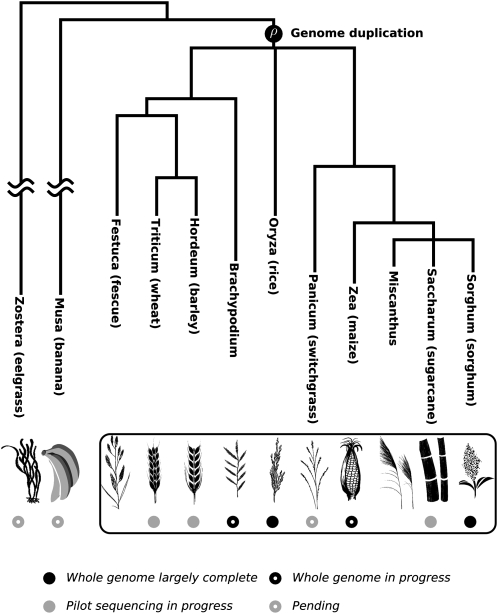
Building on a rich history of comparative genomics, scientists are making rapid progress toward a comprehensive framework for comparative genomics of the grass family (Poaceae) that will permit comparative studies at new levels of intricacy. The sequences of each of the two rice (Oryza sativa) subspecies (Goff et al., 2002; Yu et al., 2002, 2005b; Matsumoto et al., 2005), the recent completion of a high-quality genetically and physically anchored whole-genome sequence for sorghum (Sorghum bicolor; Paterson et al., 2009), and the imminent completion of a whole-genome shotgun sequence for Brachypodium distachyon together sample three important grass clades (Fig. 1). The rapidly progressing maize (Zea mays) genome sequence (www.maizegenome.org) provides a second panicoid, distinguished from sorghum by about 12 million years of evolution, including a whole-genome duplication (Swigonova et al., 2004). Comparison of these genomes with one another and with other genomes promises to clarify the grass gene set, providing phylogenetic data helpful to resolving what are sometimes striking differences between independent annotations of the same genome sequence, for example, rice RAP2 (Tanaka et al., 2008) versus TIGR5 (Ouyang et al., 2007), which differ substantially. Grass plastome sequencing is also advancing (Bortiri et al., 2008).
Figure 1.
The phylogenetic relationship between selected cereals and two outgroup species is illustrated as described elsewhere (http://www.mobot.org/MOBOT/research/APweb/welcome.html). The branches of the outgroups are approximated and shortened in length. Current progress in genome sequencing of relevant taxa is also indicated.
The generally similar gene content and order of cereals and related grasses, especially those such as Oryza, Sorghum, and Brachypodium that have not incurred lineage-specific genome duplications, provide a starting point for accelerating progress in the study and improvement of many additional taxa. Indeed, the vast majority of monocots lack sufficient genomic tools to investigate pertinent problems in agricultural productivity, conservation biology, ecology, invasion biology, population biology, and systematic biology. For example, orphan crops, which are collectively planted to 250 million ha year−1 and yield US$100 billion per year farm gate value in the developing world (Naylor et al., 2004), are largely without genomics resources. Many noncultivated monocots that are important as weeds, invasives, or potential new crops are likewise unexplored at the DNA level. Deductions about gene arrangement based on alignments of fully sequenced genomes (Tang et al., 2008) together with use of conserved exon sequences and intron locations to develop generalized monocot genomic tools (Feltus et al., 2006; Lohithaswa et al., 2007) promise to benefit research on monocot model and nonmodel systems alike.
DUPLICATION AND DIPLOIDIZATION OF GRASS GENOMES
Their similar gene content and order begs the question of what accounts for the tremendous morphological and physiological diversity among the cereals. One of the bigger surprises deriving from angiosperm genomes sequenced to date is the prominence of ancient genome duplication, and the grasses are no exception. Even in clearly diploid taxa with relatively low chromosome numbers and largely bivalent pairing at meiosis, RFLP maps hinted at large-scale duplications (Chittenden et al., 1994; Kishimoto et al., 1994; Nagamura et al., 1995). These interpretations were subsequently borne out by the rice genome sequence, revealing a genome duplication that occurred in a common grass ancestor (Paterson et al., 2004). A brief controversy about the scope of this duplication (Vandepoele et al., 2003; Wang et al., 2005) was reconciled to be largely attributable to differences in the stringency of statistical thresholds (Paterson et al., 2005).
