Abstract
Abscisic acid (ABA) is an important plant hormone for a wide array of growth and developmental processes and stress responses, but the mechanism of ABA signal perception on the plasma membrane remains to be dissected. A previous GeneChip analysis revealed that a member of the A4 subfamily of lectin receptor kinases (LecRKs) of Arabidopsis (Arabidopsis thaliana), At5g01540 (designated LecRKA4.1), is up-regulated in response to a low dose of ABA in the rop10-1 background. Here, we present functional evidence to support its role in ABA response. LecRKA4.1 is expressed in seeds and leaves but not in roots, and the protein is localized to the plasma membrane. A T-DNA knockout mutant, lecrka4.1-1, slightly enhanced ABA inhibition of seed germination. Interestingly, LecRKA4.1 is adjacent to two other members of the A4 subfamily of LecRK genes, At5g01550 (LecRKA4.2) and At5g01560 (LecRKA4.3). We found that loss-of-function mutants of LecRKA4.2 and LecRKA4.3 exhibited similarly weak enhancement of ABA response in seed germination inhibition. Furthermore, LecRKA4.2 suppression by RNA interference in lecrka4.1-1 showed stronger ABA inhibition of seed germination than lecrka4.1-1, while the response to gibberellic acid was not affected in lecrka4.1-1 and lecrka4.1-1; LecRKA4.2 (RNAi) lines. Expression studies, together with network-based analysis, suggest that LecRKA4.1 and LecRKA4.2 regulate some of the ABA-responsive genes. Taken together, our results demonstrate that the A4 subfamily of LecRKs has a redundant function in the negative regulation of ABA response in seed germination.
Abscisic acid (ABA) is a hormone that modulates a wide spectrum of growth and developmental processes (such as seed maturation, dormancy, and germination) and responses to biotic and abiotic stresses (Finkelstein et al., 2002; Himmelbach et al., 2003; Xie et al., 2005). Although ABA signal perception and transduction pathways have not been clearly demonstrated yet, nearly 50 genes have been shown to affect ABA responses through forward and reverse genetics (Finkelstein et al., 2002). Among them, the ABA signaling proteins that are associated with the plasma membrane (PM) are of particular interest (Wang and Zhang, 2008; Wasilewska et al., 2008; Zheng and Xin, 2008). It is well recognized that PM is an important site for ABA perception (Finkelstein et al., 2002; Yamazaki et al., 2003; Wang and Zhang, 2008; Zheng and Xin, 2008), although direct biochemical evidence has been reported that ABA can also be sensed in the plastid (Shen et al., 2006) or the nucleus (Razem et al., 2006).
Two major types of PM-associated proteins have been increasingly recognized as important ABA perception or signaling components. The first one is the PM-integral receptor-like kinases. Among more than 600 receptor-like kinases in Arabidopsis (Arabidopsis thaliana), RPK1 was shown to be induced starting 1 h after the ABA treatment (Hong et al., 1997). Recently, the RPK1 mutants and antisense transgenic plants were shown to reduce seed dormancy and the sensitivity to ABA inhibition of seed germination (Osakabe et al., 2005). Although RPK1 controls the expression of many ABA-inducible genes, it remains to be determined whether RPK1 is a PM-localized ABA receptor.
The second type of PM-associated ABA signaling proteins includes heterotrimeric GTP-binding proteins such as GPA1 and AGB1 (Wang et al., 2001; Ullah et al., 2002; Pandey et al., 2006) and ROP monomeric GTPases such as ROP2, ROP6, and ROP10 (Lemichez et al., 2001; Li et al., 2001; Zheng et al., 2002; Xin et al., 2005). Both of these groups of GTP-binding proteins turn on/off the signaling switch by cycling between GTP-bound and GDP-bound forms. Furthermore, these GTP-binding proteins likely act through their associations with the PM-localized receptors. For example, G-proteins can be activated by the G-protein-coupled receptors (GPCRs), based on the animal signaling paradigm. A putative GPCR called GCR1 has been reported to modulate ABA responses in seed dormancy, root growth, and stomatal behavior (Colucci et al., 2002; Pandey and Assmann, 2004). More recently, GCR2 was claimed to be another putative GPCR and to act as an ABA receptor (Liu et al., 2007), with contrasting arguments reported by other groups regarding the mutant phenotypes in ABA responses and the biochemical identity of the gene (Gao et al., 2007; Illingworth et al., 2008). Therefore, the PM-localized ABA receptors remain cryptic.
