Abstract
Plant disease resistance governed by quantitative trait loci (QTL) is predicted to be effective against a broad spectrum of pathogens and long lasting. Use of these QTL to improve crop species, however, is hindered because the genes contributing to the trait are not known. Five disease resistance QTL that colocalized with defense response genes were accumulated by marker-aided selection to develop blast-resistant varieties. One advanced backcross line carrying the major-effect QTL on chromosome (chr) 8, which included a cluster of 12 germin-like protein (OsGLP) gene members, exhibited resistance to rice (Oryza sativa) blast disease over 14 cropping seasons. To determine if OsGLP members contribute to resistance and if the resistance was broad spectrum, a highly conserved portion of the OsGLP coding region was used as an RNA interference trigger to silence a few to all expressed chr 8 OsGLP family members. Challenge with two different fungal pathogens (causal agents of rice blast and sheath blight diseases) revealed that as more chr 8 OsGLP genes were suppressed, disease susceptibility of the plants increased. Of the 12 chr 8 OsGLPs, one clustered subfamily (OsGER4) contributed most to resistance. The similarities of sequence, gene organization, and roles in disease resistance of GLP family members in rice and other cereals, including barley (Hordeum vulgare) and wheat (Triticum aestivum), suggest that resistance contributed by the chr 8 OsGLP is a broad-spectrum, basal mechanism conserved among the Gramineae. Natural selection may have preserved a whole gene family to provide a stepwise, flexible defense response to pathogen invasion.
Protection of agronomic crops from losses due to disease has largely relied on the use of genetic resistances in plant breeding programs. In major food crops such as rice (Oryza sativa), single gene-based (R gene-mediated) resistance is effective for some diseases. However, highly variable pathogens, such as Magnaporthe oryzae, can adapt rapidly to overcome R gene-mediated resistances (Bonman et al., 1992). A viable solution to the vulnerability of single gene resistance is to build a basal level of quantitative resistance, which, because of its multigenic nature, is predicted to delay the evolution of pathogens to virulence (Johnson, 1984). Quantitative resistance is particularly essential for important diseases like sheath blight, caused by Rhizoctonia solani, in which no single gene resistances are available (Lee and Rush, 1983; Rush and Lindberg, 1984; Li et al., 1995; Pinson et al., 2005; Liu et al., 2008). Unlike most R gene resistances, quantitative resistance may also be broad spectrum and effective against multiple pathogens, although direct evidence of this is limited. Incorporating quantitative trait loci (QTL) into germplasm, however, is hindered by the lack of knowledge of what genes are contributing to the QTL. As no disease resistance QTL have been cloned from rice to date, plant breeders cannot develop the precise molecular markers needed to track and select for the functional genes in crop improvement programs.
To understand the molecular basis for QTL-governed disease resistance in plants and determine its utility to control diseases in cropping systems, we and others have accumulated substantial correlative evidence that defense response (DR) genes contribute to quantitative resistance. DR genes are predicted to function in plant disease resistance, and their mRNAs and/or enzymatic activities often are induced after pathogen challenge (Dixon and Harrison, 1990). Using various mapping populations derived from rice cultivars with demonstrated variation in multigenic resistance, chromosomal regions conferring quantitative resistance to several important rice diseases, such as bacterial blight, sheath blight, and rice blast, were identified, and these disease resistance QTL were shown to colocalize with candidate DR genes (Ramalingam et al., 2003; Liu et al., 2004; Wu et al., 2004). We used five DR genes as markers to demonstrate that the more QTL accumulated into lines, the more rice blast resistance we observed in multilocation trials (Liu et al., 2004). However, causal effects of the DR genes were difficult to demonstrate due to the relatively small effects of individual genes and the presence of multiple gene family members that may play different roles in defense. Consequently, plant breeders still lacked sufficient confidence to apply the DR genes as selection markers in crop improvement programs.
To establish a causal effect between DR gene function and QTL, we have focused on a major-effect rice blast resistance QTL on rice chromosome (chr) 8 (log of the odds = 7.1–10; contributing over 30% of the phenotypic effect) that colocalized with a barley (Hordeum vulgare) oxalate oxidase-like gene marker (HvOXOLP) in several rice mapping populations (Ramalingam et al., 2003; Liu et al., 2004). Minor QTL for sheath blight resistance have also been identified in this chromosomal region (Pinson et al., 2005). Oxalate oxidase-like genes, now referred to as germin-like protein (GLP) genes, belong to the functionally diverse cupin superfamily and have been identified in Arabidopsis (Arabidopsis thaliana), grapevine (Vitis vinifera), and many Gramineae species (Membre et al., 2000; Lane, 2002; Godfrey et al., 2007; Dunwell et al., 2008). Several lines of evidence suggest that GLPs are involved in general plant defense responses (Lane, 2002), including the observation that expression of certain GLPs is enhanced after infection with pathogens, feeding of insects, or application of chemicals such as salicylic acid, hydrogen peroxide (H2O2), or ethylene (Dumas et al., 1995; Zhang et al., 1995; Wei et al., 1998; Zhou et al., 1998; Federico et al., 2006; Lou and Baldwin, 2006; Zimmermann et al., 2006; Godfrey et al., 2007). Transient overexpression of certain barley GLP subfamilies resulted in enhanced resistance to the powdery mildew fungus, and for some subfamilies, silencing resulted in enhanced susceptibility to the pathogen (Zimmermann et al., 2006). Silencing of a Nicotiana GLP increased the performance of an herbivore (Lou and Baldwin, 2006).
The mechanism by which GLPs influence plant defense is likely related to their generation of active oxygen species. They are targeted to the cell wall and apoplast, and while their functions are largely unknown, some members related to the barley HvGER4 subfamily exhibit superoxide dismutase activity (Christensen et al., 2004; Zimmermann et al., 2006; Godfrey et al., 2007). Superoxide produced by NADPH oxidase or peroxidases in response to pathogen attack is predicted to be dismutated to H2O2 by the GLP, accounting for the accumulation of H2O2 (Bolwell and Wojtaszek, 1997). H2O2 is an important component of plant defense responses, with possible roles in basal defense responses such as the oxidative cross-linking of cell wall proteins and lignin precursors as well as in papillae formation (Olson and Varner, 1993; Wei et al., 1998). H2O2 also is involved in hypersensitive cell death, signaling in systemic acquired resistance, and the induction of DR gene expression (Chen et al., 1993; Lamb and Dixon, 1997; Alvarez et al., 1998).
Underlying the chr 8 QTL, we predicted 12 putative rice GLPs (OsGLPs) clustered within 2.8 Mb. Expression profiling studies and gene and promoter sequence analyses suggest that a combination of these OsGLP family members contributes to defense responses in rice (R. Davidson, unpublished data). In this study, we use RNA interference (RNAi) silencing of the chr 8 OsGLP gene family members to confirm their contribution to resistance against two different diseases, rice blast and sheath blight. Our data show that as more OsGLP gene family members, particularly those in the OsGER4 subfamily, are suppressed, susceptibility of the transgenic plants to the pathogens causing rice blast (M. oryzae) and sheath blight (R. solani) increases. A rice line carrying the effective chr 8 QTL was grown in the field for over 14 cropping seasons and still exhibits excellent blast resistance. This unique combination of QTL mapping, gene function analysis, and field evaluations provides confidence for selecting the OsGLP gene family as a complex QTL in breeding programs.
