Axillary meristems, which form in the axils of leaves, play an essential role in plant architecture and reproduction. During vegetative development, axillary meristems give rise to branches, called tillers in grasses, while during reproductive development, axillary meristems give rise to flowering branches or to flowers. The control of branching by axillary meristems is under hormonal, environmental, developmental, and genetic control. In this Update I review the role of hormones in regulation of axillary meristem initiation and outgrowth during both vegetative and inflorescence branching.
Hormones play a critical role in regulating branching (McSteen and Leyser, 2005; Beveridge, 2006; Ongaro and Leyser, 2008). Auxin is required for axillary meristem initiation during both vegetative and inflorescence development. In addition, basipetal movement of auxin from the shoot apex suppresses axillary bud outgrowth in the phenomenon known as apical dominance. Cytokinin regulates meristem size and hence indirectly affects branching (Shani et al., 2006; Kyozuka, 2007). In addition, acropetal movement of cytokinin coming from the roots promotes axillary bud outgrowth. It has long been proposed that an additional hormone travels acropetally from the root to inhibit bud outgrowth. The highlight of last year was the identification of this new plant growth hormone, strigolactone (Gomez-Roldan et al., 2008; Umehara et al., 2008).
The environment also plays a significant role in regulating branch outgrowth. It is commonly known, especially in grasses, that increased planting density leads to reduced branching (Doust 2007a, 2007b; Kebrom and Brutnell, 2007). This could be due to shading, which is known to inhibit branching, or competition for resources, as fertilizer and nutrients are known to promote branching.
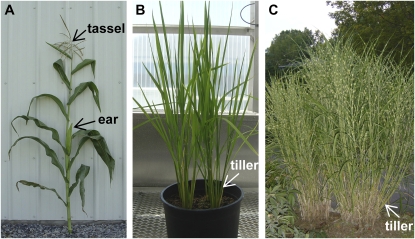
Developmental control of axillary meristems is evident from the different fates of axillary meristems during vegetative and reproductive development in different species (Steeves and Sussex, 1989; McSteen and Leyser, 2005). During vegetative development in Arabidopsis (Arabidopsis thaliana), axillary meristems appear late during leaf ontogeny, produce a few leaf primordia, and then arrest until signaled to grow. In maize (Zea mays), axillary buds remain suppressed during development (except for the ear shoot), leading to a single axis of growth (Fig. 1A; Kiesselbach, 1949). In rice (Oryza sativa), axillary meristems grow out to produce a highly tillered plant, though the buds are still under hormonal and environmental control (Fig. 1B; Shimamoto and Kyozuka, 2002).
Figure 1.
Divergent mechanisms of tillering in the grasses. A, Maize with a single axis of growth. One or two axillary shoots containing the female inflorescence (the ear) grow out. Tillers are generally suppressed. B, Several young rice plants showing that tillers grow out early in vegetative development. C, Ornamental Miscanthus. Tillers grow out from rhizomes beneath the soil surface. A triploid variety, Miscanthus × giganteus, is being investigated as a potential biofuel crop. [See online article for color version of this figure.]
During inflorescence development in Arabidopsis, floral meristems arise in the axils of reduced leaves called bract leaves (Long and Barton, 2000; Grbic, 2005). In grass inflorescence development, axillary meristems give rise to branches and spikelets before they give rise to flowers (McSteen et al., 2000; Bommert et al., 2005). The identity and determinacy of these different meristem types are controlled by transcription factors (Bortiri and Hake, 2007; Update by Thompson and Hake, this issue [Thompson and Hake, 2009]). Although hormones have been implicated in regulation of inflorescence branching, the exact mechanism is unknown (Barazesh and McSteen, 2008).
The differences in the activity of axillary meristems produced during development imply that all axillary meristems do not respond similarly to the same stimuli. For example, in Arabidopsis, there is acropetal outgrowth of buds during vegetative development and basipetal outgrowth of axillary buds during reproductive development (Hempel and Feldman, 1994; McSteen and Leyser, 2005). In grasses, the basal nodes are the ones from which tillers arise (Fig. 1B). Heterochronic mutations that extend the juvenile phase lead to increased tiller number (Poethig, 1988; Chuck et al., 2007). In foxtail millet (Setaria italica), branches are also produced from the upper nodes of the plant, in addition to tillers from basal nodes, and these are under separable genetic and environmental control (Doust et al., 2004; Doust, 2007a). These studies indicate that genetic, hormonal, and environmental signals intersect with developmental signals.
Many genes have been identified that regulate axillary meristem initiation and outgrowth during vegetative and reproductive development (Schmitz and Theres, 2005; Bennett and Leyser, 2006; Doust 2007b). Most of these genes have been identified as mutants that have fewer branches, because axillary meristems fail to initiate, or that are bushier than normal due to constitutive outgrowth of axillary buds. Some of these genes encode integral components of hormone biosynthesis, perception or signaling pathways providing a direct link to hormone control. Here, I highlight the conservation and diversification of the mechanisms that control branching within grasses and between grasses and eudicots.
AXILLARY MERISTEM INITIATION
Many of the genes regulating axillary meristem initiation affect both vegetative and reproductive development (Table I). In some cases, genes have been reported to affect only one stage of development, but often additional roles in either vegetative or reproductive development have been discovered by constructing double mutants. Thus, how much this distinction is due to redundancy with related genes remains to be seen.
Table I.
