If one considers the Gordon Conference in Plant Genetics in 1997 as the origin of the international effort to sequence the rice (Oryza sativa) genome and the publication of the data in 2005 as the completion of production and analysis, it took about 8 years and 14 major laboratories from nine countries (International Rice Genome Sequencing Project, 2005). In the case of sorghum (Sorghum bicolor) with nearly twice the size of the rice genome, its origin dates back to the Plant Animal Genome Meeting in 2005 and it has a completion date in 2008, with only three production laboratories and a smaller group of analysts than for rice (Paterson et al., 2009). This comparison clearly demonstrates the acceleration and efficiency in genome analysis. While sequencing cDNAs of any particular species has been and will be important for our understanding of gene expression and the annotation of genes, it has been insufficient to gain insight in the organization of genes in their chromosomal context. Protein and mRNA accumulation does not distinguish the contribution of highly homologous gene copies. Furthermore, we know now that major traits are manifested by sequences not transcribed. For instance, the domestication of maize (Zea mays) is linked to a sequence 60 to 90 kb upstream of the coding region of the Tb1 gene (Clark et al., 2004), indicating that coding regions are a poor predictor of the sizes of genes and that transposable elements in introns and nontranscribed regions are filtered by expression. Therefore, the foundation for the genetic and evolutionary analysis of any organism will be ultimately its genomic sequence. One of the arguments has been that the coding portion of complex genomes represents only a small fraction of the entire genome, and this subset, rather than the rest, should be sequenced because of cost and time (Whitelaw et al., 2003). Based on such arguments, the sorghum genome has been sequenced by applying methylation filtration technology with fewer than 550,000 reads (Bedell et al., 2005). However, distribution of presumably genetically inert sequences appears to be uneven, preventing simple fractionation methods to separate these types of DNA material for selective sequencing (Haberer et al., 2005). Such attempts also would fail to provide us with an understanding of spacing and linear order of sequences. Moreover, increasing knowledge of repetitive DNA sequences, their role, and divergence not only shines new light on chromosome evolution and gene regulation, but also improves the resolution of genetic maps and the algorithms to assemble contiguous sequences from shotgun reads. For instance, not only SSR markers, but also nested retrotransposons, have become invaluable genetic markers (Devos et al., 2005).
METHODS FOR SEQUENCING WHOLE GENOMES
Critical to the sequencing of large chromosomes has been the DNA shotgun sequencing method and the use of single and paired synthetic universal primers (Messing et al., 1981; Vieira and Messing, 1982). The method is based on fragmenting DNA into small sizes, purifying them by cloning, and defining the start of sequencing with a short oligonucleotide. Because fragmentation produces overlapping fragments, sequences can be concatenated by overlapping sequence information (Larson and Messing, 1982), thereby reconstructing contiguous sequences (contigs), which was first exemplified by the complete structure of a plant DNA virus (Gardner et al., 1981). In principle, there is no limit to the reiteration of contig building and, indeed, large chromosomes have been assembled (International Human Genome Sequencing Consortium, 2004). There are two valuable experimental validations to properly assemble contigs. One is restriction mapping because the sizes of restriction fragments have to match those predicted from the assembled sequence, which was already used in the case of the plant DNA virus (Gardner et al., 1981). The second is recombination maps because the genetic map has to be collinear with the sequence. These types of validations have also resulted in strategies to sequence genomes in increments and by large consortia, as it was the case for the rice genome (International Rice Genome Sequencing Project, 2005). Restriction fragment length patterns can be used to construct clone maps from bacterial artificial chromosomes (BACs) containing genomic DNA of 100 to 150 kb. Because genomic libraries are formed from partially digested genomic DNA, complete digests produce common-sized restriction fragments from overlapping clones, facilitating their concatenation into BAC contigs, called finger-printed contigs. Moreover, the process can be adapted to high-throughput robotics so that even large genomes can be mapped in relatively short periods of time (Nelson et al., 2005). Taking advantage of genetically mapped DNA sequences, these finger-printed contigs can be ordered according to their chromosomal position. In turn, sequences linked to individual BAC clones also become mapped in silico.
Such physical map resources are critical for any genome sequencing approach, whether it is a concerted effort sequencing a selection of ordered overlapping clones or whole-genome shotgun (WGS) sequencing. Furthermore, if one genome is sequenced, alignment of BAC-linked sequences of a second closely related genome can be used to order clone libraries of the second genome (Gregory et al., 2002). Indeed, both the sequencing of sorghum and maize are based on physical maps that have been constructed based on the rice sequence (Wei et al., 2007; Paterson et al., 2009). Another major difference of sequencing sorghum by WGS compared to the human genome by WGS (Venter et al., 2001) was the generation of shotgun libraries with different insert sizes of 2.44, 6.4, 6.88, 8.6, 34.7, 91, and 108 kb and sequencing its ends at different depths (Paterson et al., 2009). By careful selection of sheared DNA fragments within a narrow size range and long paired high-quality reads of over 700 bp, sequence assembly algorithms can rely on the distance of two paired reads (Jaffe et al., 2003), resulting in better concatenation and fewer supercontigs.