RESULTS
The Chr 8 QTL Contributes to Disease Resistance
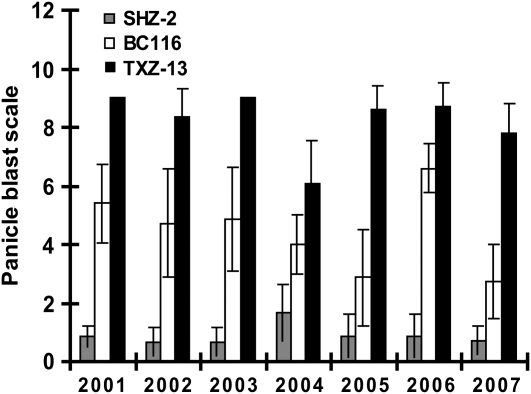
Five QTL from cv Sanhuangzhan 2 (SHZ-2), including the major-effect chr 8 QTL that is associated with OsGLP genes, were introgressed into the susceptible commercial cv Texianzhan 13 (TXZ-13) using marker-assisted selection, resulting in backcross line BC116 (Liu et al., 2004). The presence of the chr 8 QTL in line BC116 was confirmed using three marker-based mapping techniques, (Supplemental Fig. S1). Compared with the recurrent parent TXZ-13, line BC116 exhibited superior resistance to blast over 14 cropping seasons at two locations (Yangjiang [Fig. 1; Supplemental Table S1] and Conghua [data not shown]) in Guandong Province, China. Although there are other contributing QTL regions in this line, these results are consistent with the hypothesis that the chr 8 QTL contributes to effective and stable disease resistance in the field.
Figure 1.
Rice line BC116 containing the chr 8 disease resistance QTL shows consistent panicle blast resistance over 7 years (14 cropping seasons). BC116, TXZ-13 (rice blast-susceptible recurrent parent), and SHZ-2 (rice blast-resistant parent) were planted in Yangjiang, Guangdong, China, two seasons per year over 7 consecutive years. Panicle blast, the most severe form of rice blast, was evaluated in three replicates (60 individuals per replicate) and is presented as average values from two seasons per year. Differences in disease were observed between BC116 and TXZ-13 in all years (t test, P < 0.001) except 2004 (P = 0.0035).
RNAi Silencing of Rice Chr 8 OsGLP Genes
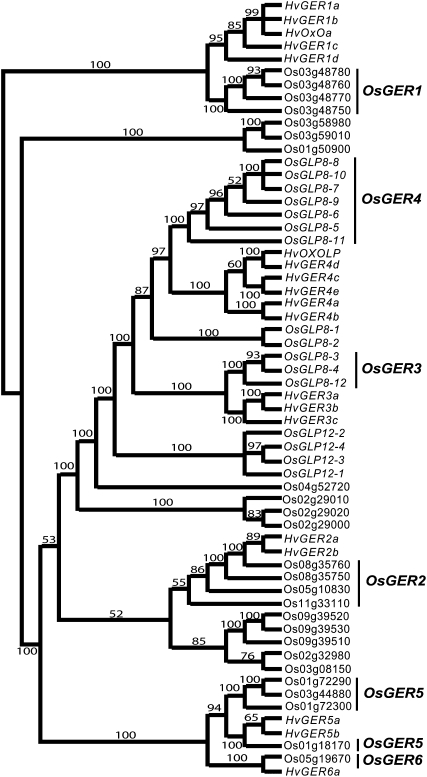
A cluster of 12 highly conserved GLP gene members was predicted within the rice chr 8 disease resistance QTL region (data not shown). GLP is a general term used to indicate proteins that are not true germins or oxalate oxidase, but they contain a germin motif and their enzyme activities may not be known (Carter et al., 1998). We designated the rice genes OsGLP8-1 to OsGLP8-12, for their 5′ to 3′ order on chr 8. Our approach to determine the contribution of individual and collective rice OsGLPs to the resistance contributed by the chr 8 QTL was by gene silencing. However, our first challenge was to identify sequences unique to chr 8 OsGLPs. We predicted 41 gene members of these families on rice chr 1, 2, 3, 4, 5, 8, 9, 11, and 12 using the barley cDNA sequences HvOXOLP and HvOXOA to scan the rice genome (Fig. 2). All predicted proteins contained exact or slight variations of the characteristic germin box sequence, PHIHPRATEI (data not shown; Lane, 2002). Most predicted OsGLPs were classified based on amino acid similarities using the nomenclature previously established for barley germins and GLPs (Zimmermann et al., 2006; Fig. 2). In keeping with the barley nomenclature, the rice subfamilies were named OsGER1 to OsGER6 (Fig. 2; Supplemental Table S2; Druka et al., 2002; Zimmermann et al., 2006). The majority of the 12 chr 8 OsGLP genes were classified in the rice OsGER4 and OsGER3 subfamilies (Fig. 2; Supplemental Table S2). The OsGER4 subfamily contains seven OsGLP members (OsGLP8-5 to -11) and is most closely related to the barley subfamily (HvGER4) that is associated with defense responses (Christensen et al., 2004; Zimmermann et al., 2006).
Figure 2.
Phylogenetic relationships of germin box-containing proteins from rice and barley. Amino acid sequence similarities among predicted GLP proteins from rice were compared with known barley HvGER proteins (Supplemental Table S2). Rice GLP gene members were classified as known subfamilies OsGER1 to OsGER6, based on relationships with the barley HvGER proteins. Inferred amino acid sequences of 60 GLP proteins were aligned using ClustalX version 1.83. The phylogenetic tree was reconstructed using Bayesian MCMC analysis (Ronquist and Huelsenbeck, 2003). Posterior probabilities (scaled to 100) are indicated at nodes.
For selective RNAi-mediated silencing of OsGLP genes on chr 8, we used a 500-bp region of OsGLP8-3 (Supplemental Fig. S2). We predicted that this region had sufficient identity to cosilence all chr 8 OsGLPs but not more distantly related OsGLPs. Silencing experiments were performed in the japonica cv Kitaake, which has no R gene-mediated resistance against M. oryzae isolate Che86061. Ideally, silencing would have been performed in SHZ-2, the chr 8 QTL donor, which is predicted to contain a highly effective combination of OsGLP genes. However, as SHZ-2 is an indica cultivar and is recalcitrant to transformation, we used the more easily transformed japonica cv Kitaake. T0 and T1 OsGLP-suppressed transgenic plants were phenotypically indistinguishable from untransformed Kitaake plants, but some failed to produce seeds. The genome insertion of the transgene was confirmed by PCR using primers to the vector and transgene (data not shown).