Genes regulating axillary meristem initiation in monocots and eudicots
| Protein | Rice | Maize | Arabidopsis | Pea | Tomato |
|---|---|---|---|---|---|
| Auxin efflux carrier | OsPIN1 | ZmPIN1 | PIN1 | PsPIN1 | |
| Ser/Thr kinase | OsPID/OsBIF2 | BIF2 | PID | PsPID | |
| Flavin mono-oxygenase | OsYUC1 | SPI1 | YUC1,2,4,6 | ToFZY | |
| Basic helix-loop-helix transcription factor | LAX | BA1 | |||
| GRAS transcription factor | MOC1/SPA | LAS | LS | ||
| NAC transcription factor | OsTIL1/OsNAC2 | ZmCUC3 | CUC1,2,3 | ||
| ZmNAC | |||||
| HD ZIP transcription factor | OsHB3 | REV/IFL | |||
| MYB transcription factor | RAX1,2,3 | BLIND |
Role of Auxin in Axillary Meristem Initiation during Vegetative and Inflorescence Development
Auxin plays a fundamentally important role in polar growth of all organ primordia, including floral meristems (Cheng and Zhao, 2007; Benjamins and Scheres, 2008; Delker et al., 2008). Multiple mutants have been identified that fail to make flowers in Arabidopsis, resulting in a pin-shaped inflorescence phenotype (Bennett et al., 1995; Fig. 2, A and B). PINFORMED1 (PIN1) encodes one of the auxin efflux carriers (Galweiler et al., 1998), while PINOID (PID) encodes a Ser/Thr protein kinase that phosphorylates and regulates the localization of PIN1 (Christensen et al., 2000; Benjamins et al., 2001; Friml et al., 2004; Michniewicz et al., 2007). Knockout of multiple genes in the YUCCA (YUC) family of flavin monooxygenases also causes a pin phenotype (Cheng et al., 2006). YUC genes are involved in localized auxin biosynthesis, indicating that auxin biosynthesis is also required for floral meristem initiation (Zhao, 2008). Recent work in maize and rice suggests that the role of auxin transport and auxin biosynthesis in axillary meristem initiation is conserved in monocots.
Figure 2.
Auxin plays a role in axillary meristem initiation in the inflorescence. A, Arabidopsis inflorescence with secondary branches (br) and flowers (fl). B, pid mutant in Arabidopsis showing the pin-shaped inflorescence due to lack of flower production. C, Normal maize tassel with long branches (br) at the base of the main spike. Short branches called spikelet pairs (sp) cover the branches and the central spike. D, bif2 mutant of maize with very few branches and spikelets. [See online article for color version of this figure.]
Three PIN1 loci have been identified in maize and rice (Xu et al., 2005; Carraro et al., 2006; Gallavotti et al., 2008b). The maize ZmPIN1a gene complements the Arabidopsis pin1 mutant, restoring its ability to make flowers, indicating that ZmPIN1a likely functions in auxin transport in maize (Gallavotti et al., 2008b). ZmPIN1a is localized in all axillary meristems and lateral organ primordia in maize similar to Arabidopsis (Carraro et al., 2006; Gallavotti et al., 2008b). Therefore, even though maize makes multiple types of axillary meristems in the inflorescence, all are characterized by expression of PIN1. Antisense knockdown of the rice OsPIN1 gene supports the conserved function of PIN1 genes in auxin transport in monocots (Xu et al., 2005).
barren inflorescence2 (bif2) mutants in maize have a phenotype analogous to the pin phenotype in Arabidopsis (Fig. 2, C and D). In bif2 mutants, fewer branches and spikelets form; however, these arise from the first axillary meristems produced by the inflorescence (McSteen and Hake, 2001). The maize BIF2 gene encodes a Ser/Thr protein kinase co-orthologous to PID (McSteen et al., 2007). BIF2 phosphorylates ZmPIN1a and affects ZmPIN1a localization, indicating that BIF2 regulates PIN1 in maize, similar to the function of PID in Arabidopsis (A. Skirpan and P. McSteen, unpublished data; Friml et al., 2004; Michniewicz et al., 2007). Overexpression of rice OsPID has effects on seedling development similar to the effects of treatment with an auxin transport inhibitor, providing further evidence for a function in auxin transport (Morita and Kyozuka, 2007). Therefore, it is likely that PID-like proteins have conserved roles in regulating auxin transport in monocots and eudicots.
On the other hand, BIF2 is localized in the nucleus as well as at the cell periphery, indicating that it plays additional roles in development (Skirpan et al., 2008). Yeast two-hybrid screening with PID identified calcium-binding proteins that act upstream of PID (Benjamins et al., 2003). In contrast, a yeast two-hybrid screen with BIF2 identified BARREN STALK1 (BA1), a nuclear localized basic helix-loop-helix putative transcription factor as an interacting partner with BIF2 (Gallavotti et al., 2004; Skirpan et al., 2008). In vitro kinase assays showed that BIF2 phosphorylates BA1 (Skirpan et al., 2008). bif2 and ba1 mutants both have defects in axillary meristem initiation, indicating that the in vitro interaction may have in vivo relevance (McSteen and Hake, 2001; Ritter et al., 2002; Skirpan et al., 2008). Therefore, in maize, BIF2 also functions in the nucleus. Whether the same is true for PID in Arabidopsis or OsPID/OsBIF2 in rice is not yet known. Although PID has not been reported to be nuclear localized, other PID-like proteins in Arabidopsis have been reported to be localized in the nucleus (Zegzouti et al., 2006). There is an ortholog of BA1 in rice, LAX PANICLE (LAX; Komatsu et al., 2003), but an ortholog of BA1 has not been reported in Arabidopsis.
While the exact molecular mechanism of the interaction of BA1/LAX with auxin is not clear, there does appear to be a connection with auxin (Gallavotti et al., 2008b; Skirpan et al., 2008). As already mentioned, BA1 has been shown to physically interact with BIF2 (Skirpan et al., 2008). BA1 may be auxin induced, as it is not expressed following treatment with an auxin transport inhibitor (Wu and McSteen, 2007). In addition, the auxin reporter DR5 indicates that auxin maxima do not form in the ba1 mutant (Gallavotti et al., 2008b). Overexpression of LAX in rice leads to defects in development, indicating that LAX functions in auxin-mediated development (Komatsu et al., 2003). Therefore, it is likely that BA1/LAX plays a role in auxin-mediated axillary meristem initiation.