Recent advances in using solid support as opposed to capillary electrophoresis (CE) to increase throughput of sequence reads have introduced a new level of massive parallelization of sequencing, also referred to as nextgen (next generation) sequencing, although the concept of single-nucleotide extension by polymerase represents the oldest sequencing method (Wu and Kaiser, 1968). While these advances have permitted deep sequencing of RNA mixtures and single-nucleotide polymorphism analysis of sequenced genomes (Brenner et al., 2000), it has been less clear whether they would also be a new leap in allelic sequencing or even sequencing entire genomes. The main force would be cost and speed. Recently, we tested a SOLiD (sequence by oligonucleotide ligation and detection) nextgen system and it appears to be feasible to achieve 1 Gb/d for $1,000. The technical challenge, however, is the read length of the nextgen systems. It may be conceivable to apply a hybrid approach and reduce redundancy of CE sequencing from 8 to 10 times to a much lower level by mixing them with short paired reads from nextgen systems. Still, in addition to read length, accuracy of the nextgen sequencing technology has to go into the equation as well. Although 454 sequencing achieves longer sequencing reads than other nextgen sequencing methods, it also appears to have higher error rates up to 10% depending on sequence composition (Brockman et al., 2008). It also has a lower throughput than the Solexa system, but Solexa has very short reads of about 35 nucleotides and an error rate of 2% to 3%, which is compensated by the higher redundancy of sequences (Dohm et al., 2008). The new SOLiD system differs from the 454 and Solexa systems because it uses DNA ligase for querying the order of nucleotides and claims to have an error rate of only 0.1% (http://appliedbiosystems.cnpg.com/lsca/webinar/rhodes/chemistry/20070618), but it also has very short reads like Solexa, although ABI claims that read length of more than 100 could be achieved with improved dyes and emulsion PCR. Nevertheless, a hybrid approach of CE and nextgen sequencing, particularly if combined with paired reads, should provide accelerated access to genomic sequences and be a catalyst for comparative genomics.
If hybrid sequencing approaches are applied in the future for additional genomes of the Poaceae, the use of reference genomes of high quality will have an even greater impact. As demonstrated by various controls, the physical map of sorghum, the synteny with rice, and its new sequencing strategy contributed to a high-quality sorghum genome sequence in record time and great efficiency. Therefore, one can consider both the rice and sorghum genome sequence as a synergistic reference. Indeed, alignments of genes from rice and sorghum with maize has led to the identification of genes conserved through ancestry, also called orthologous gene copies, whereas genes in either of the three genomes that deviate from a common order are inserted in chromosomal regions by illegitimate recombination similar to transposable elements after speciation and are referred to as paralogous gene copies (Messing and Bennetzen, 2008). In this article, we can see how orthologous and paralogous sequences have been identified in rice, sorghum, and maize.
RETROTRANSPOSONS
Sequence alignments have played a critical role in defining related DNA sequences (Maizel and Lenk, 1981). In most complex genomes, like those of plants, amplification of small genomic sequences and dispersal of sequence copies by illegitimate recombination have been the most common mode to generate a large proportion of repeat sequence families. Because of the transition through RNA intermediates (class I transposable elements), the increase in copy number of specific sequences is very high compared to DNA transposition (class II transposable elements), which occurs either through single copies or without replication by straight transposition. Therefore, it is not surprising that the major force in genome size variation in plants is the differential burst of retrotransposition (Messing and Bennetzen, 2008). Because at the time of insertion the retrotranscript forms long terminal repeats (LTRs), the insert is flanked by identical sequences that can undergo recombination before meiosis so that either a single retroelement is lost in the next generation or even unrelated sequences between two neighboring elements. This activity can then reverse chromosome expansion and the remaining LTR is called a solo LTR. However, detection of retroelements in the genome based on paired flanking LTRs is hampered by layered insertions and mechanisms that disrupt the intact structure of retroelements like chromosome breaks. Such fractured elements may simulate solo LTRs, but do not reduce chromosome expansion as rapidly as unequal crossovers of LTRs do.