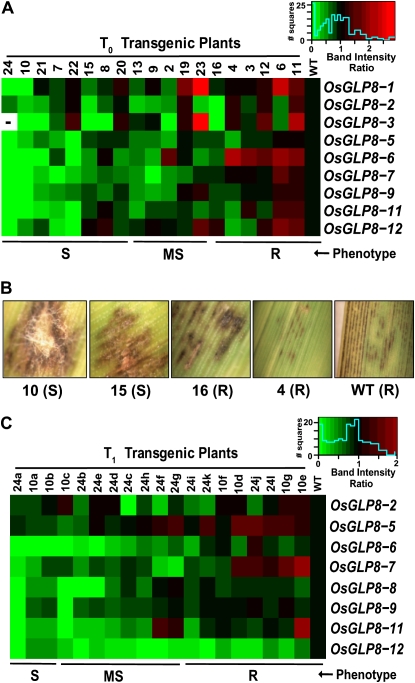
The single RNAi construct suppressed all chr 8 OsGLP genes transcribed by Kitaake, with different efficiencies among the T0 transgenic plants, as demonstrated by semiquantitative reverse transcription (RT)-PCR analysis (Fig. 3A; Supplemental Fig. S3A). Two T0 plants, 10 and 24, in which all expressed chr 8 OsGLP genes were suppressed, were advanced to the T1 generation. Most genes silenced in T0 parental plants were also differentially silenced among the T1 progeny (Fig. 3C; Supplemental Fig. S3B). Fewer genes were suppressed in T1 plants than in T0 parents, suggesting reduced silencing in the T1 generation. A further reduction of silencing was observed in the T2 generation (data not shown). Transcription of all 12 genes was assessed, but expression of OsGLP8-4 and OsGLP8-10 was not observed in Kitaake under our experimental conditions. Transcripts of OsGLP8-8 were not observed in T0, and transcripts of OsGLP8-1 and OsGLP8-3 were not observed in T1 plants, possibly due to developmental differences between generations (data not shown).
Figure 3.
Rice transgenic plants silenced for chr 8 OsGLP gene expression show increased rice blast disease relative to wild-type (WT) Kitaake. Silencing of OsGLP gene expression in independent uninoculated T0 (A) and T1 (C) transgenic plants, as determined by semiquantitative RT-PCR, is indicated as heat maps. Each square in the heat maps indicates band intensity ratio (transgenic-wild type) for a single chr 8 OsGLP gene family member (row) in an independent transgenic plant (column). Color keys for each map show the range of expression (relative to the wild type; green = maximal suppression; red = maximal expression; − = missing data) and histograms with distributions of data points. Rice blast disease phenotypes for individual plants (S, susceptible; MS, moderately susceptible; R, resistant) are indicated below the heat maps. B shows the range of blast disease symptoms on individual T0 and wild-type plants at 7 d after inoculation.
Because of the close relationship of the OsGLP family members, and because silencing of multiple gene family members with one construct had not been widely reported, we confirmed the specificity of silencing to closely related gene members. The silencing construct cosilenced three chr 12 genes (OsGLP12-1, -2, and -3) that Kitaake expressed in the T1 plants (Supplemental Fig. S3C) but did not suppress more distantly related chr 3 oxalate oxidase genes (data not shown).
OsGLP-Suppressed Transgenic Plants Are More Susceptible to Two Different Diseases
Disease phenotype was assessed using a detached leaf spot inoculation assay (Jia et al., 2003). This assay reliably measured quantitative resistance to M. oryzae isolate Che86061 in Kitaake, the host for our silencing studies, compared with the susceptible control Nipponbare (Supplemental S4A). The quantitative resistance of Kitaake was also confirmed by spray inoculation, a widely used inoculation method for rice blast studies (Supplemental Fig. S4B; Valent et al., 1991).
Nineteen independent T0 plants with differential gene silencing and confirmed presence of the transgene were inoculated with M. oryzae, and disease phenotypes, ranging from susceptible to resistant, were observed (Fig. 3, A and B). In the T1 generation, 60 plants were first screened by inoculation with M. oryzae, and 19 plants that exhibited a range of disease phenotypes were preselected based on extreme phenotypes and evaluated for gene silencing and transgene presence (Fig. 3C). All of the plants preselected by phenotype contained the transgene (Supplemental Fig. S3B).
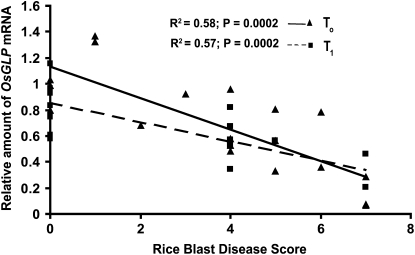
In both the T0 and T1 generations, the transgenic plants with more OsGLP gene members silenced were more susceptible to infection with M. oryzae (Fig. 3; Supplemental Fig. S3). To test the collective effect of OsGLP gene expression, rice blast disease scores for the transgenic silenced plants were correlated with the sums of OsGLP band intensity ratios (transgenic line/wild type) in T0 and T1 generations (Fig. 4). Significant negative relationships (P = 0.0002) indicated that as the total amount of silencing increased, more susceptibility to M. oryzae was observed in T0 and T1 plants.
Figure 4.
Reduced expression of rice chr 8 OsGLP gene members correlates with increased rice blast disease in both T0 and T1 plants. Rice blast disease score was assessed in individual T0 and T1 transgenic plants at 7 d after inoculation using a scale from 0 (no mycelia or colonization) to 7 (extensive mycelial growth and colonization). Total chr 8 OsGLP gene expression for each T0 and T1 independent plant was the sum of the relative amounts of mRNA for each constitutively expressed OsGLP (band intensity ratio of transgenic-wild type) and normalized with the band intensity of the internal control EF1-α for each plant.
To test the relationship of each chr 8 OsGLP gene silencing to blast disease in the T0 and T1 plants, single linear regressions comparing disease scores with band intensity ratios were computed (Table I). Negative relationships (P < 0.05) were repeated for OsGER4 subfamily members (OsGLP8-5, -6, -7, -9, and -11) in both generations. Estimates of relative contributions of individual family members as predictors were tested using multiple regressions (full models) incorporating all OsGLP band intensity ratios (Table I). These tests, however, showed few significant relationships due to high collinearity among independent variables. Collinear variables convey repetitive information (Farrar and Glauber, 1967), suggesting coregulated expression and functional redundancy among these gene family members. The overall P values for both full models were significant (Table I), similar to the total OsGLP regressions (Fig. 4). This supports the hypothesis that the cluster of chr 8 OsGLP genes contributes collectively to disease resistance. Among all statistical tests, relationships between gene silencing and blast disease occurred mostly for OsGER4 subfamily members, in particular OsGLP8-6, which likely contributes more to disease resistance against M. oryzae than other family members, as it showed the lowest P value in both generations.
Table I.