It remains to be determined how much influence this ancient pan-grass duplication had on grass genomic diversity. Estimates based on the divergence of gene sequences suggest that the duplication occurred about 70 million years ago, followed by about 20 million years of evolution as a common lineage before the divergence of the panicoid, pooid, and oryzoid lineages (Paterson et al., 2004). This time period is long enough that most gene loss and neofunctionalization or subfunctionalization should have occurred in the common ancestor (Lynch et al., 2001). However, morphologically distinct tissues found in coprolites dated to approximately 65 million years ago (Prasad et al., 2005) suggest that the major grass lineages had diverged much earlier than indicated by gene sequences. Detailed comparison of the fates of duplicated genes in Oryza, Sorghum, and Brachypodium will help to distinguish between the possibilities that (1) the duration of the period between duplication and taxon divergence was shorter than previously thought, as reflected in the substantial degree of divergence among these lineages regarding which genes were retained/lost, or (2) the duplication event itself was more ancient than is indicated by clock-like analysis of DNA sequences.
A host of more recent duplications may have contributed more than the shared ancient duplication to structural and functional diversification of the cereals. The first of these likely to be accessible on a genome-wide scale will be in maize. The sorghum sequence will provide an attractive outgroup for inferences about the ancestral states of maize homologs in that the duplication occurred in the maize lineage since the two taxa diverged from a common ancestor about 12 million years ago (Swigonova et al., 2004). In a manner similar to the maize-sorghum comparison, comparison of Brachypodium to tetraploid and hexaploid wheats (Triticum spp.) may offer insight into the early stages of adaptation to the duplicated state in that these formed about 300,000 to 500,000 and approximately 10,000 years ago, respectively (Levy and Feldman, 2002).
The potential merits of cellulosic biofuels (Farrell et al., 2006) have invigorated research into several taxa of current (Saccharum) or potential (Miscanthus and Panicum) importance in which polyploidy and associated heterozygosity complicate the genetic analysis. For example, Saccharum genotypes are characterized by numerous (from 36 to more than 200) variably sized chromosomes thought to have shared common ancestry with the 2n = 20 chromosomes of sorghum between 5 and 9 million years ago (Aljanabi et al., 1994). Many regions of the Sorghum genome correspond to four or more homologous regions of Saccharum, suggesting that in the short period since their divergence from a common ancestor, Saccharum has been through at least two whole-genome duplications (Ming et al., 1998). The members of individual homologous groups of Saccharum chromosomes reveal a remarkable range of affinities, from high levels of preferential pairing (although never true allelism) to frequent univalence (Jannoo et al., 2004). Better understanding of the levels and patterns of variation among homologous chromosomes in complex autopolyploid genomes may complement findings from allopolyploid wheat and perhaps help to clarify which mode of polyploidization occurred in maize and other cereals.
EUCHROMATIN VERSUS HETEROCHROMATIN: TALES OF TWO GENOMES
The generally similar gene content and order of grasses are in stark contrast to their remarkable differences in genome size, spanning at least a 50-fold range from the approximately 300 million bp of B. distachyon to the approximately 16,000 million bp of Triticum aestivum (hexaploid wheat; Arumuganathan and Earle, 1991). Building on a long list of prior studies comparing orthologous bacterial artificial chromosomes (BACs) in different taxa, comparison of whole-genome sequences, or many thousands of genetically and/or physically characterized sequence-tagged sites now permit the exploration of levels and patterns of microsynteny on a genome-wide scale.
Despite the large changes in overall genome sizes in the grasses, the size of gene-rich regions in these genomes remains similar (Feuillet and Keller, 1999). Detailed comparison of the rice sequence with a genetically anchored sorghum physical map showed cytologically distinct euchromatin versus heterochromatin to correlate closely with gene versus repeat abundance, patterns of rice-sorghum synteny, and the frequency of recombination (Bowers et al., 2005). Synteny is highest and retroelement abundance is lowest in distal portions of the chromosomes, where recombination rate per unit of DNA has generally been highest. Preferential preservation of microsynteny in recombinogenic regions suggests that gene rearrangement is generally deleterious, an intuitive hypothesis that has previously lacked empirical support. “Muller's ratchet” (Muller, 1932) predicts that deleterious mutations may accumulate in, and contribute to the degeneration of, nonrecombinogenic regions, classical examples being mammalian Y chromosomes or incipient plant sex chromosomes (Liu et al., 2004). This is consistent with a strong negative correlation between repetitive DNA content and the recombinational length of rice chromosomes and with the much greater abundance of repetitive DNA than genes in the pericentromeric regions (Bowers et al., 2005). Accelerated gene loss in recombination-poor regions of wheat (Akhunov et al., 2003) and a propensity for small insertions in centromeric regions of mammalian genomes (Bailey et al., 2001) lend further support to this idea.