Localized auxin biosynthesis is also required for axillary meristem initiation in maize as in Arabidopsis, but, interestingly, there is a higher level of redundancy in Arabidopsis than in maize. The sparse inflorescence1 (spi1) mutant of maize has fewer branches and spikelets due to the absence of axillary meristems (Gallavotti et al., 2008a). SPI1 encodes a monocot-specific YUC gene family member required for localized auxin biosynthesis (Gallavotti et al., 2008a). Knockout of four YUC genes is required in Arabidopsis to generate a phenotype as severe as the spi1 mutant in maize (Cheng et al., 2006). Phylogenetic analysis shows that this is due to massive expansion of the gene family in both monocots and eudicots. Interestingly, the expression of SPI1 differs from the expression of OsYUC1, the ortholog of SPI1 in rice (Yamamoto et al., 2007). Furthermore, antisense knockdown of OsYUC1 in rice has severe defects in root and stem elongation, but a defect in the inflorescence was not reported (Yamamoto et al., 2007). Therefore, there has been functional diversification of the YUC gene family even within the grasses.
BIF2, BA1/LAX, and SPI1 also play a role in axillary meristem initiation during vegetative development (Ritter et al., 2002; Komatsu et al., 2003; McSteen et al., 2007; Gallavotti et al., 2008a). As maize does not normally tiller, these studies took advantage of the teosinte branched1 (tb1) mutant of maize (discussed further in the outgrowth section). tb1 mutants are highly tillered, because all axillary buds that are normally suppressed grow out producing a bushy plant (Doebley et al., 1997). Double mutant combinations between ba1 and tb1 produced no tillers, indicating that BA1 is required for axillary meristem initiation during vegetative development (Ritter et al., 2002). bif2;tb1 and spi1;tb1 double mutants also produced fewer tillers than tb1 single mutants, indicating that they too function in axillary meristem initiation (McSteen et al., 2007; Gallavotti et al., 2008a). In rice, double mutants between lax and small panicle (spa) lead to an absence of tillers, indicating that LAX and SPA play overlapping roles in tiller development (Komatsu et al., 2003).
In contrast, pid1, pin1, and yuc mutants in Arabidopsis do not apparently produce fewer side branches (Bennett et al., 1995; Cheng et al., 2006). In fact, plants containing a knockout of multiple YUC genes have a bushy appearance due to reduced apical dominance. These differences could be due to genetic redundancy, but it is interesting that the initiation of vegetative axillary meristems in Arabidopsis does not appear to be as sensitive to reductions in auxin biosynthesis or transport as in maize.
Other transcription factors that regulate axillary meristem initiation in monocots and eudicots include the GRAS-type transcription factor LAS1/LS/MOC1 (Schumacher et al., 1999; Greb et al., 2003; Li et al., 2003) and the HD ZIP class III transcription factor REV/OsHB3 (Otsuga et al., 2001; Itoh et al., 2008; Table I). The NAC domain transcription factors CUC1,2,3 play a role in axillary meristem initiation in Arabidopsis, but in rice, overexpression of OsTIL1 enhances axillary meristem outgrowth rather than initiation (Vroemen et al., 2003; Hibara et al., 2006; Mao et al., 2007; Raman et al., 2008). R2 R3 Myb transcription factors RAX1,2,3/BLIND (Schmitz et al., 2002; Keller et al., 2006; Muller et al., 2006) play a role in eudicots, but a homolog in monocots has not yet been identified. Therefore, the roles of these transcription factors still need to be clarified in monocots. Furthermore, the relationship of these transcription factors with hormonal regulation is not clear.
Role of Cytokinin in Regulating Apical Meristem Size
Cytokinin also regulates branch and spikelet number in grasses, but in this case, the effect on branching is a secondary effect due to a defect in the shoot apical meristem. Cytokinin plays a fundamental role in regulation of apical meristem size (Shani et al., 2006; Kyozuka, 2007; Zhao, 2008). In Arabidopsis, increased cytokinin levels lead to increased meristem size and reduced cytokinin levels lead to reduced meristem size (Nogue et al., 2000; Werner et al., 2003).
Two recent articles have demonstrated the fundamental importance of cytokinin in branching and, hence, yield in rice. CYTOKININ OXIDASE (CKX) is an enzyme that degrades cytokinin (Sakakibara, 2006). Mutations in the rice CKX gene led to increased panicle branch and spikelet number in the inflorescence and increased yield (Ashikari et al., 2005). Conversely, mutations in the rice LONELY GUY gene, which encodes an enzyme that catalyzes the last step in cytokinin biosynthesis, led to the production of fewer branches and spikelets and decreased yield (Kurakawa et al., 2007). Therefore, the role of cytokinin in controlling meristem size appears to be conserved in monocots and eudicots.
AXILLARY MERISTEM OUTGROWTH
Role of Auxin and Cytokinin in Control of Bud Outgrowth
Once axillary buds have initiated, the outgrowth of axillary buds is under hormonal as well as environmental control (Ongaro and Leyser, 2008). Apical dominance is a well-known phenomenon in which auxin traveling basipetally from the shoot apex suppresses the outgrowth of axillary buds (Thimann and Skoog, 1933; Leyser, 2003; Fig. 3). If a shoot is decapitated, the axillary buds are activated. If auxin is applied to the cut end of the shoot, suppression of buds reoccurs. Apical dominance occurs in many plants, leading horticulturists to prune plants to promote branching. Arabidopsis exhibits some apical dominance, as decapitation causes one additional branch to grow out (Aguilar-Martinez et al., 2007). Even though rice is quite bushy, it was recently demonstrated to exhibit apical dominance. Removal of the panicle (inflorescence) caused an increase in tiller elongation, which was suppressed by the addition of auxin (Arite et al., 2007).
Figure 3.