Expression/silencing of OsGER4 subfamily members correlates with rice blast disease (P ≤ 0.05; boldface)
Regressions of rice blast disease score by OsGLP gene band intensity ratio; n = 19 individuals per generation. –, Not expressed.
| OsGLP | T0 Transgenic Plants
|
T1 Transgenic Plants
|
||||||
|---|---|---|---|---|---|---|---|---|
| Slopea | r2a | Pa | Pb | Slopea | r2a | Pa | Pb | |
| 8-1 | −1.30 | 0.14 | 0.133 | 0.382 | – | – | – | – |
| 8-2 | −2.51 | 0.19 | 0.074 | 0.289 | −1.51 | 0.04 | 0.417 | 0.189 |
| 8-3 | −0.92 | 0.12 | 0.160 | 0.257 | – | – | – | – |
| 8-5 | −4.06 | 0.25 | 0.035 | 0.587 | −5.09 | 0.41 | 0.003 | 0.253 |
| 8-6 | −2.67 | 0.72 | <0.0001 | 0.067 | −5.79 | 0.66 | <0.0001 | 0.014 |
| 8-7 | −4.20 | 0.62 | 0.0001 | 0.771 | −3.64 | 0.47 | 0.001 | 0.032 |
| 8-8 | – | – | – | – | −3.95 | 0.29 | 0.018 | 0.570 |
| 8-9 | −4.90 | 0.48 | 0.001 | 0.625 | −5.61 | 0.39 | 0.004 | 0.449 |
| 8-11 | −3.50 | 0.39 | 0.006 | 0.876 | −2.38 | 0.22 | 0.043 | 0.397 |
| 8-12 | −1.57 | 0.11 | 0.182 | 0.425 | −1.40 | 0.01 | 0.697 | 0.099 |
| Overall P value for the full model | 0.025 | 0.003 | ||||||
Single linear regression.
Multiple regression.
The hypothesis of collinearity of expression/silencing patterns among OsGLP genes was tested by pairwise correlation analyses (Table II). In T0 plants, expression patterns for six OsGLP genes significantly predicted the expression of five or more other genes. Likewise, in T1 plants, expression patterns of five genes predicted the expression at least four other OsGLPs. Only correlations between OsGER4 family members OsGLP8-5, -6, -9, and -11 were repeated in both T0 and T1 generations. The consistent pairwise correlations among the OsGER4 genes suggest cosilencing and, therefore, indicate coregulated expression of these particular OsGLP genes. These results confirm the collinearity of independent variables in the multiple regressions (Table I).
Table II.
Pairwise correlations of OsGLP gene expression ratios in silenced transgenic lines (n = 19)
Using band intensity ratios for the T0 and T1 plants, correlation tests of all pairwise combinations of OsGLP genes were computed using the SAS program PROC CORR. r2 values are reported in the matrices, and significant values are indicated: * P < 0.05, ** P < 0.0001. Gene expression ratios refer to transgenic gel band intensity-to-wild-type gel band intensity, normalized with the gel band intensity of the internal control EF1-α.
| T0 Transgenic Plants
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 8-1 | 8-2 | 8-3 | 8-5 | 8-6 | 8-7 | 8-9 | 8-11 | 8-12 | OsGLP Genes |
| 1.00 | 0.22 | 0.72* | 0.61* | 0.34 | 0.64* | 0.80** | 0.48* | 0.63* | 8-1 |
| 1.00 | 0.44 | 0.14 | 0.50* | 0.33 | 0.43 | 0.19 | −0.18 | 8-2 | |
| 1.00 | 0.20 | 0.15 | 0.33 | 0.61* | 0.1 | 0.26 | 8-3 | ||
| 1.00 | 0.52* | 0.72* | 0.80** | 0.88** | 0.81** | 8-5 | |||
| 1.00 | 0.81** | 0.63* | 0.70* | 0.37 | 8-6 | ||||
| 1.00 | 0.86** | 0.75* | 0.63* | 8-7 | |||||
| 1.00 | 0.73* | 0.65* | 8-9 | ||||||
| 1.00 | 0.72* | 8-11 | |||||||
| 1.00 | 8-12 | ||||||||
| T1 Transgenic Plants
| ||||||||
| 8-2 | 8-5 | 8-6 | 8-7 | 8-8 | 8-9 | 8-11 | 8-12 | OsGLP Genes |
| 1.00 | 0.08 | 0.01 | 0.43 | −0.23 | −0.03 | 0.04 | 0.54* | 8-2 |
| 1.00 | 0.81** | 0.49* | 0.70* | 0.70* | 0.66* | 0.01 | 8-5 | |
| 1.00 | 0.53* | 0.74* | 0.72* | 0.69* | 0.11 | 8-6 | ||
| 1.00 | 0.26 | 0.44 | 0.44 | 0.48* | 8-7 | |||
| 1.00 | 0.77* | 0.69* | 0.06 | 8-8 | ||||
| 1.00 | 0.63* | 0.21 | 8-9 | |||||
| 1.00 | 0.28 | 8-11 | ||||||
| 1.00 | 8-12 | |||||||
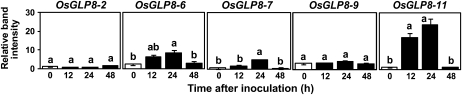
Gene expression of OsGER4 members that showed high (OsGLP6, -7, -9, and -11) and low (OsGLP8-2) correlation to blast resistance in gene silencing studies was evaluated in wild-type Kitaake rice plants at 0, 12, 24, and 48 h after inoculation with M. oryzae. Consistent with predictions from the silencing results, three (OsGLP8-6, -7, and -11) of the four OsGER4 gene members tested were up-regulated after M. oryzae inoculation. Inoculation with M. oryzae did not induce OsGLP8-2 or OsGLP8-9 gene expression, but there was a measurable basal level of OsGLP8-9 at all time points (Fig. 5).
Figure 5.
Induction of OsGLP genes after inoculation with M. oryzae. Three-week-old wild-type Kitaake plants were inoculated with M. oryzae isolate Che86061 (105 spores mL−1), and leaves were sampled for RNA at 12, 24, and 48 h after inoculation (x axis). Plants at time 0 were not inoculated. Expression of selected OsGLP genes was screened by RT-PCR, and gel band intensities were quantified and normalized against the reference gene, EF1-α (y axis; relative band intensities are in arbitrary units). Time point means (n = 3 biological repetitions) for each gene were compared with SAS and Proc GLM using the lsd method with a Student-Newman-Keuls test.
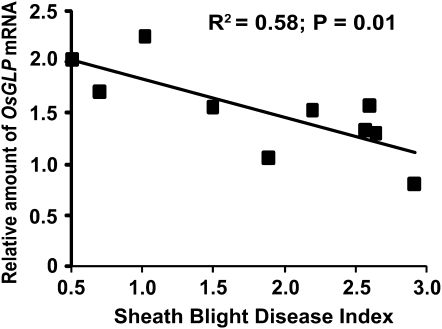
Silencing of chr 8 OsGLP members also correlated with disease susceptibility to another major rice disease, sheath blight caused by R. solani. Thirty T1 progeny were inoculated with R. solani, and 10 plants containing the transgene and showing contrasting phenotypes were tested for silencing of OsGLP family members (Supplemental Fig. 5, A and B). The more OsGLP family members silenced, the more susceptible the transgenic lines were to sheath blight (Fig. 6). Single linear regressions of disease index on individual gene band intensity ratios showed that OsGLP8-6, -7, -9, and -11 contribute most to sheath blight resistance (Table III). These are among the same OsGER4 subfamily members that contribute most to rice blast resistance (Table I), with one exception (OsGLP8-5; P = 0.184). Similar to observations in the rice blast experiments, expression patterns of the five OsGER4 subfamily members in the sheath blight data set were correlated for all pairwise combinations (P < 0.15; P values ranged from 0.0004 to 0.13).