Because LTRs are identical at the time of insertion, nucleotide substitution rates of LTR pairs have served as a measure to assign relative insertion dates to elements (SanMiguel et al., 1998). Based on these measures, large bursts of retrotranspositions appeared to have occurred long after speciation, mostly in the last million years (Du et al., 2006; Paterson et al., 2009). Not surprisingly, the size of the rice genome compared to sorghum and maize is largely due to the lower percentage of retroelements, 39.5%, 62%, and 82.1%, respectively. Based on some estimates, differential contraction might have influenced these differences as well. Interestingly, the increased activity of retrotransposition in sorghum and maize shaped their chromosomes quite differently (Paterson et al., 2009). In sorghum, retrotransposition has uniformly increased heterochromatic regions, whereas in maize retrotransposition occurred unevenly (Bruggmann et al., 2006). Furthermore, maize has a higher ratio of copia-like elements, surprisingly extensively hypomethylated, in contrast to gypsy-like elements (Messing et al., 2004). Alignments of orthologous regions then indicate that copia-like elements have penetrated gene-rich regions in maize, whereas those are relatively low in gene-rich regions of sorghum. A small sample of orthologous genomic sequences from gene-dense regions showed 45.1% in maize, 21.1% in rice, and 3.7% in sorghum of sequence occupied by retroelements (Du et al., 2006). Interestingly, the pericentromeric regions in sorghum are dominated by a single retroelement, retrosor6, which amounts to 6.9% of the sorghum genome (Messing and Llaca, 1998; Peterson et al., 2002). It appears that, in sorghum, retroelements have expanded the pericentromeric regions and concentrated recombinogenic regions to the ends of chromosomes (Paterson et al., 2009), whereas in maize the uneven distribution of retroelements acted as a countermeasure to the WGD by the two progenitors of maize (Messing, 2009). Because meiosis selects against the presence of homoeologous chromosomes, uneven retrotransposition in maize might have reduced the pairing of homoeologous chromosomes and contributed to the stability and diploidization of the maize genome. With respect to rice, the proportion of solo LTRs is very high in the centromeric region relative to retroelements (Ma and Bennetzen, 2004). It could well be that rice has contracted in pericentromeric regions and has a more even, albeit low, distribution of retroelements throughout its genome. An overview of these hypothetical chromosome dynamics in the different lineages of maize, sorghum, and rice is illustrated in Figure 1. More recently, it has been shown in orthologous regions of these three species that centromeres in rice and sorghum disrupt collinearity relative to maize chromosomes and insertion of centromeric sequences might have occurred in ancient chromosomal fragile sites. Furthermore, disruption of collinearity by formation of new centromeres indicates a larger insertion in sorghum of 12 Mb and in rice of only 4.5 Mb, consistent with the proposal of contraction in rice centromeric regions (Xu and Messing, 2008b).
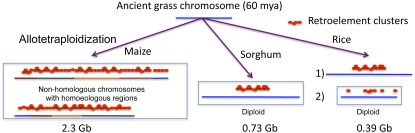
Figure 1.
Expansion and contraction of chromosomes in progenitors of rice, sorghum, and maize. The assumption is made that expansion and contraction of chromosomes occurred in recent times long after speciation, perhaps mostly in the last 1 million years (Du et al., 2006). In rice, (1) expansion in pericentromeric regions was followed by (2) contraction and low retrotransposition throughout the chromosome (Ma and Bennetzen, 2004). Sorghum experienced mainly expansion in pericentromeric regions (Paterson et al., 2009). Maize underwent WGD of two diverged progenitors, then early on chromosome breakage and fusion, resulting in a mosaic of syntenous chromosome blocks (shown in different colors), which was followed by uneven expansion and contraction of those blocks (Bruggmann et al., 2006). Therefore, distribution of retroelement clusters is more even than in sorghum. The size of maize is 2.3 Gb or billion bases (Wei et al., 2007), of sorghum 0.73 (Paterson et al., 2009), and of rice 0.39 (International Rice Genome Sequencing Project, 2005).
DNA TRANSPOSABLE ELEMENTS
Other forms of replicative transpositions have been proposed via a rolling circle mechanism, where the intermediate is single-stranded DNA (Kapitonov and Jurka, 2001). Because the trans-acting function involves a helicase, these elements have been called helitrons. Although helitrons are quite common, the detection of helitrons with internally replaced sequences has been difficult because, in contrast to conventional transposable elements, the terminal sequences vary among different species (Lai et al., 2005; Morgante et al., 2005; Du et al., 2008). Before genome-wide analysis of these elements can be undertaken, it becomes necessary to align allelic sequences from the same species to build individual catalogs of consensus sequences for each species. Still, the degree to which they influence chromosomal expansion is very small because in sorghum they amount to about 1% and in maize 1.5% of total genomic DNA (Paterson et al., 2009). On the other hand, they presumably contribute to noncollinearity of genes and gene fragments because they can copy orthologous gene copies (Xu and Messing, 2006). Interestingly, a similar function can be contributed to Mu-like transposable elements in rice (Jiang et al., 2004) and CACTA elements in sorghum (Paterson et al., 2009). Therefore, transposable elements might function to some degree and in some instances as vectors for paralogous gene copies.