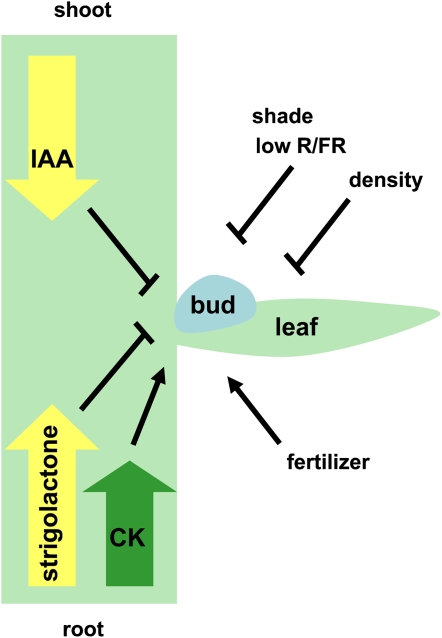
Model for endogenous and exogenous factors controlling bud outgrowth. The blue ball represents an axillary bud in the axil of a leaf. Both endogenous and exogenous factors determine whether or not the axillary bud grows out. Endogenous hormones regulate branching. Auxin (IAA) traveling basipetally inhibits bud outgrowth, cytokinin (CK) moving acropetally promotes outgrowth, while acropetal movement of strigolactone inhibits bud outgrowth. Exogenous factors such as shading and density inhibit bud outgrowth, while fertilizer and nutrients promote bud outgrowth.
The role of auxin in apical dominance is also illustrated by mutants with defects in auxin signaling, biosynthesis, and transport. For example, auxin-resistant mutants in Arabidopsis, such as auxin resistant1 (axr1), are bushy (Lincoln et al., 1990). Reduction of auxin biosynthesis by multiple yuc mutants causes reduced apical dominance (Cheng et al., 2006). Similarly, reductions in auxin transport led to increased tillering in monocots. OsPIN1 knockdown mutants almost doubled the number of tillers, indicating that reduced auxin transport results in outgrowth of tillers even in rice, which has a large number of tillers (Xu et al., 2005). OsPIN1 knockdown mutants also affected tiller angle such that tillers were at a wider angle than normal. This phenotype is also seen in other mutants that affect auxin transport in rice and indicates that auxin transport is required for tiller gravitropism as well as tiller outgrowth (Li et al., 2007).
As auxin does not enter the bud to inhibit bud outgrowth, a second messenger was proposed (Booker et al., 2003). Cytokinin was a good candidate for the second messenger, as cytokinin travels from the root to the shoot and enters the bud where it promotes outgrowth (Fig. 3; Ongaro and Leyser, 2008), and auxin has been shown to inhibit cytokinin biosynthesis (Nordstrom et al., 2004; Tanaka et al., 2006). However, there is also evidence that cytokinin acts independently of auxin (Chatfield et al., 2000). Mutants that overproduce cytokinin are bushy (Helliwell et al., 2001; Tantikanjana et al., 2001). The identification of another class of bushy mutants led to the discovery of the new branching hormone, strigolactone (Fig. 3).
Role of Strigolactones in Suppressing Axillary Bud Outgrowth
A novel compound required for regulation of branching was proposed based on the identification of the more axillary meristem (max) mutants in Arabidopsis, ramosus (rms) mutants in pea (Pisum sativum), and decreased apical dominance (dad) mutants in petunia (Petunia hybrida; Beveridge, 2006; Ongaro and Leyser, 2008; Table II). Grafting experiments demonstrated the existence of a long-distance signal traveling from the root, that instead of promoting outgrowth like cytokinin, suppressed outgrowth like auxin (Fig. 3). Cloning the MAX/RMS genes indicated that the compound was likely derived from the carotenoid biosynthetic pathway.
Table II.
Genes regulating axillary bud outgrowth in monocots and eudicots
| Protein | Rice | Maize | Arabidopsis | Pea | Petunia |
|---|---|---|---|---|---|
| Carotenoid cleavage dioxygenase 7 | HTD1/D17 | MAX3 | RMS5 | ||
| Carotenoid cleavage dioxygenase 8 | D10 | MAX4 | RMS1 | DAD1 | |
| Cyt P450 | MAX1 | ||||
| F box | D3 | MAX2 | RMS4 | ||
| TCP transcription factor | FC1 | TB1 | BRC1 |
MAX3/RMS5 encodes carotenoid cleavage dioxygenase CCD7 (Booker et al., 2004; Johnson et al., 2006), and MAX4/RMS1/DAD1 encode carotenoid cleavage dioxygenase CCD8 (Sorefan et al., 2003; Snowden et al., 2005). These enzymes are proposed to act in tandem to cleave β-carotene into an unknown product (Schwartz et al., 2004). MAX1 encodes a Cyt P450 enzyme that acts downstream of MAX3 and MAX4 in the synthesis of this unknown carotenoid-derived compound (Booker et al., 2005). Cloning of a number of highly tillered mutants from rice showed that this pathway is conserved in grasses. HIGH TILLERING DWARF1 (HTD1) encodes an ortholog of MAX3/RMS5 (Zou et al., 2006) and DWARF10 (D10) encodes an ortholog of MAX4/RMS1 (Arite et al., 2007). The dwarfing in the rice mutants is a secondary effect of the formation of tillers, as removal of tillers restores plant height (Zou et al., 2006).
The search was on for the elusive compound. The answer, strigolactone, was completely unexpected. To understand how the breakthrough was made, I will first explain what strigolactones are. Strigolactones were discovered as a group of compounds released from plant roots that promote the germination of seeds of parasitic plants (Bouwmeester et al., 2007). Parasitic Striga species infect monocots and are a major cause of crop loss in Africa. Similarly, Orobanche species infect eudicots. Recently, strigolactones were also discovered to be involved in colonization of roots by symbiotic arbuscular mycorrhyzal (AM) fungi (Akiyama et al., 2005). In this case, the interaction is beneficial to the plant, leading to increased uptake of nutrients. Another clue to the breakthrough was the publication of an article showing that strigolactones are synthesized from the carotenoid pathway (Matusova et al., 2005).