The patterns of gene and repeat organization and other features that distinguish euchromatin and heterochromatin have been substantially preserved since the divergence of “paleologous” (homologous) chromosomes that resulted from the duplication of a common ancestral chromosome approximately 70 million years ago, remaining correlated in the modern chromosome pairs (Bowers et al., 2005). The exciting finding that the heterochromatic state of Arabidopsis (Arabidopsis thaliana) is determined by transposable elements and related tandem repeats, under the control of the chromatin-remodeling ATPase DDM1 and guided by small interfering RNAs (Lippman et al., 2004), suggests a starting point for better understanding of the mechanisms responsible for the establishment and preservation of the two “genomes within a genome” in the cereals.
DISTINGUISHING FEATURES OF THE CEREALS
What genes (if any) distinguish the grasses from other clades, or from one another? Comparisons of rice with Arabidopsis and other dicots reveal thousands of rice gene models for which no homolog can be discerned. While some of these may be annotation errors, early studies suggest that an appreciable population of genes may be lineage specific in the grasses (Campbell et al., 2007). This finding, while important, is not unprecedented. For example, the major seed storage proteins of maize (zein) and sorghum (kafirin) are encoded by genes that appear to have evolved since the divergence of the panicoids from the oryzoids, being absent in rice (Song and Messing, 2003). Mechanisms that may lead to the creation of new genes by juxtaposition of existing gene fragments are now well known. Mutator-like “Pack-MULE” elements appear to be the predominant gene-transducing element (Jiang et al., 2004) in rice, and intact “helitrons” (i.e. that potentially encode a helicase) are implicated in maize gene movement (Brunner et al., 2005). Sequences of additional grass genomes will permit researchers to reciprocally improve gene annotations for each and help to clarify which gene models are lineage specific at various levels. For example, which genes are shared by the Oryza-Sorghum-Brachypodium group but not dicots, and which are shared by panicoids (such as sorghum and maize) but not oryzoids (rice)? An important artifactual explanation of lineage-specific genes is that they are unrecognized pseudogenes or transposons. Such instances may sometimes be discerned by identifying cases that are incongruous with surrounding synteny/colinearity relationships.
Are specific groups of genes held in close proximity by selection? The grasses have long been noted for their seemingly slow structural evolution, preserving gene content and order over many tens of millions of years and despite enormous changes in genome size. While the Oryza-Sorghum-Brachypodium group is still viewed as having evolved relatively slowly, grasses that have been through recent genome duplications have undergone substantial fractionation of ancestral gene orders across multiple chromosomes. For example, there is greater similarity of gene order between the genomes of sorghum and rice, diverged by 40 to 50 million years (Paterson et al., 2004; Bowers et al., 2005), than between the genomes of sorghum and maize, diverged by 10 to 15 million years (Bowers et al., 2003). As yet unknown factors other than polyploidy also influence grass genome stability. Some grass lineages show singularly rapid chromosomal rearrangements independent of polyploidization (Devos et al., 1993), and others appear to have undergone substantial numbers of chromosomal condensations over short time periods (Tang et al., 2008).
Preferential preservation of microsynteny in recombinogenic regions suggests that gene rearrangement in grasses is generally somewhat deleterious (Bowers et al., 2005), but it provides little if any evidence regarding why. In mammals, gene order conservation has been associated with the regulation of gene expression (Semon and Duret, 2006; Gierman et al., 2007), replication, and chromatin structure (Huvet et al., 2007). Nonrandom patterns of gene expression are found among proximally located rice (Ma et al., 2005) and Arabidopsis (Ren et al., 2007) genes. While a comparative study to date found no microsynteny between the two species in these domains (Ren et al., 2007), inferences about monocot-dicot synteny are much more complicated than was realized before the availability of whole-genome sequences (Paterson et al., 1996), due largely to multiple paleopolyploidies and associated gene losses. While monocot-dicot synteny is difficult to discern, synteny blocks in the grasses often comprise entire chromosomes or chromosome arms and offer little resolution to discern chance persistence from the preservation of gene order by selection. Additional genomes and new approaches are needed to investigate the importance of specific gene arrangements, particularly across long periods of angiosperm evolution, such as those separating monocots and dicots (Wang et al., 2006), which have included multiple genome duplications (Tang et al., 2008).