From a genome point of view, class II elements constitute only a small percentage of genomic sequences. One reason could be that they are more mutagenic because they, in contrast to retroelements, preferentially insert into genes and have even been called a search engine for genes (Cowperthwaite et al., 2002). Another reason is that they usually are not replicative and undergo cut-and-paste mechanisms. A third reason is that they are relatively small in size and can be quite parasitic with respect to genes. In particular, many genes carry miniature inverted-repeat transposable elements (MITES) without imposing on gene function (Bureau and Wessler, 1992). MITES are almost as abundant as retroelements in rice, 33% versus 43%, but in sorghum represent only 19% versus 74% and in maize 5% versus 90% of the total repetitive DNA content, respectively (Haberer et al., 2005). This distribution is consistent with the decreasing gene density in these genomes. Furthermore, in sorghum, where retroelements are concentrated in pericentromeric and genes in telomeric regions, MITES are nearly collinear with the telometric regions (Paterson et al., 2009).
GENE DUPLICATIONS AND LOSSES
A thorough catalog of repeat elements customized for each genome simplifies the detection of genes because searches can be performed with masked repeat sequences. Because repeat sequences are also transcribed, it prevents false gene calls from aligning the transcriptome with genomic sequences (Messing et al., 2004). An important confirmation of gene calls can come from collinear arrangements of genes. If repeat sequences were amplified very recently after speciation, the likelihood that the same sequence inserted into the same order in one species compared to another is extremely low. Therefore, one can make the assumption that predicted genes conserved in the same order in ancestral chromosomal regions (orthologous) do not belong to either class of transposable elements and are probably functionally selected. Still, within the same genome, orthologous genes can be copied and might have a function as a paralogous gene copy. Four mechanisms may lead to gene-copying events. Tandem gene copies most likely arose by unequal crossing over. Another mechanism involves segmental duplication or WGD. A third is duplication of a short genomic sequence containing a single gene inserted somewhere else. The fourth is transduction of a gene by a transposable element.
Relative to rice and sorghum, maize chromosomal regions match them 1:2, indicating that maize underwent a WGD event (Swigonova et al., 2004). Interestingly, after all genes were duplicated, more than one-half of the time the second copy was lost (Messing et al., 2004). It has been suggested that unequal crossing over between LTRs led to losses of the second copy because the presence of the other copy rendered it redundant, and increased dissimilarity of homoeologous chromosomal regions provided greater stability to meiosis (Messing, 2009). In addition to unequal crossing over of LTRs, class II elements could have played an even greater role in chromosome contraction because of the ability of transposase to act on the ends of two elements separated by long stretches of nonrelated sequences (Zhang and Peterson, 2004; Huang and Dooner, 2008). The expansion/contraction was favored by meiosis to prevent pairing of nonhomologous chromosomes. Still, the remaining copies facilitate alignment of chromosomal regions of maize with sorghum and rice. Because sorghum is largely collinear with rice despite 50 million years of divergence, nearly the age of the grass family, we can assume that the rice genome resembles the ancestral grass genome except perhaps for the positions of centromeres and chromosome numbers. Genomes of grass species that deviate from chromosomal collinearity with rice and sorghum were presumably formed by breaks and fusions of chromosomal fragments. Therefore, alignment of rice chromosomes with the high-density gene map of maize can be used to reconstruct the chromosome sets of the two progenitors of maize, which each appear to have had 10 chromosomes (Wei et al., 2007). Indeed, the progenitor of sorghum also had 10 chromosomes and split from the two progenitors of maize about the same time, 11.9 million years ago (Swigonova et al., 2004). While the sorghum lineage did not undergo WGD, the two other progenitors hybridized to form maize. Therefore, maize had first 20 chromosomes, but selection against polyploidy during meiosis triggered massive rearrangements, a loss of 10 centromeres, and the formation of 10 larger chromosomes instead of 20 smaller ones. This was accomplished with 62 breaks and fusions. Furthermore, insertions of retroelements further contributed to the divergence of homoeologous regions and a second doubling of genome size of the maize genome after allotetraploidization (Fig. 1).
INTRAGENOMIC COLLINEARITY
When a genome is aligned to itself, a dot plot can visualize duplications of chromosomal segments (Maizel and Lenk, 1981). Rice has 18 such major duplications (Yu et al., 2005) that range in size from 1 to 20 Mb, with a mean of 6.4 Mb, and cover about two-thirds of the genome. Therefore, there are many duplicated genes represented by these segmental duplications. Alternatively, one could argue that the progenitor of the grasses has also arisen by WGD. However, based on nucleotide substitution rates of duplicated gene copies, it is difficult to determine the time that this occurred because 17 of the 18 segmental duplications are spread over a 15-million-year time period before the speciation of rice (Yu et al., 2005), except for the one duplication of telomeric regions of rice chromosome 11 and 12, which could have occurred as recently as 7.7 million years ago (Rice Chromosomes 11 and 12 Sequencing Consortium, 2005). Clarification could come from sequencing a genome that relates to rice as Saccharomyces cerevisiae to Kluyveromyces waltii. Sequencing of the latter organism showed a 2:1 genetic relationship that had deteriorated over time by extensively eliminating the second copy of duplicated genes in the S. cerevisiae genome (Kellis et al., 2004).