Figure 6.
Reduced expression of rice chr 8 OsGLP gene members in individual silenced T1 plants correlates with increased sheath blight disease. Sheath blight disease index was assessed at 14 d after inoculation as described (Jia et al., 2007), and total relative OsGLP mRNA values were determined as in Figure 4.
Table III.
Expression/silencing of OsGER4 subfamily members correlates with sheath blight disease in T1 transgenic plants (P ≤ 0.05; boldface)
Linear regression of sheath blight disease index by OsGLP gene band intensity ratio; n = 10 individuals.
| OsGLP | Slope | r2 | P |
|---|---|---|---|
| 8-5 | −1.12 | 0.21 | 0.184 |
| 8-6 | −0.78 | 0.40 | 0.049 |
| 8-7 | −1.28 | 0.55 | 0.014 |
| 8-9 | −3.07 | 0.40 | 0.048 |
| 8-11 | −0.99 | 0.60 | 0.008 |
DISCUSSION
We demonstrate that an OsGLP gene cluster, which physically colocalizes to a rice blast resistance QTL, functions as a complex locus in disease resistance in rice. Furthermore, this resistance is effective against two distinct important rice pathogens. Field evaluations of rice line BC116, which contains the major effect chr 8 QTL, confirm that presence of the QTL correlates with enhanced resistance to rice blast disease for over 7 years of planting (14 cropping seasons). Thus, the resistance provided by the chr 8 QTL, which contains the OsGLP cluster, is broad spectrum, and trends to date suggest that the resistance will be effective for a long time.
While correlation of the presence of the chr 8 OsGLP cluster with resistance is useful for plant selection purposes, it does not constitute proof of function because BC116 has other introgression segments from the donor of resistance (Liu et al., 2004). Given the multigenic nature of this QTL and the inherent difficulty of making isogenic lines with and without OsGLP, we conducted gene silencing of all 12 related chr 8 OsGLP members using a highly conserved region. We observed cosilencing of closely related gene family members to variable degrees with a single highly conserved trigger sequence. The phenomenon of silencing multiple gene family members to different levels with a single construct was demonstrated previously for another rice gene family, the OsRac family (Miki et al., 2005).
In some suppressed T0 and T1 plants, the level of expression of some OsGLP genes was higher than the expression in the untransformed wild-type plants (Fig. 3; Supplemental Fig. S3). This could be due to the induction of some OsGLP family members as a compensatory measure for the suppression of others (Kafri et al., 2006) or because of developmental variation in expression. These options were not explored. We observed considerable variation in OsGLP mRNA levels among T0 and T1 generation plants (Fig. 3; Supplemental Fig. S3), possibly due to plant-to-plant variation rather than to gene suppression. However, expression of OsGLPs in Kitaake wild-type plants showed that variation among biological replications is very low (Fig. 5). This is most obvious for the expression of OsGLP8-2, which is not induced after inoculation with M. oryzae and remains the same during the time course of infection (Fig. 5). Thus, expression variation among transgenic plants is likely due to the presence of the transgene and suppression of the OsGLP members rather than to plant-to-plant variation.
By determining the number of genes cosilenced and the relative amounts of silencing in both T0 and T1 lines, we demonstrated that the chr 8 OsGLP genes contribute collectively to disease resistance, because as more genes were cosilenced the amount of disease increased (Figs. 3, 4, and 6). Closely related OsGLP family members on chr 12 were cosilenced in some lines; however, their silencing did not alone increase rice blast susceptibility, suggesting that their contributions to resistance are negligible if any (data not shown). It is possible that genes other than the OsGLPs that reside within the QTL interval on chr 8 may also contribute to the resistance phenotype. However, because suppressed expression of the chr 8 OsGLPs, and particularly the OsGER4 family members, rendered the plants more susceptible to both rice blast and sheath blight infection, we conclude that these genes are major contributors to disease resistance and may explain resistance governed by the chr 8 QTL.
Contributions of individual chr 8 OsGLP gene family members to the resistance phenotype varied, as shown in regression analyses, with certain gene family members contributing more than others. Orthologous GLP members in barley and grapevine are implicated in basal defense responses (Zimmermann et al., 2006; Godfrey et al., 2007). Indeed, in rice, OsGER4 subfamily members were consistently major contributors to resistance against rice blast compared with OsGER3 subfamily members, because their level of silencing was most significantly correlated with increased susceptibility in the transgenic plants. The importance of the OsGER4 subfamily was also observed for sheath blight. Interestingly, most of the OsGER4 genes that were correlated with resistance against rice blast were also correlated with resistance against sheath blight, with the exception of OsGLP8-5 (Tables I and III).
It is not clear from wild-type expression data whether resistance depends on constitutive or induced expression of OsGER4 genes. Transgenic plants used for experiments to identify OsGLP genes important for resistance were not inoculated. Therefore, our experiments measured the silencing of constitutive gene expression. In separate experiments using wild-type plants inoculated with M. oryzae, some OsGER4 genes identified as important by silencing (OsGLP8-6, -7, and -11) were induced above basal levels after infection with M. oryzae (Fig. 5). On the other hand, OsGLP8-9 showed basal expression but was not further induced after inoculation. Basal levels of gene expression could be important for resistance by creating a preformed resistant state in the plant. Furthermore, activation of the constitutively expressed enzymes could result in increased production of H2O2, which has been shown to induce HvGLPs in barley (Zimmermann et al., 2006).
Many OsGLPs, particularly the OsGER4 members, showed correlated expression/silencing patterns among transgenic plants, as indicated by the multiple regression and pairwise correlation analyses (Table II). This suggests coregulation and functional redundancy, as has been speculated for barley GLPs (Zimmermann et al., 2006). The close proximity and redundancy of the OsGLP gene family members, as well as their high sequence similarity, are suggestive of gene amplification through duplication followed by diversification (Kafri et al., 2006). In other studies, we have shown that the OsGLPs exhibit different induction patterns during development, wounding, and pathogen invasion and that the most commonly shared promoter motifs occur among OsGER4 family members (R. Davidson, unpublished data). Alternatively, diversification of the coding sequence could have created gene members that encode proteins with different enzymatic properties and protein activation and localization differences.
While our results show that the OsGLP genes confer broad-spectrum resistance, how they function to inhibit pathogens remains unknown. The different contributions observed for some of the OsGER4 gene family members against rice blast and sheath blight may result from tissue-specific induction of these genes rather than pathogen-specific induction, considering that both pathogens have different infection strategies with respect to tissue specificity. However, both pathogens directly penetrate the plant cuticle with distinct structures that may release general elicitors that will activate common defense responses (Marshall and Rush, 1980; Talbot, 2003).