One striking compositional feature of the cereal genes is the distinctly bimodal GC content distribution. Such bimodality is evident in all major clades of cereals investigated, suggesting a common origin that predates the cereal diversification and possibly even before the Musa-grasses divergence (Carels and Bernardi, 2000; Lescot et al., 2008). Additionally, a subset of the cereal genes shows a unique negative GC gradient from the 5′ untranslated region and extending about 1 kb into the coding region (Wong et al., 2002). The causes of such features are largely elusive, but their effects are large enough to warrant extra caution for gene annotations.
DIVERSITY WITHIN GRASS SPECIES
Rice is particularly advanced with regard to analysis of within-species diversity, enjoying both a BAC-based sequence of one subspecies (Matsumoto et al., 2005) and a whole-genome shotgun sequence of another (Yu et al., 2005b). Comparisons of the two subspecies (Feltus et al., 2004; Yu et al., 2005b) and a growing mass of within-subspecies data (McNally et al., 2006) provide early insight into the nature and distribution of intraspecific polymorphism. Massively parallel resequencing approaches (Margulies et al., 2005; Shendure et al., 2005) provide for the economical surveying of a diverse sampling of field-evaluated germplasm for functionally significant alleles, increasing the breadth and depth of our knowledge of diversity within individual grass gene pools. Single nucleotide polymorphism (SNP) identification in plants is borrowing heavily from algorithms used in human (Marth et al., 1999), which appear to be adaptable to calling SNPs in the next-generation sequence (Barbazuk et al., 2007). Indeed, even in taxa that lack a reference sequence, reduced representation approaches are being employed to discover large SNP sets useful in DNA marker-assisted studies. With completed sequences in hand, diversity analysis may target genes directly, rather than via the proxy markers that have been necessities for the past 20 years. New clone-free DNA sequencing methods that provide increased cost efficiency and speed (Margulies et al., 2005; Shendure et al., 2005) present the possibility of “transcriptome shadowing.” The rapid sequencing of large populations of cDNA to a deep level of coverage for a diverse sampling of germplasm is expected to shed light on the levels and patterns of functional polymorphism.
The merits of association genetics approaches (Remington et al., 2001; Thornsberry et al., 2001; Yu et al., 2005a), using linkage disequilibrium and/or identification of functional nucleotide variants to link DNA sequences to their phenotypic consequences, are motivating the development of “diversity panels” in many crops for key phenotypes and their characterization for genome-wide sets of DNA markers (usually simple sequence repeats) needed to provide background information about population structure (relatedness). For example, 230 accessions that represent most of the genetic diversity present in sorghums from Africa (the center of diversity), and 148 genotypes sampling elite public breeding lines and other lines of genetic and historical interest, have been phenotyped for eight traits (flag leaf length and width, plant height, terminal branch length, flowering time [measured as time to midanthesis], panicle length, and flag leaf height and exsertion). Additionally, levels of population structure and familial relatedness were assessed with 47 simple sequence repeat loci (Casa et al., 2008). In maize, the development of 25 recombinant inbred line populations from a carefully planned set of crosses promises very precise delineation of associations by “nested association mapping” (Yu et al., 2005a).
FURTHER SEQUENCING AND RESEQUENCING(S)
A singularly important need to round out our knowledge of grass genomic diversity is the genome sequence of an outgroup taxon for the pan-grass genome duplication. The Oryza-Sorghum-Brachypodium group appears to have diverged perhaps 20 million years or more after the duplication (Paterson et al., 2004; but see above). Since most postduplication change is likely to have been completed by this time (Lynch et al., 2001), their comparison may offer only modestly more insight into types, levels, and patterns of postduplication evolution than can already be gleaned from comparing duplicated regions within any one of them. Ideally, an outgroup for the pan-grass duplication would be sufficiently close to the grasses that nonfunctional sequences would be largely diverged but conserved noncoding sequences (Inada et al., 2003) would still be discernible. The marine angiosperm Zostera marina (eelgrass), targeted by the Joint Genome Institute Community Sequencing Program (www.jgi.doe.gov/sequencing/cspseqplans2009.html), and Musa (banana), for which a draft sequence is planned by Genoscope (N. Roux, personal communication), may each be too distant evolutionarily to discern conserved noncoding sequences, although gene order similarities are clear for Musa (Lescot et al., 2008).