The discovery that strigolactones are involved in shoot branching came from two directions. Researchers working on the colonization of roots by AM fungi wanted to identify the genes regulating the biosynthesis of strigolactone. As the rms mutants are defective in carotenoid cleavage enzymes, they contacted the researchers working on the pea rms mutants. The first clue that the rms genes may be involved in synthesis of a strigolactone came when it was discovered that the rms mutants failed to interact with AM fungi (Gomez-Roldan et al., 2008). Coming from a biochemical direction, the rice researchers determined that the rice dwarf mutants were deficient in strigolactones (Umehara et al., 2008).
In both pea and rice, it was shown that root exudates of the rms/dwarf mutants are deficient in strigolactones (Gomez-Roldan et al., 2008; Umehara et al., 2008). Importantly, the rice group also showed that dwarf mutants have lower endogenous levels of strigolactone (Umehara et al., 2008). In the case of pea, orbanchyl acetate is one of the strigolactones that are missing (Gomez-Roldan et al., 2008), while in rice, 2′-epi-5-deoxystrigol and another compound are absent (Umehara et al., 2008). The availability of GR24, a synthetic analog of strigolactone, facilitated the research. The direct addition of GR24 to the buds of pea rms or Arabidopsis max mutants inhibited branching (Gomez-Roldan et al., 2008). The rice group used hydroponics to show that feeding of GR24 to rice dwarf or Arabidopsis max mutants caused complete suppression of branching (Umehara et al., 2008). To complete the story, it was shown that root exudates from the pea mutants were unable to promote germination of Orbanche seed (Gomez-Roldan et al., 2008), while the rice mutants were unable to be parasitized by Striga (Umehara et al., 2008). Hence, a strigolactone may be the elusive second messenger for auxin in the control of apical dominance.
How are strigolatones perceived? Some of the max/rms mutants did not respond to grafting and were proposed to be involved in perception of the max hormone. MAX2/RMS4 encode an F box Leu-rich repeat protein that is a component of the SCF complex (Stirnberg et al., 2002; Johnson et al., 2006). The rice ortholog is D3 (Ishikawa et al., 2005). The F box provides substrate specificity to the SCF complex, which promotes the ubiquitination and subsequent degradation of target proteins. SCF complexes are involved in multiple hormone signaling pathways, where they usually degrade a transcription factor required for signaling (McSteen and Zhao, 2008). A major question that remains is: What is the target of MAX2/RMS4/D3? It must be a protein, perhaps a transcription factor, that is involved in promoting bud outgrowth so that its degradation by the SCFMAX2/RMS4/D3 complex inhibits bud outgrowth. Another question that remains unresolved is: How does the SCFMAX2/RMS4/D3 complex perceive strigolactones? Do strigolactones bind the F box protein MAX2/RMS4/D3 in the same way that auxin binds the auxin receptor F box protein TIR1 (Tan et al., 2007)? An interesting evolutionary question is: How is the strigolactone signal perceived by AM fungi and Striga seed? Is there an F box protein expressed in Striga seed or in AM fungal hyphae?
The discovery of strigolactones may also explain some of the environmental effects of branching. As strigolactone biosynthesis is induced by low phosphorous and low nitrogen, it is proposed that this induces AM fungi to help scavenge these important nutrients (Bouwmeester et al., 2007). The rice group speculates that strigolactone would provide a mechanism for the plant to communicate nutrition status underground to the shoot, to increase branching when nutrients are not limited, or to decrease branching when nutrients are limited (Umehara et al., 2008). Furthermore, MAX2 is also involved in perception of red/far-red light signals providing a mechanism to integrate light signals with branching (Shen et al., 2007).
Although strigolactone appears to be a conserved regulator of branching in monocots and eudicots, some differences in wiring between components have been identified (Bainbridge et al., 2005; Dun et al., 2006). For example, RMS1 is induced by auxin in the shoot in pea and D10 is induced by auxin in the shoot in rice (Foo et al., 2005; Arite et al., 2007), but max4 is induced by auxin in the root in Arabidopsis (Bainbridge et al., 2005). Furthermore, RMS1 in pea and D10 in rice are up-regulated in other rms/d mutants (Foo et al., 2005; Arite et al., 2007), while MAX4 is not appreciably up-regulated in the max mutants (Bainbridge et al., 2005). As pea, rice, and Arabidopsis have different morphology with respect to branch outgrowth, it is not surprising that there should be differences in the strength of the interaction between components. Gene regulatory network analysis in these species may determine which nodes are conserved and which differ. Analysis of more species is required to determine if differences in the strength of interactions between auxin, cytokinin, and strigolactone could explain differences in plant architecture.
Role of tb1 in Integration of Responses to Hormones and the Environment
One of the best understood examples of domestication in plants is the discovery that selection on the expression of the TB1 locus was involved in the domestication of maize from its wild ancestor teosinte (Zea mays ssp. parviglumis; Doebley, 2004). Teosinte plants are highly branched. Vegetative branches grow out at every node (except the last few), resulting in a very bushy plant. Maize, on the other hand, has a single axis of growth (Fig. 1A). TB1 encodes a putative transcription factor expressed in axillary buds (Doebley et al., 1997; Hubbard et al., 2002). TB1 is expressed in organs that are suppressed in both maize and teosinte (Hubbard et al., 2002). In maize, there is an increase in the expression of TB1 relative to teosinte, causing buds to be suppressed, perhaps by affecting the cell cycle (Kosugi and Ohashi, 1997; Hubbard et al., 2002; Li et al., 2005). The increase in expression is proposed to be due to selection at a distant enhancer upstream of the TB1 locus (Clark et al., 2004, 2006).