Defense responses to these two pathogens share pathways, as shown in large-scale expression profiling experiments in rice (Venu et al., 2007; Zhao et al., 2008). We hypothesize that the chr 8 OsGLP genes contribute to resistance through enhancement of basal defense responses (Chisholm et al., 2006). Although rice enzyme function has not been tested, the OsGLPs on chr 8 are predicted to encode enzymes with superoxide dismutase activity based on high amino acid similarity to the barley HvGER4 member (HvOXOLP; Fig. 2) and the wheat (Triticum aestivum) TaGLP4 gene (Christensen et al., 2004; data not shown). These superoxide dismutases are proposed to be involved in basal defense responses, specifically through H2O2 generation (Christensen et al., 2004; Zimmermann et al., 2006).
The chr 8 OsGLP genes are highly related in sequence, structure, and organization to GLP genes in divergent cereals such as barley and wheat (Druka et al., 2002). In rice, seven of 12 putative OsGLPs are tightly clustered on chr 8 (R. Davidson, unpublished data). The orthologous barley HvGER4 subfamily contains at least nine clustered duplicated gene members including HvGERa, -b, -c, -d, and -e (Wei et al., 1998; Druka et al., 2002; Zimmermann et al., 2006). The chr 8 OsGER4 family members that contribute most clearly to disease resistance are the closest related rice members to barley HvGER4s, which are associated with defense responses in fungus-barley interactions (Wei et al., 1998). By comparing markers reported in different studies, we have found that the HvGER4s colocalize with barley QTL for fungal resistance (Chen et al., 2003), and the barley markers flanking this QTL were physically mapped to rice chr 8 (data not shown). Taken together, the evidence from rice, barley, and wheat implicates these cereal genes as contributors to an ancient plant basal defense mechanism (Lane, 2002; Christensen et al., 2004; Zimmermann et al., 2006; Godfrey et al., 2007). Coordinated function among members of the gene family could be an evolutionarily advantageous strategy by providing a stepwise, flexible response in proportion to the severity of infection.
The fact that several chr 8 OsGLP genes function together to confer resistance supports the emerging concept that QTL may not necessarily resolve to a single locus but instead may be controlled by several contiguous loci with small additive effects. QTL are predicted to provide broad-spectrum resistance, or resistance against multiple types of the same pathogen and/or diverse pathogen types. Consistent with this, the chr 8 QTL was originally identified in multiple-location trials in China and the Philippines, with vastly different populations of M. oryzae (Liu et al., 2004), suggesting that it confers resistance to many races of M. oryzae. Additionally, OsGLP-suppressed plants are more susceptible to sheath blight, which is particularly significant, because so far no simply inherited resistance has been identified for sheath blight. The broad-spectrum nature of the OsGLP-containing complex QTL may be responsible for the highly effective resistance observed in the deployed rice line BC116. Overall, the identification of multiple OsGLP loci conferring quantitative resistance has broad implications for the deployment of defense genes in breeding. If multiple loci are involved, single-gene transformation experiments may lead to the erroneous conclusion that the gene is not important for resistance. Future selection may need to take into consideration the allelic states of multiple loci in a gene family, and selection for a specific OsGLP cluster may be necessary to capture the collective effect of the specific gene family members.
MATERIALS AND METHODS
Field Studies
Five QTL from rice (Oryza sativa) SHZ-2, including the major-effect chr 8 QTL that is associated with OsGLP genes, were introgressed into the susceptible rice TXZ-13 using marker-assisted selection to develop the line BC116 (Liu et al., 2004). The field experiment (natural infestation) was performed for 7 years with two cropping seasons per year (total of 14 cropping seasons). Plots used a randomized complete block design and three replicates. The experiment was replicated in a second rice blast disease nursery at Conghua, Guangdong Province, China, with similar results (data not shown). Panicle blast symptoms were evaluated on each rice line using the International Rice Research Institute Standard Evaluation System for Rice (http://www.knowledgebank.irri.org/ses/SES.htm).
The presence of the chr 8 QTL in BC116 was confirmed using single sequence repeat analysis (Temnykh et al., 2000), single nucleotide polymorphism detection by TILLING analysis (Raghavan et al., 2007), and single feature polymorphism analysis using the University of Arizona rice genotyping array (Galbraith, 2006). TILLING analysis was used to detect mismatches between the two different alleles for OsGLP8-8 and OsGLP8-9 from SHZ-2 and TXZ-13 by heteroduplex cleavage and was performed as described (Raghavan et al., 2007; Supplemental Fig. S1). Primers for TILLING were OsGLP8-8F (5′-CTTGTTCTCCATCACAAGTTTACG-3′), OsGLP8-8R (5′-ATGCACGCCAAATAATTGATAGTA-3′), OsGLP8-9F (5′-AGAGAAGATAGCAGAAACCCAAAG-3′), and OsGLP8-9R (5′-AGCTTGCAAGTATGCATAACAAGT-3′).
Bioinformatics and Phylogenetic Analysis
Barley (Hordeum vulgare) cDNA sequences HvOXOA and HvOXOLP (accession nos. Y142203 and X93171, respectively) were used as queries for tBLASTx searches (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) using the HTGS database. FGENESH (http://www.softberry.com/berry.phtml) was used to predict putative oxalate oxidase and OsGLP from significant rice bacterial artificial chromosome hits. All nucleotide and inferred amino acid sequences corresponding to different predicted members were aligned using ClustalW. 1.83 (Thompson et al., 1997) in the BCM Search Launcher Interface (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). Ambiguous regions were removed, and 214 amino acid characters were used in Bayesian MCMC analysis (Ronquist and Huelsenbeck, 2003) to generate the tree. A mixed amino acid model was specified, and four chains were allowed to run for 1.5 × 107 generations (repeated four times), after which 5,000 trees were sampled from each run to determine the final consensus tree and posterior probabilities for each clade.
RNAi Silencing
The OsGLP RNAi construct (pTSi-OsGLP) was generated by cloning an antisense 500-bp PCR product corresponding to the second exon of OsGLP8-3, which is highly conserved among all OsGLP gene members on rice chr 8, into XcmI-digested pTSi vector (pTSi-OsGLP; Zhao, 2004; Supplemental Fig. S2). This fragment was amplified from IR64 genomic DNA using the primers OXOF2 (5′-TGGGTTTCCTTGCAAGAACC-3′) and OXOR2 (5′-TTCTTCTCCACTTGAAATGCC-3′). The two NheI restriction sites in the pTSi-1 RNAi vector were used to clone the pTSi-OsGLP RNAi construct into XbaI-digested pCAMBIA 1305 binary vector (http://www.cambia.org/daisy/cambia/materials/vectors/585.html) and used to transform Agrobacterium tumefaciens EHA105. Kitaake rice was used for Agrobacterium transformation (Hiei et al., 1994; Zhao et al., 2005). The presence of the transgene was confirmed in T0 plants by PCR amplification with a reverse primer specific to the 2xCaMV35S promoter and a forward primer specific to the transgene (OXOF2R2; Supplemental Table S3).