More Poaceae genomes are also needed. The chloridoid, bambusoid, and arundonoid clades each lack a sequenced genome and have only limited EST resources, despite containing species important as food, feed, turf, forage, biofuel, and revegetation/remediation crops, among others. Moreover, it would enhance our ability to employ phylogenetic inference to analyze distinguishing features of individual genomes if we have multiple genomes per clade. For example, Setaria italica (www.jgi.doe.gov/sequencing/cspseqplans2008.html) and maize will provide additional representatives of the panicoid clade, complementing sorghum.
How will the next wave of grass genomes be sequenced? The costs and benefits of BAC-based versus whole-genome shotgun sequencing remained controversial following a comparison of both approaches in rice (Matsumoto et al., 2005; Yu et al., 2005b), perhaps partly motivating the BAC-based approach being taken in maize (www.maizesequence.org). However, relatively high contiguities have been achieved in whole-genome shotgun assemblies of sorghum (www.phytozome.net/sorghum) and Brachypodium (www.brachybase.org/), albeit aided in sorghum by detailed genetic and physical maps and the ability to use rice synteny as supporting evidence to reject unlikely assemblies and corroborate equivocal ones (Bowers et al., 2003, 2005).
Among the grass genomes that are high priorities for sequencing are several that pose new challenges, partly in terms of size (wheat) but equally if not more importantly in terms of organization (Saccharum, Miscanthus, Panicum). How these large, heterozygous polyploid genomes will be tackled remains an open question. Reduced-representation approaches, begun some time ago for Saccharum (Vettore et al., 2003) and now in progress for Panicum (www.jgi.doe.gov), will permit “overlaying” of genes from these systems onto appropriate outgroups for their respective duplications (Sorghum for Saccharum and Miscanthus; Setaria for Panicum), providing a foundation for some applications. However, genome structural information, which will be largely lacking from these reduced-representation approaches, would provide insight into the types, levels, and patterns of evolution associated with adaptation to polyploidy. Such information may be of importance to the productivity of these grasses and may also provide fundamental insights into different paths by which a genome adapts to the duplicated state by comparison with allopolyploid wheat. One suggestion (Paterson, 2006) has been to try to simplify such genomes by identifying a BAC tiling path that covers their basic chromosome set(s) once, deferring the issue of variation among different homologous members of each chromosome in the basic set to later resequencing studies. Continuing improvements in primary sequencing efficiency might render this idea obsolete, making economical the possibility of whole-genome shotgun approaches even in these large autopolyploid genomes. The feasibility of accurate assembly will be determined by the degree of nucleotide-level divergence among homologous alleles, superimposed on the nature of the repetitive DNA and levels of single-gene duplicates that are considerations in all genomes. Targeted comparisons of homologous BACs are beginning to provide the information needed to evaluate the feasibility of assembling whole-genome shotgun data for such genomes (Jannoo et al., 2007).
NEW FUNCTIONAL TOOLS AND INFORMATION
The sequencing of each new grass genome will be a strong enticement to attract scientists with new skills and ideas, offering new opportunities to fill important gaps in relating grass genes to their functions. Extensive functional genomics efforts in progress for Oryza and Zea will no doubt reveal the functions of many grass genes. However the preferred system in which to study a particular gene, gene family, or trait will vary on a case-by-case basis due to complicating factors such as single gene duplication as well as novel features such as unusual expression patterns or nucleotide-based evidence of positive selection. For example, recent whole-genome duplication in Zea may make sorghum a simpler system in which to study many genes. On the other hand, subfunctionalization or neofunctionalization of duplicated genes following its polyploidy may be unique to Zea. For each successive genome sequenced, it will become more important to develop functional tools that can be readily targeted to the analysis of subsets of genes that show distinctive features relative to those already characterized in previous models. Approaches such as TILLING (McCallum et al., 2000; Till et al., 2003; Comai and Henikoff, 2006), virus-induced gene silencing (Robertson, 2004), and small RNA-based gene silencing (Ossowski et al., 2008) are attractive examples.