Isolation of the TB1 locus from rice (OsTB1) has shown that TB1 also controls tillering in rice even though rice is already tillered (Takeda et al., 2003). Loss of function of OsTB1 causes the fine culm1 (fc1) mutant, which has more tillers and a thinner stem than normal (Takeda et al., 2003). Whether the thin stem is a secondary effect of increased tillering is not known, but overexpression of OsTB1 also leads to reduced tiller number and increased culm thickness. Overexpression of maize TB1 also leads to a reduction in tiller number in wheat (Lewis et al., 2008).
Tillering is regulated by many environmental components, including planting density, shading, and fertilizer treatment (Doust, 2007b; Kebrom and Brutnell, 2007). fc1 mutants in rice are still affected by planting density, indicating that OsTB1/FC1 is not entirely responsible for this response (Takeda et al., 2003). In sorghum (Sorghum bicolor), it has been shown that SbTB1 responds to red/far-red light signaling (Kebrom et al., 2006). Red light is absorbed by plants and far-red light is reflected, so when plants are grown at high density, there is a decreased red to far-red ratio. This leads to the shade avoidance syndrome, one aspect of which is decreased branching (Kebrom and Brutnell, 2007). The red to far-red ratio is sensed by the PHYTOCHROMEs (PHYs). In sorghum, phyB mutants have reduced branching and increased SbTB1 expression (Kebrom et al., 2006). Therefore, TB1 integrates light signals as part of the shade avoidance pathway.
There are several genes related to TB1 in Arabidopsis, but TCP18/BRC1/TBL1 appears to play a similar role in Arabidopsis as TB1 in grasses (Aguilar-Martinez et al., 2007; Finlayson, 2007). Similar to maize, BRC1 is expressed in axillary buds, and loss-of-function mutants have more branches in particular from rosette leaves. Furthermore, extensive analysis of BRC1 interactions showed that BRC1 acts as an integrator of hormonal and environmental signals to regulate whether or not a branch grows out (Aguilar-Martinez et al., 2007; Finlayson, 2007). BRC1 acts downstream of the MAX pathway as the levels of BRC1 are significantly reduced in max mutants (Aguilar-Martinez et al., 2007; Finlayson, 2007). Furthermore, double mutants between max and brc1 mutants are as bushy as either single mutant, indicating that BRC1 acts in the same pathway as the MAX genes. It was proposed that the MAX pathway positively regulates BRC1 at the transcriptional level to suppress branching. BRC1 also interacts with auxin. yuc1D mutants that overexpress YUC have fewer branches, and yuc1D;brc1 double mutants have many branches, indicating that BRC1 is required for auxin-mediated apical dominance. The effect of auxin on BRC1 levels was not as clear. One research group did not detect a statistically significant change in BRC1 levels in the yuc1D or axr1 mutants using real-time reverse transcription-PCR (Aguilar-Martinez et al., 2007), but another lab did find that BRC1 is auxin regulated using semiquantitative reverse transcription-PCR on isolated buds (Finlayson, 2007). BRC1 levels were also affected by decapitation and planting density. Hence, in eudicots, BRC1 is proposed to act downstream of auxin and the MAX genes.
However, the interaction of OsTB1/FC1 with auxin and the strigolactone pathway differs in rice. Unlike Arabidopsis, the expression of FC1 was not down-regulated in the first node of dwarf mutants even though tillers grow out (Arite et al., 2007). Therefore, FC1 is proposed to act independently of the max pathway in rice (Arite et al., 2007). However, double mutants between the dwarf mutants and fc1 have not yet been reported. Furthermore, FC1 is not induced by auxin, although the mutant was found to be hypersensitive to auxin (Arite et al., 2007).
It is apparent that TB1/FC1/BRC1 is an integrator of hormonal and environmental signals in both monocots and eudicots. However, the wiring between components appears to be different, perhaps reflecting the different growth habits of maize, rice, and Arabidopsis. As selection on the TB1 promoter has occurred in maize, it would be interesting to compare the regulatory regions of TB1/FC1/BRC1 in different plant species to determine if these differences in wiring are due to changes in the regulatory region of TB1/FC1/BRC1.
The Role of Hormones in the Regulation of Rhizomes in Perennial Grasses
There is great interest at the moment in the development of perennial grasses as biofuels (Heaton et al., 2004; Bouton, 2007). There are multiple mechanisms for perennialism, but the one used by many perennial grasses is the production of an over-wintering, underground stem called the rhizome. The rhizome arises from an axillary meristem from the basal part of the stem. Axillary meristems on the rhizome give rise to tillers (Fig. 1C). At the end of the growing season, nutrients from the plant are relocated to the rhizome, which has the capacity to overwinter. The following spring, tillers sprout anew from the rhizome. An understanding of the regulation of axillary meristem initiation and outgrowth from rhizomes will be critical for efforts to manipulate bioenergy grasses such as switchgrass (Panicum virgatum) and Miscanthus for use in biofuel production.
Interestingly, it appears that many of the mechanisms controlling rhizome function are similar to those already known to regulate axillary meristems. Auxin is involved in rhizome initiation as auxin is required to produce rhizomes in culture, and treatment of plants with auxin transport inhibitors prevents the initiation of rhizomes (Kapoor and Rao, 2006). The levels of various hormones, including auxin and cytokinin, vary in the rhizome during the life cycle of the plant (Maslova et al., 2007). For example, during summer when rhizomes are active, there are high auxin and low cytokinin levels, while during the autumn when rhizomes are dormant, there are high cytokinin and low auxin levels. Therefore, hormones play a fundamental role in the control of tillering by rhizomes.
Furthermore, even though tillers in perennial grasses arise from rhizomes, they are still under the control of the same environmental conditions that regulate tillering in annual grasses (Ma et al., 2001; Heaton et al., 2004). For example, increased planting density reduces tiller number and increased fertilizer treatment increases tiller number. Genetic studies show that perennialism is controlled by relatively few genes (Hu et al., 2003; Westerbergh and Doebley, 2004). The identification of these genes will be essential for understanding the regulation of branching in perennial grasses.