Plant and Fungal Growth and Inoculation Methods
For rice blast assays, plants were grown with a photoperiod of 16 h of light/8 h of dark in a growth chamber with photon flux of 135 μmol m−2 s−1 and day/night temperatures of 28°C/26°C. Magnaporthe oryzae was grown on oatmeal agar medium under constant light at 26°C for 2 weeks. T0 transgenic plants were inoculated 2 weeks after transfer from tissue culture to soil. The phenotype was validated by inoculation of leaves from the same T0 plants at an older growth stage. T1 transgenic and Kitaake wild-type plants were inoculated 2 weeks after planting in soil. Disease phenotypes were assessed using a detached leaf spot inoculation assay (Jia et al., 2003) that distinguished QTL-governed resistant and susceptible responses and that we confirmed to produce results in agreement with a commonly used spray inoculation assay (Valent et al., 1991; Supplemental Fig. S4). The second youngest rice leaves were spotted with a 5 × 104 spores mL−1 suspension of M. oryzae isolate Che86061, and disease was scored by visual assessment of the amount of mycelia present and colonization at the site (R = little or no mycelia or colonization, score 0–2; MS = moderate levels of mycelia and colonization, score 3–4; S = extensive mycelial growth and colonization, score 5–7) at 7 d after inoculation. Sheath blight assays of T1 lines derived from T0 plant 10 were performed in a greenhouse using a microchamber screening method (Jia et al., 2007). Plants were inoculated with Rhizoctonia solani isolate RM0140-1 at 14 d after seed germination and were scored at 14 d after inoculation (Jia et al., 2007).
For expression experiments of wild-type plants after inoculation with M. oryzae isolate Che86061, tissue from 21-d-old plants was harvested by combining the three most fully expanded leaves pooled from two plants per cultivar. Three rounds of RT-PCR were performed with three independently isolated total RNA samples (from three independent plant inoculation experiments). Plants were inoculated with 5 × 105 spores mL−1 at 20 pounds per square inch using an artist's air brush (Valent et al., 1991). Plants were kept in a mist chamber at 100% relative humidity for 24 h after inoculation and then returned to the growth chamber under the conditions described above.
DNA and RNA Analyses
Rice leaf genomic DNA was isolated (Murray and Thompson, 1980) and quantified by UV absorbance. Silencing patterns of the OsGLP gene family members were determined in independent T0 and T1 transgenic lines using RT-PCR. RT-PCR was performed using gene-specific primers for each OsGLP gene family member (Supplemental Table S3) in uninoculated T0 and T1 transgenic plants. Leaf tissue for RNA extraction was harvested from T0 transgenic plants 2 weeks after transfer from tissue culture to soil and from 2-week-old T1 transgenic plants and Kitaake wild-type plants. Total RNA for RT-PCR was isolated with Trizol reagent (Invitrogen) and treated with DNase (1 unit μg−1 total RNA; Promega). cDNA was synthesized using the SuperScript III reverse transcriptase kit (Invitrogen) and was amplified using HotStar Taq DNA Polymerase (Qiagen) and gene-specific primers (10 pmol of each primer) for each OsGLP gene on chr 3, 8, and 12 (Supplemental Table S3). EF1-α and ubiquitin genes were amplified as internal controls (Supplemental Table S3). Optimized cycles for unsaturated PCR, determined by a PCR cycle gradient with internal control primers, were 25 and 30 cycles for ubiquitin and EF1-α, respectively (data not shown). Hygromycin primers (Supplemental Table S3) and construct-specific primers (Supplemental Fig. S2) were used to determine the transgene presence. Ethidium bromide-stained gels were digitally photographed using the Gene Genius Bioimaging System and associated Gene Tools Gel Analysis software (Syngene). Band intensity values were calculated by subtracting the signal of the negative control on a given gel and were normalized with the band intensity of the EF1-α internal control. Band intensity ratios of the transgenic line to the wild type were calculated for each constitutively expressed OsGLP gene. Heat maps were drawn using R (http://www.R-project.org). Single linear and multiple regressions of disease scores on OsGLP band intensity ratios were performed using SAS software version 9.1.3 for Windows (SAS Institute) and PROC REG, and correlations of OsGLP band intensity ratios were computed using PROC CORR. Time point means (n = 3 biological replications) for wild-type expression of each gene were compared using PROC GLM and the lsd method with a Student-Newman-Keuls multiple testing correction.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Backcross rice line BC116 contains the chr 8 QTL harboring the OsGLP8 gene cluster from the resistant parent SHZ-2.
Supplemental Figure S2. RNAi silencing vector pTSi-1 and the OsGLP RNAi construct.
Supplemental Figure S3. Silencing patterns of the OsGLP in independent T0 and T1 transgenic plants.
Supplemental Figure S4. Disease phenotypes of Kitaake and Nipponbare after M. oryzae (Mo) inoculation.
Supplemental Figure S5. OsGLP-silenced T1 plants show higher levels of sheath blight disease.
Supplemental Table S1. Disease ratings for SHZ-2 (donor resistant parent), BC116 (backcross line harboring QTL from SHZ-2), and TXZ-13 (recurrent susceptible parent) after 7 years of field evaluation in a rice blast nursery in Yangjiang, Guangdong, China.
Supplemental Table S2. Gene members of the HvGER subfamilies used for the phylogenetic analysis of the OsGER subfamilies in rice.
Supplemental Table S3. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank L. Yan and P. Reeves for technical assistance and G. Mosquera for reviewing the manuscript.