NEW ECOSYSTEM SERVICES AND AGRICULTURAL SYSTEMS
The expansion of agriculture to increase plant biomass available for the production of fuels without detracting from the growing need for food production may stimulate new interest in aspects of grass biology that are currently underexplored. Today, more than two-thirds of global cropland is sown to monocultures of annual crops. Soil erosion has followed tillage agriculture as it spread across the earth's surface, by some estimates sacrificing one-third of the planet's arable land in the past few decades (Pimentel et al., 1995). Before agriculture, nearly all terrestrial plant communities were diverse mixtures dominated by perennial species, which offer more protection against soil erosion (Gantzer et al., 1990), manage water and nutrients more effectively (Randall and Mulla, 2001), store more carbon below ground, and are more resilient to abiotic stresses (Glover, 2005). It has been suggested that diverse mixtures dominated by perennial species may have been approximately 5% more productive, averaged worldwide, than the highly managed croplands that have displaced them (Field, 2001).
The need to develop new cropping systems that preserve marginal land while maximizing the biomass yield of promising new crop species, such as the highly productive Miscanthus (Heaton et al., 2008), is a considerable challenge that may add new applications of comparative grass genomics to the ongoing need to increase a high-quality food supply. Indeed, food production, too, might benefit from more serious consideration of alternative production systems in some environments (Tilman et al., 2002). We have only limited understanding of the genetics of perenniality in grasses (Cox et al., 2002; DeHaan et al., 2005) and even less information about the optimal balance between harvestable biomass and perennation organs for biofuels crops, although classical forage breeding provides an obvious guide. Better understanding of the basis for the extraordinary biomass yields of a select few plants (Heaton et al., 2008) may also offer benefits in the improvement of a host of cereals.
Acknowledgments
We thank several members of the Paterson laboratory for valuable comments.
This work was supported by the U.S. National Science Foundation (grant nos. DBI–9872649, DBI–0115903, and MCB–0450260), the National Sorghum Producers, the International Consortium for Sugarcane Biotechnology, and a John Simon Guggenheim Foundation fellowship to A.H.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew H. Paterson (paterson@uga.edu).
References
- Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin, Gustafson JP, Lazo G, Chao S, et al (2003) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res 13 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljanabi SM, McClelland M, Petersen C, Sobral BWS (1994) Phylogenetic analysis of organellar DNA-sequences in the Andropogoneae, Saccharinae. Theor Appl Genet 88 933–944 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9 208–218 [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current Human Genome Project assembly. Genome Res 11 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS (2007) SNP discovery via 454 transcriptome sequencing. Plant J 51 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri E, Coleman-Derr D, Lazo G, Anderson O, Gu Y (2008) The complete chloroplast genome sequence of Brachypodium distachyon: sequence comparison and phylogenetic analysis of eight grass plastomes. BMC Res Notes 1 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Abbey C, Anderson S, Chang C, Draye X, Hoppe AH, Jessup R, Lemke C, Lennington J, Li Z, et al (2003) A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165 367–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Arias MA, Asher R, Avise JA, Ball RT, Brewer GA, Buss RW, Chen AH, Edwards TM, Estill JC, et al (2005) Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci USA 102 13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17 343–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Zhu W, Jiang N, Lin H, Ouyang S, Childs KL, Haas BJ, Hamilton JP, Buell CR (2007) Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol 145 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Bernardi G (2000) Two classes of genes in plants. Genetics 154 1819–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48 30–40 [Google Scholar]
- Chittenden LM, Schertz KF, Lin YR, Wing RA, Paterson AH (1994) A detailed RFLP map of Sorghum bicolor X S. propinquum, suitable for high-density mapping, suggests ancestral duplication of sorghum chromosomes or chromosomal segments. Theor Appl Genet 87 925–933 [DOI] [PubMed] [Google Scholar]
- Comai L, Henikoff S (2006) TILLING: practical single-nucleotide mutation discovery. Plant J 45 684–694 [DOI] [PubMed] [Google Scholar]
- Cox TS, Bender M, Picone C, Van Tassel DL, Holland JB, Brummer EC, Zoeller BE, Paterson AH, Jackson W (2002) Breeding perennial grain crops. Crit Rev Plant Sci 21 59–91 [Google Scholar]