Examination of an ancient duplication, dating back to 56 million years ago, before the progenitors of rice, sorghum, and maize split 50 million years ago (Kellogg, 2001) and present on rice chromosomes 7 and 3, showed that it contains the ortholog of the maize endosperm-specific transcription factor OPAQUE2 (O2). Despite this chromosomal duplication, orthologous gene copies in sorghum can unambiguously be assigned (Xu and Messing, 2008a). Rice 7 is orthologous to sorghum 2, whereas rice 3 is orthologous to sorghum 1, consistent with the conclusion that gene copies arose already in a progenitor of these species. Therefore, despite extensive segmental duplications, the rice and sorghum genomes appear largely collinear (Paterson et al., 2009). Furthermore, because maize arose from hybridization of two progenitors, rice 7 is orthologous to maize 2 and maize 7, and rice 3 is orthologous to maize 5 and maize 1 (see figure 1 in Xu and Messing, 2008a). The O2 gene on maize chromosome 7 therefore corresponds to a gene copy on sorghum 2 and rice 7. While the ortholog OHP of the O2 gene on maize chromosome 1 is duplicated again on maize 5, O2 itself was duplicated by allotetraploidization on maize 2, but the duplicate was then deleted. On the other hand, OHP on maize 1 got tandemly duplicated after allotetraploidization and O2 on sorghum chromosome 2 also has two tandem copies.
Various alleles of the maize O2 gene give opaque kernel phenotypes, indicating that the ancient segmental duplication has diverged sufficiently so that the descending gene copies give rise to normal Mendelian factors. This is consistent with the assumption that, in most cases, gene duplications result in subfunctionalization (Rodriguez-Trelles et al., 2003). Indeed, the alignment of these orthologous chromosomal segments indicates that insertion and deletion of genes occurred before and after the progenitor of rice, sorghum, and maize split so that the orthologous gene sets differ between any pairwise combination of two species and each interval has now unique sets of noncollinear genes (Xu and Messing, 2008a). Phylogenetic analysis also indicates that nonhomologous chromosome pairing must have occurred via these intervals within the same lineage, leading to gene conversions. Such a mechanism might also explain the concerted evolution of gene pairs. On the other hand, one can envision counterbalances to such a mechanism because of selection of homologous chromosome pairs during meiosis. In this respect, rapid deterioration of homology between the segmental duplications might be favored. Clearly, insertions and deletions would be consistent with such structural divergence. The duplication on rice 7 is larger than on rice 3. Rice 7 has more genes than rice 3 in the same interval. Interestingly, the bias is even more pronounced in maize than in rice and sorghum. Whereas maize 7 and 1, orthologs to rice 7 and 3, are expanded severalfold, the maize homoeologs 2 and 5 are hardly expanded compared to rice and sorghum. This difference in size of maize homoeologs also exhibits a bias that one homoeolog has preferentially lost a gene duplicate. As discussed already above, the loss could be explained by contraction through unequal crossing over of neighboring LTRs or action of a transposase on a pair of DNA transposable elements. In addition, the alignments also visualize inversions and translocations, probably exemplifying the entire array of sequence dynamics of plant chromosomes.
SINGLE GENE DUPLICATIONS
The duplication mechanisms discussed above, however, do not explain the abundance and sizes of gene families. The most common mechanism for the amplification of gene copies is unequal crossing over during meiosis, resulting in tandemly duplicated genes. Still, layered single gene insertions or transpositions into intergenic space can disrupt perfect tandem arrays or obscure the size of such clusters. Probably, the most commonly known gene clusters comprise ribosomal RNA genes (Messing et al., 1984). However, when the first plant genomes had been sequenced, it became apparent that about one-fourth of the genes in Arabidopsis (Arabidopsis thaliana) and one-third of the genes in rice have been tandemly duplicated (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005). The number appears to be lower with only one-fifth of the genes in sorghum as tandem arrays (Paterson et al., 2009), but, in each genome study, the definition of genes and window of alignments varies so that direct comparison is difficult. In any case, whole-genome analysis confirms a large degree of tandem gene amplification in plant genomes. Tandem arrays of genes are not always stable and contract again, giving rise to variation in the size of gene clusters, ranging in size from two to 134 members in the rice genome (International Rice Genome Sequencing Project, 2005). The largest gene cluster in sorghum appears to encode proteins with a P-450 motif, probably reflecting the stress tolerance of sorghum (Paterson et al., 2009). The most rapidly evolving gene clusters are disease resistance genes. For instance, the rp1 locus in maize undergoes unequal crossing within single generations (Hulbert and Bennetzen, 1991).
