004 [DOI] [PubMed] [Google Scholar]
- Ding XS, Schneider WL, Chaluvadi SR, Mian MA, Nelson RS (2006) Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol Plant Microbe Interact 19 1229–1239 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols 2 953–971 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914 [DOI] [PubMed] [Google Scholar]
- Freialdenhoven A, Scherag B, Hollricher K, Collinge DB, Thordal-Christensen H, Schulze-Lefert P (1994) Nar-1 and Nar-2, two loci required for Mla12-specified race-specific resistance to powdery mildew in barley. Plant Cell 6 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T (2003) Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 3 710–720 [DOI] [PubMed] [Google Scholar]
- Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, Xia Y (2007) AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Maldonado J, Avila C, Torre F, Canas R, Canovas FM, Campbell MM (2004) Functional interactions between a glutamine synthetase promoter and MYB proteins. Plant J 39 513–526 [DOI] [PubMed] [Google Scholar]
- Graham MA, Silverstein KAT, VandenBosch KA (2008) Defensin-like genes: genomic perspectives on a diverse superfamily in plants. Crop Sci 48 S-3–S-11 [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P (2001) The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J 25 335–348 [DOI] [PubMed] [Google Scholar]
- Halterman DA, Wei F, Wise RP (2003) Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol 131 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman DA, Wise RP (2004) A single-amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J 38 215–226 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby I, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30 315–327 [DOI] [PubMed] [Google Scholar]
- Horton P, Park K, Obayashi T, Nakai K (2006) Protein subcellular localization prediction with WoLF PSORT. In Proceedings of Asian Pacific Bioinformatics Conference, Taipei, Taiwan. Imperial College Press, London, pp 39–48
- Huang G, Dong R, Allen R, Davis EL, Baum TJ, Hussey RS (2006) A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol Plant Microbe Interact 19 463–470 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R (2005) Powdery mildew susceptibility and biotrophic infection strategies. FEMS Microbiol Lett 245 9–17 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Dechert C, Kogel KH (2003) Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis. Proc Natl Acad Sci USA 100 5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Jørgensen JH (1994) Genetics of powdery mildew resistance in barley. Crit Rev Plant Sci 13 97–119 [Google Scholar]
- Jørgensen JH (1996) Effect of three suppressors on the expression of powdery mildew resistance genes in barley. Genome 39 492–498 [DOI] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416 447–451 [DOI] [PubMed] [Google Scholar]
- Kwon C, Bednarek P, Schulze-Lefert P (2008) Secretory pathways in plant immune responses. Plant Physiol 147 1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C, Hrubikova K, Hein I (2003) Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J 34 543–553 [DOI] [PubMed] [Google Scholar]
- Lam E (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5 305–315 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Casson S, Chilley P (2002) Peptides: new signalling molecules in plants. Trends Plant Sci 7 78–83 [DOI] [PubMed] [Google Scholar]
- Liu Y, Luo J, Xu C, Ren F, Peng C, Wu G, Zhao J (2000) Purification, characterization, and molecular cloning of the gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiol 122 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC (2003) Virus-induced gene silencing in plants. Methods 30 296–303 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Yang H, Sakagami Y (2001) Peptide signals and their receptors in higher plants. Trends Plant Sci 6 573–577 [DOI] [PubMed] [Google Scholar]
- Meng Y, Patel G, Heist M, Betts MF, Tucker SL, Galadima N, Donofrio NM, Brown D, Mitchell TK, Li L, et al (2007) A systematic analysis of T-DNA insertion events in Magnaporthe oryzae. Fungal Genet Biol 44 1050–1064 [DOI] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33 D284–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman JG (1972) Isogenic barley lines for reaction to Erysiphe graminis f.sp. hordei. Crop Sci 12 681–682 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10 1–6 [DOI] [PubMed] [Google Scholar]
- Nielsen K, Olsen O, Oliver R (1999) A transient expression system to assay putative antifungal genes on powdery mildew infected barley leaves. Physiol Mol Plant Pathol 54 1–12 [Google Scholar]