- DeHaan LR, Van Tassel DL, Cox TS (2005) Perennial grain crops: a synthesis of ecology and plant breeding. Renewable Agriculture and Food Systems 20 5–14 [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, Liu CJ, Masojc P, Xie DX, Gale MD (1993) Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet 85 673–680 [DOI] [PubMed] [Google Scholar]
- Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM (2006) Ethanol can contribute to energy and environmental goals. Science 311 506–508 [DOI] [PubMed] [Google Scholar]
- Feltus FA, Singh HP, Lohithaswa HC, Schulze SR, Silva T, Paterson AH (2006) Conserved intron scanning primers: targeted sampling of orthologous DNA sequence diversity in orphan crops. Plant Physiol 140 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Keller B (1999) High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA 96 8265–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CB (2001) Sharing the garden. Science 294 2490–2491 [DOI] [PubMed] [Google Scholar]
- Gantzer CJ, Anderson SH, Thompson AL, Brown JR (1990) Estimating soil erosion after 100 years of cropping on Sanborn Field. J Soil Water Conserv 45 641–644 [Google Scholar]
- Gierman HJ, Indemans MHG, Koster J, Goetze S, Seppen J, Geerts D, van Driel R, Versteeg R (2007) Domain-wide regulation of gene expression in the human genome. Genome Res 17 1286–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J (2005) The necessity and possibility of perennial grain crops. Renewable Agriculture and Food Systems 20 1–4 [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang RL, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296 92–100 [DOI] [PubMed] [Google Scholar]
- Heaton EA, Dohleman FG, Long SP (2008) Meeting US biofuel goals with less land: the potential of Miscanthus. Glob Change Biol 14 2000–2014 [Google Scholar]
- Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, Thermes C (2007) Human gene organization driven by the coordination of replication and transcription. Genome Res 17 1278–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada DC, Bashir A, Lee C, Thomas BC, Ko C, Goff SA, Freeling M (2003) Conserved noncoding sequences in the grasses. Genome Res 13 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannoo N, Grivet L, Chantret N, Garsmeur O, Glaszmann JC, Arruda P, D'Hont A (2007) Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J 50 574–585 [DOI] [PubMed] [Google Scholar]
- Jannoo N, Grivet L, David J, D'Hont A, Glaszmann JC (2004) Differential chromosome pairing affinities at meiosis in polyploid sugarcane revealed by molecular markers. Heredity 93 460–467 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao ZR, Zhang XY, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573 [DOI] [PubMed] [Google Scholar]
- Kishimoto N, Higo H, Abe K, Arai S, Saito A, Higo K (1994) Identification of the duplicated segments in rice chromosomes 1 and 5 by linkage analysis of cDNA markers of known functions. Theor Appl Genet 88 722–726 [DOI] [PubMed] [Google Scholar]
- Lescot M, Piffanelli P, Ciampi AY, Ruiz M, Blanc G, Leebens-Mack J, da Silva FR, Santos CMR, D'Hont A, Garsmeur O, et al (2008) Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics 9 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AA, Feldman M (2002) The impact of polyploidy on grass genome evolution. Plant Physiol 130 1587–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black MA, Vaughn M, Dedhia N, McCombie WR, Lavine K, Mittal V, May BP, Kasschau K, et al (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471–476 [DOI] [PubMed] [Google Scholar]
- Liu ZY, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu QY, Pearl HM, Kim MS, Charlton JW, Stiles JI, et al (2004) A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427 348–352 [DOI] [PubMed] [Google Scholar]
- Lohithaswa HC, Feltus FA, Singh HP, Bacon CD, Bailey CD, Paterson AH (2007) Leveraging the rice genome sequence for comparative genomics in monocots. Theor Appl Genet 115 237–243 [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LG, Chen C, Liu XG, Jiao YL, Su N, Li L, Wang XF, Cao ML, Sun N, Zhang XQ, et al (2005) A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res 15 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiva S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu ZJ, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR (1999) A general approach to single-nucleotide polymorphism discovery. Nat Genet 23 452–456 [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M, Namiki N, Mizuno H, Yamamoto K, Antonio BA, Baba T, et al (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol 123 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K, Bruskiewich R, Mackill D, Buell CR, Leach J, Leung H (2006) Sequencing multiple and diverse rice varieties: connecting whole-genome variation with phenotypes. Plant Physiol 141 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Liu SC, Lin YR, da Silva J, Wilson W, Braga D, van Deynze A, Wenslaff TF, Wu KK, Moore PH, et al (1998) Detailed alignment of Saccharum and Sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150 1663–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ (1932) Some genetic aspects of sex. Am Nat 66 118–138 [Google Scholar]