FUTURE PERSPECTIVE
Different plants have different architecture regulated by the extent of branching from axillary meristems. Research on monocots and eudicots has shown that similar mechanisms control branching in these divergent species. As axillary meristems arose at the base of the seed plants, it is not surprising to see so many similarities in monocots and eudicots. Much research remains to be done to understand the function of additional components and how the components are integrated with each other. In particular, an understanding of how axillary meristems at different stages of development respond to genetic, environmental, and hormonal factors is lacking. A challenge for the future will be to understand how changes in the wiring or strength of interaction between components have led to the diversity of plant morphology seen today. An exciting area of research will be to determine how axillary meristems arose by understanding the function of these genes in emerging non-seed plant model systems.
Acknowledgments
I thank Klaus Theres (Max Planck Institute, Cologne) and Elizabeth Dun (University of Queensland, Australia) for stimulating discussions about axillary meristems, Yinong Yang (Penn State University) for providing the rice plants to photograph, and two anonymous reviewers for their helpful comments on the manuscript. This review is dedicated to the memory of Don Kaplan (Professor Emeritus, University of California, Berkeley), who inspired a generation of plant biologists to appreciate the power of comparative morphology for understanding plant development.
This work was supported by the National Science Foundation and the U.S. Department of Agriculture.
Note on genetic nomenclature: Arabidopsis and rice use the same nomenclature, but the nomenclature differs for maize. For consistency, the Arabidopsis/rice nomenclature was used throughout this article even for maize and other grasses.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paula McSteen (pcm11@psu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Aguilar-Martinez JA, Poza-Carrion C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51 1019–1029 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309 741–745 [DOI] [PubMed] [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O (2005) Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J 44 569–580 [DOI] [PubMed] [Google Scholar]
- Barazesh S, McSteen P (2008) Hormonal control of grass inflorescence development. Trends Plant Sci 13 656–662 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Ampudia CSG, Hooykaas PJJ, Offringa R (2003) PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol 132 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59 443–465 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8 505–520 [Google Scholar]
- Bennett T, Leyser O (2006) Something on the side: axillary meristems and plant development. Plant Mol Biol 60 843–854 [DOI] [PubMed] [Google Scholar]
- Beveridge CA (2006) Axillary bud outgrowth: sending a message. Curr Opin Plant Biol 9 35–40 [DOI] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46 69–78 [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Chatfield S, Leyser O (2003) Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8 443–449 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Hake S (2007) Flowering and determinacy in maize. J Exp Bot 58 909–916 [DOI] [PubMed] [Google Scholar]
- Bouton JH (2007) Molecular breeding of switchgrass for use as a biofuel crop. Curr Opin Genet Dev 17 553–558 [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G (2007) Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci 12 224–230 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O (2000) The hormonal regulation of axillary bud growth in Arabidopsis. Plant J 24 159–169 [DOI] [PubMed] [Google Scholar]
- Cheng YF, Dai XH, Zhao YD (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YF, Zhao YD (2007) A role for auxin in flower development. J. Integr. Plant Biol. 49 99–104 [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478 [DOI] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39 544–549 [DOI] [PubMed] [Google Scholar]
- Clark RM, Linton E, Messing J, Doebley JF (2004) Pattern of diversity in the genomic region near the maize domestication gene TB1. Proc Natl Acad Sci USA 101 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J (2006) A distant upstream enhancer at the maize domestication gene TB1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38 594–597 [DOI] [PubMed] [Google Scholar]
- Delker C, Raschke A, Quint M (2008) Auxin dynamics: the dazzling complexity of a small molecule's message. Planta 227 929–941 [DOI] [PubMed] [Google Scholar]
- Doebley J (2004) The genetics of maize evolution. Annu Rev Genet 38 37–59 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Doust A (2007. a) Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 100 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN (2007. b) Grass architecture: genetic and environmental control of branching. Curr Opin Plant Biol 10 21–25 [DOI] [PubMed] [Google Scholar]
- Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA (2004) Genetic control of branching in foxtail millet. Proc Natl Acad Sci USA 101 9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, Ferguson BJ, Beveridge CA (2006) Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol 142 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SA (2007) Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol 48 667–677 [DOI] [PubMed] [Google Scholar]
- Foo E, Buillier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, et al (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P (2008. a) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008. b) The relationship between auxin transport and maize branching. Plant Physiol 147 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter M, Doebley JF, Pe ME, Schmidt RJ (2004) The role of barren stalk1 in the architecture of maize. Nature 432 630–635 [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan CH, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al (2008) Strigolactone inhibition of shoot branching. Nature 455 189–194 [DOI] [PubMed] [Google Scholar]
- Grbic V (2005) Comparative analysis of axillary and floral meristem development. Can J Bot 83 343–349 [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton E, Voigt T, Long SP (2004) A quantitative review comparing the yields of two candidate C4 perennial biomass crops in relation to nitrogen, temperature and water. Biomass and Bioenergy 27 21–30 [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A (2001) The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13 2115–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Feldman LJ (1994) Bidirectional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192 276–286 [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka KI, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FY, Tao DY, Sacks E, Fu BY, Xu P, Li J, Yang Y, McNally K, Khush GS, Paterson AH, et al (2003) Convergent evolution of perenniality in rice and sorghum. Proc Natl Acad Sci USA 100 4050–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46 79–86 [DOI] [PubMed] [Google Scholar]