However, this type of amplification does not explain the insertion of noncollinear genes. Whereas postspeciation gene-copying events seemed to employ transposable elements and helitrons, as discussed above, gene-copying events that occurred in the progenitor of species do not exhibit the DNA footprints associated with these types of events. Alignment of orthologous regions of rice, sorghum, and maize containing the prolamin storage protein genes show that the α-prolamin genes arose after the divergence of the Panicoideae subfamily 21 to 26 million years ago (Xu and Messing, 2008b). The founder gene was copied and inserted into a different chromosomal location 20 to 24 million years ago, which then was tandemly amplified. Therefore, the progenitors of maize and sorghum have both loci, but rice does not. It is interesting that one of three progenitors for maize and sorghum produced another unlinked copying event about 9 million years ago, making the three progenitors different in prolamin gene loci. As a consequence, this gene copy is absent in sorghum and one of the progenitors of maize. After allotetraploidization, this gene copy is absent in the homoeologous chromosomal region, not because of gene loss, but because it was never duplicated. Another unlinked copying event occurred 2.4 million years ago in maize, which is also absent in the orthologous region in sorghum and the homoeologous region in maize, demonstrating that a similar copying mechanism without the apparent use of transposable elements persisted also after speciation. In addition, with one exception, all gene-copying events in sorghum and maize underwent tandem duplications. A similar pattern of gene insertion and tandem duplication without the apparent use of transposable elements has also been observed in rice (Lai et al., 2004). This analysis shows that unlinked gene copies arose at different stages of divergence of lineages within the same family of species and for maize contributed to allotetraploidization instead of straight polyploidization (Xu and Messing, 2008b).
GENE EXPRESSION OF GENE COPIES
While little is known about what triggers gene amplification, it is clear that gene copies are not expressed developmentally and quantitatively equally. The simplest examples are the Mendelianization of gene copies derived from WGD events. For instance, a gene regulating anthocyanin accumulation in the tissues of maize got duplicated during allotetraploidization. Allelic variants of each gene copy gave rise to different phenotypes. Mapping of these phenotypes resulted in the discovery of the R and B genes in maize that encode a transcription factor with the same function, but different regulation (McClintock and Hill, 1931; Coe, 1959). Because pigment accumulation is not essential for plant survival, other examples of diverged regulation of the same function have been discovered in allotetraploid maize, confirming that subfunctionalization is a common cause for gene duplication (Rodriguez-Trelles et al., 2003).
In the case of the storage protein genes described above, it is interesting to note that, of 41 copies of α-prolamin genes in inbred B73, only 16 appear to be expressed. Interestingly, in sorghum, of 23 genes, 19 are expressed, exhibiting a lower level of gene silencing than in maize (Xu and Messing, 2008b). One mechanism of gene silencing appears to be the introduction of premature stop codons (Llaca and Messing, 1998). It appears that those genes can still be transcribed, but their mRNAs accumulate at a very reduced level (Liu and Rubenstein, 1993; Song and Messing, 2003). It has been suggested that premature termination of translation triggers accelerated turnover of mRNA (van Hoof and Green, 1996). Furthermore, truncated proteins are probably less stable as well and lose their function. Another mechanism is gene truncation, resulting in the same effect as premature stop codons. However, in the former case, gene conversion of intact genes could counteract gene silencing (Llaca and Messing, 1998). Yet another one is epigenetic silencing. This mechanism is probably similar to the silencing of transposable elements and reversible (Chomet et al., 1987; Peschke et al., 1987). Epigenetic changes of the state of a gene that are heritable usually arise from changes in chromatin structure, which can coincide with the modification of DNA by methylation. For instance, it has been shown that the epiallele P-pr in contrast to the normal allele P-rr has DNAse-hypersensitive sites in the enhancer region that stay closed in the tissue, where gene expression should occur (Lund et al., 1995). Furthermore, the same site is methylated in the P-pr allele, but not in the P-rr allele.