- Panstruga R (2003) Establishing compatibility between plants and obligate biotrophic pathogens. Curr Opin Plant Biol 6 320–326 [DOI] [PubMed] [Google Scholar]
- Panstruga R (2004) A golden shot: how ballistic single cell transformation boosts the molecular analysis of cereal-mildew interactions. Mol Plant Pathol 5 141–148 [DOI] [PubMed] [Google Scholar]
- Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM (2003) Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol Biol 53 597–608 [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr (2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253 895–897 [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33 W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JDG, Brown JKM (2006) Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18 2402–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Sablowski RW, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J 13 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB (1999) The male determinant of self-incompatibility in Brassica. Science 286 1697–1700 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R (2002) A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol 128 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R (2003) Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J 36 589–601 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R (2003) Establishment of biotrophy by parasitic fungi and reprogramming of host cells for disease resistance. Annu Rev Phytopathol 41 641–667 [DOI] [PubMed] [Google Scholar]
- Schweizer P (2007) Nonhost resistance of plants to powdery mildew: new opportunities to unravel the mystery. Physiol Mol Plant Pathol 70 3–7 [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA (2005) BarleyBase: an expression profiling database for plant genomics. Nucleic Acids Res 33 D614–D618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Bieri S, Zhou F, Haizel T, Shirasu K, Schulze-Lefert P (2003) Recognition specificity and RAR1/SGT1 dependency in barley Mla disease resistance alleles to the powdery mildew fungus. Plant Cell 15 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nielsen K, Piffanelli P, Oliver RP, Schulze-Lefert P (1999) Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J 17 293–299 [Google Scholar]
- Silverstein KA, Moskal WA Jr, Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA (2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51 262–280 [DOI] [PubMed] [Google Scholar]
- Stevens JR, Doerge RW (2005) Combining Affymetrix microarray results. BMC Bioinformatics 6 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational charges of NPR1 via s-nitrosylation and thioredoxins. Science 321 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torp J, Jørgensen JH (1986) Modification of barley powdery mildew resistance gene Mla12 by induced mutation. Can J Genet Cytol 28 725–731 [Google Scholar]
- Wang CIA, Guncar G, Forwood JK, Teh T, Catanzariti AM, Lawrence GJ, Loughlin FE, Mackay JP, Schirra HJ, Anderson PA, et al (2007) Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19 2898–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14 1903–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise R, Ellingboe AH (1983) Infection kinetics of Erysiphe graminis f. sp. hordei on barley with different alleles at the Ml-a locus. Phytopathology 73 1220–1222 [Google Scholar]
- Wise RP, Caldo RA, Hong L, Shen L, Cannon EK, Dickerson JA (2007a) BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. In D Edwards, ed, Methods in Molecular Biology. Vol 406. Plant Bioinformatics: Methods and Protocols. Humana Press, Totowa, NJ, pp 347–363
- Wise RP, Ellingboe AH (1985) Fine structure and instability of the Ml-a locus in barley. Genetics 111 113–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA (2007b) Transcript profiling in host-pathogen interactions. Annu Rev Phytopathol 45 329–369 [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8 625–637 [DOI] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount NY, Yeaman MR (2004) Multidimensional signatures in antimicrobial peptides. Proc Natl Acad Sci USA 101 7363–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415 389–395 [DOI] [PubMed] [Google Scholar]
- Zhang C, Yang C, Whitham SA, Hill JH (2009) Development and use of an efficient DNA-based viral gene silencing vector for soybean. Mol Plant Microbe Interact 22 (in press) [DOI] [PubMed]
- Zhang L, Castell-Miller C, Dahl S, Steffenson B, Kleinhofs A (2008) Parallel expression profiling of barley-stem rust interactions. Funct Integr Genomics 8 187–198 [DOI] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Vale G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P (2001) Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.