- Nagamura Y, Inoue T, Antonio B, Shimano T, Kajiya H, Shomura A, Lin S, Kuboki Y, Harushima Y, Kurata N, et al (1995) Conservation of duplicated segments between rice chromosomes 11 and 12. Breed Sci 45 373–376 [Google Scholar]
- Naylor RL, Falcon WP, Goodman RM, Jahn MM, Sengooba T, Tefera H, Nelson RJ (2004) Biotechnology in the developing world: a case for increased investments in orphan crops. Food Policy 29 15–44 [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53 674–690 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35 D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH (2006) Leafing through the genomes of our major crop plants: strategies for capturing unique information. Nat Rev Genet 7 174–184 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature (in press) [DOI] [PubMed]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Vandepoele K, Van de Peer Y (2005) Ancient duplication of cereal genomes. New Phytol 165 658–661 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lan TH, Reischmann KP, Chang C, Lin YR, Liu SC, Burow MD, Kowalski SP, Katsar CS, DelMonte TA, et al (1996) Toward a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat Genet 14 380–382 [DOI] [PubMed] [Google Scholar]
- Pimentel D, Harvey C, Resosudarmo P, Sinclair K, Kurz D, McNair M, Crist S, Shpritz L, Fitton L, Saffouri R, et al (1995) Environmental and economic costs of soil erosion and conservation benefits. Science 267 1117–1123 [DOI] [PubMed] [Google Scholar]
- Prasad V, Stromberg CAE, Alimohammadian H, Sahni A (2005) Dinosaur coprolites and the early evolution of grasses and grazers. Science 310 1177–1180 [DOI] [PubMed] [Google Scholar]
- Randall GW, Mulla D (2001) Nitrate nitrogen in surface waters as influenced by climatic conditions and agricultural practices. J Environ Qual 30 337–344 [DOI] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98 11479–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XY, Stiekema W, Nap JP (2007) Local coexpression domains in the genome of rice show no microsynteny with Arabidopsis domains. Plant Mol Biol 65 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55 495–519 [DOI] [PubMed] [Google Scholar]
- Semon M, Duret L (2006) Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Mol Biol Evol 23 1715–1723 [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin XX, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM (2005) Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309 1728–1732 [DOI] [PubMed] [Google Scholar]
- Song R, Messing J (2003) Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA 100 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, Sakai H, Wu J, Itoh T, Sasaki T, et al (2008) The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res 36 D1028–D1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and colinearity in plant genomes. Science 320 486–488 [DOI] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28 286–289 [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, et al (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S (2002) Agricultural sustainability and intensive production practices. Nature 418 671–677 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Simillion C, Van de Peer Y (2003) Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15 2192–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore AL, da Silva FR, Kemper EL, Souza GM, da Silva AM, Ferro MIT, Henrique-Silva F, Giglioti EA, Lemos MVF, Coutinho LL, et al (2003) Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res 13 2725–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shi X, Hao BL, Ge S, Luo J (2005) Duplication and DNA segmental loss in rice genome and their implications for diploidization. New Phytol 165 937–946 [DOI] [PubMed] [Google Scholar]
- Wang XY, Shi XL, Li Z, Zhu QH, Kong L, Tang W, Ge S, Luo JC (2006) Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics 7 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GKS, Wang J, Tao L, Tan J, Zhang JG, Passey DA, Yu J (2002) Compositional gradients in Gramineae genes. Genome Res 12 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu SN, Wang J, Wong GKS, Li SG, Liu B, Deng YJ, Dai L, Zhou Y, Zhang XQ, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 296 79–92 [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs W, Bi IV, Yamasaki M, Doebley J, McMullen M, Gaut B, Nielsen D, Holland J, et al (2005. a) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38 203–208 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li SG, Li H, Zhou J, Ni PX, Dong W, Hu SN, Zeng CQ, et al (2005. b) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3 266–281 [DOI] [PMC free article] [PubMed] [Google Scholar]