One of the challenges in examining the expression of paralogous gene copies is highly conserved coding sequences. Hybridizations and sequence alignments frequently are ambiguous and can only be resolved by matching individual mRNAs and genomic gene copies. By knowing each member of a gene family by genomic position, one can establish a polymorphism grid prior to the analysis of cDNA sequences (Xu and Messing, 2008b). Interestingly, allelic variation of gene copies is lower than paralogous copies even if they are 99% conserved. Therefore, polymorphism grids can even be used to study gene expression in different cultivars of the same species. Lack of expression of allelic gene copies also led to the discovery of variation in gene copy number within the same species, which can be based on additional tandem sequence amplification or deletion of tandem sequence repeats (Song and Messing, 2003). Furthermore, there appear to be differences in the expression of allelic and nonallelic gene copies between different maize inbred lines. In the case of the most recently amplified gene copies, one could even find a divergence in transcriptional regulation, which could explain the frequently observed genetic background effect on certain phenotypes (Song et al., 2001). Background effects have even been observed in plant development, when heterochronic mutations are introgressed into different inbred lines (Poethig, 1988). The relative stability of noncollinear gene clusters in allelic regions of the same species is manifested in form of a few haplotypes that might have an important bearing on the potential vigor of hybrids. Indeed, when maize inbreds with different haplotypes of 22-kD α-prolamin gene clusters were crossed in both directions, gene expression of allelic and nonallelic gene copies did not always exhibit an additive effect (Song and Messing, 2003). On the contrary, sometimes there was an overdominant effect on the increase and decrease of gene expression, indicating that hybrids could produce their unique expression pattern.
Given the unequal expression of members of a gene family, one might wonder about the role of less active or silent members. The same question is apparent in the genome-wide annotation of predicted genes. In many cases, overannotation includes truncated paralogs, which belong to the group of pseudogenes and are thought to be nonfunctional. Interestingly, recent studies in mice and fruit flies have shown that pseudogenes are not only transcribed, but also processed in small interfering RNA sequences, which could play a regulatory role (Sasidharan and Gerstein, 2008). Such a mechanism might even play a role in the differential expression of the maize prolamin haplotypes in hybrid crosses as described above.
CONCLUSION
Because of the conservation of gene sequences within the same plant family, sequence analysis of entire genomes of multiple confamilial species have facilitated alignments of large chromosomal regions, here exemplified by the grass family with the rice and sorghum genome as synergistic references. The complementarity of alignments has been useful in ordering clone maps along the genetic map and raises the confidence in gene prediction analysis. In addition, the collinearity of genomes, diverged since the beginnings of the grass family, provides us with a consensus organization of ancestral grass chromosomes. Alignment of other genomes with such a consensus organization has facilitated the reconstruction of progenitor chromosomes from reshuffled genomes and analysis of gene duplications. If what we have learned from one plant family of species is representative, plant chromosomes exhibit an unexpected degree of sequence flux. Previously, it was recognized that sequences outside of genes are quite mobile because of transposable elements. We have learned now through alignments of orthologous chromosomal regions of species within the same family that gene-copying events are more copious as ever suspected. Moreover, the mechanisms underlying such copying events appear to be multiple and are probably triggered and controlled independently.
This work was supported, in part, by a grant from the Department of Energy (grant no. DE–FG05–95ER20194) and the National Science Foundation (grant no. DBI 03–20683).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Joachim Messing (messing@waksman.rutgers.edu).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Bedell JA, Budiman MA, Nunberg A, Citek RW, Robbins D, Jones J, Flick E, Rholfing T, Fries J, Bradford K, et al (2005) Sorghum genome sequencing by methylation filtration. PLoS Biol 3 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, Russ C, Lander ES, Nusbaum C, Jaffe DB (2008) Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res 18 763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, Pontaroli AC, Wei F, Haberer G, Fuks G, Du C, et al (2006) Uneven chromosome contraction and expansion in the maize genome. Genome Res 16 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR (1992) Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet PS, Wessler S, Dellaporta SL (1987) Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J 6 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Linton E, Messing J, Doebley JF (2004) Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc Natl Acad Sci USA 101 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe EH (1959) A regular and continuing conversion-type phenomenon at the B locus in maize. Proc Natl Acad Sci USA 45 828–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M, Park W, Xu Z, Yan X, Maurais SC, Dooner HK (2002) Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Ma J, Pontaroli AC, Pratt LH, Bennetzen JL (2005) Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc Natl Acad Sci USA 102 19243–19248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H (2008) Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36 e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Caronna J, He L, Dooner HK (2008) Computational prediction and molecular confirmation of Helitron transposons in the maize genome. BMC Genomics 9 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Swigonova Z, Messing J (2006) Retrotranspositions in orthologous regions of closely related grass species. BMC Evol Biol 6 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RC, Howarth AJ, Hahn P, Brown-Luedi M, Shepherd RJ, Messing J (1981) The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res 9 2871–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Sekhon M, Schein J, Zhao S, Osoegawa K, Scott CE, Evans RS, Burridge PW, Cox TV, Fox CA, et al (2002) A physical map of the mouse genome. Nature 418 743–750 [DOI] [PubMed] [Google Scholar]
- Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G, Butler E, Wing RA, Rounsley S, Birren B, et al (2005) Structure and architecture of the maize genome. Plant Physiol 139 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JT, Dooner HK (2008) Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell 20 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Bennetzen JL (1991) Recombination at the Rp1 locus of maize. Mol Gen Genet 226 377–382 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431 931–945 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP, Zody MC, Lander ES (2003) Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res 13 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573 [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J (2001) Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA 98 8714–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 617–624 [DOI] [PubMed] [Google Scholar]
- Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Li Y, Messing J, Dooner HK (2005) Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA 102 9068–9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Ma J, Swigonova Z, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park YJ, Jeong OY, Bennetzen JL, et al (2004) Gene loss and movement in the maize genome. Genome Res 14 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R, Messing J (1982) Apple II software for M13 shotgun DNA sequencing. Nucleic Acids Res 10 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CN, Rubenstein I (1993) Transcriptional characterization of an alpha-zein gene cluster in maize. Plant Mol Biol 22 323–336 [DOI] [PubMed] [Google Scholar]
- Llaca V, Messing J (1998) Amplicons of maize zein genes are conserved within genic but expanded and constricted in intergenic regions. Plant J 15 211–220 [DOI] [PubMed] [Google Scholar]
- Lund G, Prem Das O, Messing J (1995) Tissue-specific DNase I-sensitive sites of the maize P gene and their changes upon epimutation. Plant J 7 797–807 [PubMed] [Google Scholar]
- Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel JV Jr, Lenk RP (1981) Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci USA 78 7665–7669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B, Hill HE (1931) The cytological identification of the chromosome associated with the R-G linkage group in ZEA MAYS. Genetics 16 175–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J (2009) The polyploid origin of maize. In JL Bennetzen, SC Hake, eds, The Maize Handbook: Domestication, Genetics, and Genome. Springer Science + Business Media, New York, pp 221–238
- Messing J, Bennetzen J (2008) Grass genome structure and evolution. Genome Dyn 4 41–56 [DOI] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KF, et al (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Carlson J, Hagen G, Rubenstein I, Oleson A (1984) Cloning and sequencing of the ribosomal RNA genes in maize: the 17S region. DNA 3 31–40 [DOI] [PubMed] [Google Scholar]
- Messing J, Crea R, Seeburg PH (1981) A system for shotgun DNA sequencing. Nucleic Acids Res 9 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Llaca V (1998) Importance of anchor genomes for any plant genome project. Proc Natl Acad Sci USA 95 2017–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37 997–1002 [DOI] [PubMed] [Google Scholar]
- Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, Kim H, Wing RA, Messing J, Soderlund C (2005) Whole-genome validation of high-information-content fingerprinting. Plant Physiol 139 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature (in press) [DOI] [PubMed]
- Peschke VM, Phillips RL, Gengenbach BG (1987) Discovery of transposable element activity among progeny of tissue culture-derived maize plants. Science 238 804–807 [DOI] [PubMed] [Google Scholar]
- Peterson DG, Schulze SR, Sciara EB, Lee SA, Bowers JE, Nagel A, Jiang N, Tibbitts DC, Wessler SR, Paterson AH (2002) Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res 12 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS (1988) Heterochronic mutations affecting shoot development in maize. Genetics 119 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosomes 11 and 12 Sequencing Consortium (2005) The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol 3 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ (2003) Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc Natl Acad Sci USA 100 13413–13417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20 43–45 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Gerstein M (2008) Genomics: protein fossils live on as RNA. Nature 453 729–731 [DOI] [PubMed] [Google Scholar]
- Song R, Llaca V, Linton E, Messing J (2001) Sequence, regulation, and evolution of the maize 22-kD alpha zein gene family. Genome Res 11 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Messing J (2003) Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA 100 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Green PJ (1996) Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J 10 415–424 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al (2001) The sequence of the human genome. Science 291 1304–1351 [DOI] [PubMed] [Google Scholar]
- Vieira J, Messing J (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19 259–268 [DOI] [PubMed] [Google Scholar]
- Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, et al (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302 2118–2120 [DOI] [PubMed] [Google Scholar]
- Wu R, Kaiser AD (1968) Structure and base sequence in the cohesive ends of bacteriophage lambda DNA. J Mol Biol 35 523–537 [DOI] [PubMed] [Google Scholar]
- Xu JH, Messing J (2006) Maize haplotype with a helitron-amplified cytidine deaminase gene copy. BMC Genet 7 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Messing J (2008. a) Diverged copies of the seed regulatory Opaque-2 gene by a segmental duplication in the progenitor genome of rice, sorghum, and maize. Mol Plant 1 760–769 [DOI] [PubMed] [Google Scholar]
- Xu JH, Messing J (2008. b) Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proc Natl Acad Sci USA 105 14330–14335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peterson T (2004) Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167 1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]