This work was supported by the U.S. Department of Agriculture-Cooperative State Research, Extension, and Education Service-National Research Initiative (grant no. 2003–01551), the Cereal Comparative Genomics Initiative, a U.S. Agency for International Development Linkage Project, and the Kansas State and Colorado State Experiment Stations. R.M.D. was supported by the U.S. Department of Agriculture-Cooperative State Research, Extension, and Education Service-National Research Initiative (Rice-CAP grant no. 2004–35317–14867) and a Ford Foundation Diversity Fellowship. Field studies were funded by a Guangdong Academy of Agricultural Sciences (B.L., X.Z.) and International Rice Research Institute (H.L.) collaborative project.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jan Leach (jan.leach@colostate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alvarez M, Pennell R, Meijer P, Ishikawa A, Dixon R, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92 773–784 [DOI] [PubMed] [Google Scholar]
- Bolwell G, Wojtaszek P (1997) Mechanism for the generation of reactive species of oxygen species in plant defence: a broad perspective. Physiol Mol Plant Pathol 51 347–366 [Google Scholar]
- Bonman J, Khush G, Nelson R (1992) Breeding rice for resistance to pests. Annu Rev Phytopathol 30 507–528 [Google Scholar]
- Carter C, Graham RA, Thornburg RW (1998) Arabidopsis thaliana contains a large family of germin-like proteins: characterization of cDNA and genomic sequences encoding 12 unique family members. Plant Mol Biol 38 929–943 [DOI] [PubMed] [Google Scholar]
- Chen H, Wang S, Xing Y, Xu C, Hayes P, Zhang Q (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci USA 100 2544–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Silva H, Klessig D (1993) Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262 1883–1886 [DOI] [PubMed] [Google Scholar]
- Chisholm S, Coaker G, Day B, Staskawicz B (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Christensen A, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjaer M, Dudler R, Schweizer P (2004) The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact 17 109–117 [DOI] [PubMed] [Google Scholar]
- Dixon R, Harrison M (1990) Activation, structure, and organization of genes involved in microbial defense in plants. Adv Genet 28 165–234 [DOI] [PubMed] [Google Scholar]
- Druka A, Kudrna D, Kannangara G, Von Wettstein D, Kleinhofs A (2002) Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc Natl Acad Sci USA 99 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas B, Freysinet G, Pallett K (1995) Tissue-specific expression of germin-like oxalate oxidase during development and fungal infection of barley seedlings. Plant Physiol 107 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Gibbings JG, Mahmood T, Naqvi SMS (2008) Germin and germin-like proteins: evolution, structure, and function. Crit Rev Plant Sci 27 342–375 [Google Scholar]
- Farrar D, Glauber R (1967) Multicollinearity in regression analysis: problem revisited. Rev Econ Stat 49 92–107 [Google Scholar]
- Federico M, Iniguez-Luy F, Skadsen R, Kaeppler H (2006) Spatial and temporal divergence of expression in duplicated barley germin-like protein-encoding genes. Genetics 174 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW (2006) DNA microarray analyses in higher plants. OMICS 10 455–473 [DOI] [PubMed] [Google Scholar]
- Godfrey D, Able A, Dry I (2007) Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: a possible role in defense? Mol Plant Microbe Interact 20 1112–1125 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Jia Y, Correa-Victoria F, McClung A, Zhu L, Liu G, Wamishe Y, Xie J, Marchetti M, Pinson S, Rutger J, Correll J (2007) Rapid determination of rice cultivar responses to the sheath blight pathogen Rhizoctonia solani using a micro-chamber screening method. Plant Dis 91 485–489 [DOI] [PubMed] [Google Scholar]
- Jia Y, Valent B, Lee F (2003) Determination of host responses to Magnaporthe grisea on detached rice leaves using a spot inoculation method. Plant Dis 87 129–133 [DOI] [PubMed] [Google Scholar]
- Johnson R (1984) A critical analysis of durable resistance. Annu Rev Phytopathol 22 309–330 [Google Scholar]
- Kafri R, Levy M, Pilpel Y (2006) The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci USA 103 11653–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon R (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Mol Biol 48 251–275 [DOI] [PubMed] [Google Scholar]
- Lane B (2002) Oxalate, germins, and higher-plant pathogens. IUBMB Life 53 67–75 [DOI] [PubMed] [Google Scholar]
- Lee FN, Rush MC (1983) Rice sheath blight: a major rice disease. Plant Dis 67 829–832 [Google Scholar]
- Li Z, Pinson S, Marchetti M, Stansel J, Park W (1995) Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solani). Theor Appl Genet 91 382–388 [DOI] [PubMed] [Google Scholar]
- Liu B, Zhang S, Zhu X, Yang Q, Wu S, Mei M, Mauleon R, Leach J, Mew T, Leung H (2004) Candidate defense genes as predictors of quantitative blast resistance in rice. Mol Plant Microbe Interact 17 1146–1152 [DOI] [PubMed] [Google Scholar]
- Liu G, Jia Y, Correa-Victoria FJ, McClung A, Correll JC (2008) Identification of quantitative trait loci (QTLs) responsible for sheath blight resistance in rice using recombinant inbred line population of Lemont X Jasmine 85. Phytopathology 98 S92 [Google Scholar]
- Lou Y, Baldwin I (2006) Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol 140 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D, Rush M (1980) Infection cushion formation on rice sheaths by Rhizoctonia solani. Phytopathology 70 947–950 [Google Scholar]
- Membre N, Bernier F, Staiger D, Berna A (2000) Arabidopsis thaliana germin-like proteins: common and specific features point to a variety of functions. Planta 211 345–354 [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K (2005) RNAi silencing of single and multiple members in a gene family in rice. Plant Physiol 138 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Thompson W (1980) Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res 8 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P, Varner J (1993) Hydrogen peroxide and lignification. Plant J 4 887–892 [Google Scholar]
- Pinson S, Capdevielle F, Oard J (2005) Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci 45 503–510 [Google Scholar]
- Raghavan C, Naredo M, Wang H, Atienza G, Liu B, Qiu F, McNally K, Leung H (2007) Rapid method for detecting SNPs on agarose gels and its application in candidate gene mapping. Mol Breed 19 87–101 [Google Scholar]
- Ramalingam J, Vera Cruz C, Kukreja K, Chittoor J, Wu J, Lee S, Baraoidan M, George M, Cohen M, Hulbert S, et al (2003) Candidate resistance genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact 16 14–24 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rush MC, Lindberg GD (1984) Controlling sheath blight in rice. Louisiana Agriculture 27 16–18 [Google Scholar]
- Talbot N (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57 177–202 [DOI] [PubMed] [Google Scholar]
- Temnykh S, Park W, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishli T, McCouch S (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100 697–712 [Google Scholar]
- Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B, Farrall L, Chumley F (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venu R, Jia Y, Gowda M, Jia M, Jantasuriyarat C, Stahlberg E, Li H, Rhineheart A, Boddhireddy P, Singh P, et al (2007) RL-SAGE and microarray analysis of the rice transcriptome after Rhizoctonia solani infection. Mol Genet Genomics 278 421–431 [DOI] [PubMed] [Google Scholar]
- Wei Y, Zhang Z, Andersen C, Schmelzer E, Gregersen P, Collinge D, Smedegaard-Petersen V, Thordal-Christensen H (1998) An epidermis/papilla-specific oxalate oxidase-like protein in the defense response of barley attacked by the powdery mildew fungus. Plant Mol Biol 36 101–112 [DOI] [PubMed] [Google Scholar]
- Wu JL, Sinha PK, Variar M, Zheng KL, Leach JE, Courtois B, Leung H (2004) Association between molecular markers and blast resistance in an advanced backcross population of rice. Theor Appl Genet 108 1024–1032 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Collinge D, Thordal-Christensen H (1995) Germin-like oxalate oxidase, a H2O2-producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J 8 139–145 [Google Scholar]
- Zhao B (2004) Isolation and characterization of the maize nonhost resistance gene Rxo1 and the corresponding bacterial effector gene avrRxo1 from Xanthomonas oryzae pv. oryzicola. PhD thesis. Kansas State University, Manhattan, KS
- Zhao BY, Lin XH, Poland J, Trick H, Leach J, Hulbert S (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102 15383–15388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Wang A, Shi Y, Wang L, Liu W, Wang Z, Lu G (2008) Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor Appl Genet 116 501–516 [DOI] [PubMed] [Google Scholar]
- Zhou F, Zhang Z, Gregersen L, Mikkelsen J, de Neergaard E, Collinge D, Thordal-Christensen H (1998) Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol 117 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Baumlein H, Mock H, Himmelbach A, Schweizer P (2006) The multigene family encoding germin-like proteins of barley: regulation and function in basal host resistance. Plant Physiol 142 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.