- Itoh JI, Hibara KI, Sato Y, Nagato Y (2008) Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogne K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Rao IU (2006) In vitro rhizome induction and plantlet formation from multiple shoots in Bambusa bambos var. gigantea Bennet and Gaur by using growth regulators and sucrose. Plant Cell Tissue Organ Cult 85 211–217 [Google Scholar]
- Kebrom TH, Brutnell TP (2007) The molecular analysis of the shade avoidance syndrome in the grasses has begun. J Exp Bot 58 3079–3089 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA (2006) Phytochrome B represses teosinte branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesselbach TA (1949) The Structure and Reproduction of Corn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003) LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655 [DOI] [PubMed] [Google Scholar]
- Kyozuka J (2007) Control of shoot and root meristem function by cytokinin. Curr Opin Plant Biol 10 442–446 [DOI] [PubMed] [Google Scholar]
- Lewis JM, Mackintosh CA, Shin S, Gilding E, Kravchenko S, Baldridge G, Zeyen R, Muehlbauer GJ (2008) Overexpression of the maize teosinte branched1 gene in wheat suppresses tiller development. Plant Cell Rep 27 1217–1225 [DOI] [PubMed] [Google Scholar]
- Leyser O (2003) Regulation of shoot branching by auxin. Trends Plant Sci 8 541–545 [DOI] [PubMed] [Google Scholar]
- Li CX, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PJ, Wang YH, Qian Q, Fu ZM, Wang M, Zeng DL, Li BH, Wang XJ, Li JY (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17 402–410 [DOI] [PubMed] [Google Scholar]
- Li XY, Qian Q, Fu ZM, Wang YH, Xiong GS, Zeng DL, Wang XQ, Liu XF, Teng S, Hiroshi F, et al (2003) Control of tillering in rice. Nature 422 618–621 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218 341–353 [DOI] [PubMed] [Google Scholar]
- Ma Z, Wood CW, Bransby DI (2001) Impact of row spacing, nitrogen rate and time on carbon partitioning of switchgrass. Biomass and Bioenergy 20 413–419 [Google Scholar]
- Mao CZ, Ding WN, Wu YR, Yu J, He XW, Shou HX, Wu P (2007) Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol 176 288–298 [DOI] [PubMed] [Google Scholar]
- Maslova SP, Tabalenkova GN, Kurenkova SV, Plusnina SN (2007) Seasonal changes in anatomical and morphological structure and the content of phytohormones and sugars in underground shoots of a long rhizome perennial grass Plalaroides arundinacea. Russ J Plant Physiol 54 491–497 [Google Scholar]
- Matusova R, Rani K, Verstappen FWA, Franssen MCR, Beale MH, Bouwmeester HJ (2005) The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol 139 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Hake S (2001) barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5 61–66 [DOI] [PubMed] [Google Scholar]
- McSteen P, Leyser O (2005) Shoot branching. Annu Rev Plant Biol 56 353–374 [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu XT, Kellogg E, Hake S (2007) barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Zhao Y (2008) Plant hormones and signaling: common themes and new developments. Dev Cell 14 467–473 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056 [DOI] [PubMed] [Google Scholar]
- Morita Y, Kyozuka J (2007) Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol 48 540–549 [DOI] [PubMed] [Google Scholar]
- Muller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogue F, Grandjean O, Craig S, Dennis E, Chaudhury A (2000) Higher levels of cell proliferation rate and cycD3 expression in the Arabidopsis amp1 mutant. Plant Growth Regul 32 275–283 [Google Scholar]
- Nordstrom A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V, Leyser O (2008) Hormonal control of shoot branching. J Exp Bot 59 67–74 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25 223–236 [DOI] [PubMed] [Google Scholar]
- Poethig RS (1988) Heterochronic mutations affecting shoot development in maize. Genetics 119 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55 65–76 [DOI] [PubMed] [Google Scholar]
- Ritter MK, Padilla CM, Schmidt RJ (2002) The maize mutant barren stalk1 is defective in axillary meristem development. Am J Bot 89 203–210 [DOI] [PubMed] [Google Scholar]
- Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57 431–449 [DOI] [PubMed] [Google Scholar]
- Schmitz G, Theres K (2005) Shoot and inflorescence branching. Curr Opin Plant Biol 8 506–511 [DOI] [PubMed] [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA 99 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz C, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin XQ, Loewen MC (2004) The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J Biol Chem 279 46940–46945 [DOI] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9 484–489 [DOI] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E (2007) The F-Box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto K, Kyozuka J (2002) Rice as a model for comparative genomics of plants. Annu Rev Plant Biol 53 399–419 [DOI] [PubMed] [Google Scholar]
- Skirpan A, Wu X, McSteen P (2008) Genetic and physical interaction suggest that BARREN STALK1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J 55 787–797 [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ (2005) The Decreased apical dominance 1/petunia hybrida carotenoid cleavage dioxygenase8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T, Sussex I (1989) Patterns in Plant Development. Cambridge University Press, Cambridge, UK
- Stirnberg P, van de Sande K, Leyser HMO (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141 [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33 513–520 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45 1028–1036 [DOI] [PubMed] [Google Scholar]
- Tantikanjana T, Yong JWH, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V (2001) Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev 15 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K, Skoog F (1933) Studies on the growth hormone of plants III: the inhibitory action of the growth substance on bud development. Proc Natl Acad Sci USA 19 714–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BE, Hake S (2009) Translational biology: from Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 195–200 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbergh A, Doebley J (2004) Quantitative trait loci controlling phenotypes related to the perennial versus annual habit in wild relatives of maize. Theor Appl Genet 109 1544–1553 [DOI] [PubMed] [Google Scholar]
- Wu X, McSteen P (2007) The role of auxin transport during inflorescence development in maize (Zea mays, Poaceae). Am J Bot 94 1745–1755 [DOI] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou HX, Wu P (2005) A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46 1674–1681 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Li W, Lorenz TC, Xie MT, Payne CT, Smith K, Glenny S, Payne GS, Christensen SK (2006) Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J Biol Chem 281 35520–35530 [DOI] [PubMed] [Google Scholar]
- Zhao Y (2008) The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol 11 16–22 [DOI] [PubMed] [Google Scholar]
- Zou JH, Zhang SY, Zhang WP, Li G, Chen ZX, Zhai WX, Zhao XF, Pan XB, Xie Q, Zhu LH (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48 687–696 [DOI] [PubMed] [Google Scholar]