In an effort to understand the transcriptional control of ABA response by ROP10 small GTPase, a negative regulator of ABA signaling (Zheng et al., 2002), we recently performed an Arabidopsis GeneChip analysis (Xin et al., 2005). Our results led to the identification of a group of genes, including a lectin receptor kinase encoded by At5g01540, which are only up-regulated by 1 μm ABA in rop10-1 (Xin et al., 2005). For this particular group of genes, higher ABA concentrations (10 and 100 μm) did not affect their expression in both the wild type and rop10-1, indicating that ROP10 is likely involved in the gating of low-dose ABA-regulated gene expression (Xin et al., 2005). The At5g01540 gene product has been grouped into the A4 subfamily of lectin receptor kinases (LecRKs; Barre et al., 2002); therefore, it is designated LecRKA4.1 here. In this report, we characterize LecRKA4.1 expression and protein subcellular localization patterns. More importantly, we show that LecRKA4.1 and the adjacent two other members, LecRKA4.2 and LecRKA4.3, have redundant functions in the negative regulation of ABA response in seed germination. Therefore, these results increase our understanding of the PM-associated ABA signaling events.
RESULTS
LecRKA4.1 Expression and Protein Subcellular Localization Patterns
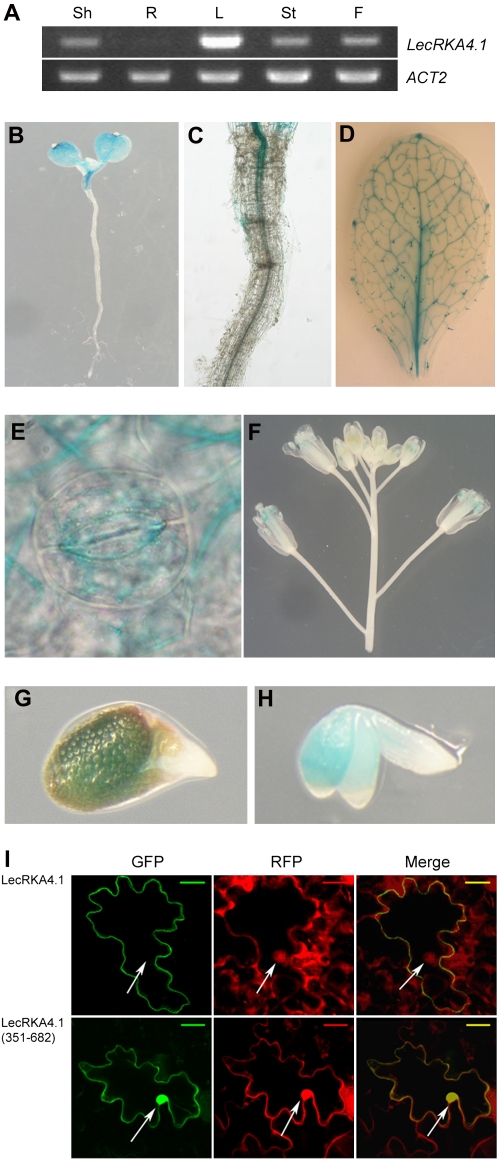
To determine where LecRKA4.1 is expressed, both reverse transcription (RT)-PCR and promoter:GUS reporter analyses were performed. RT-PCR analysis showed that LecRKA4.1 is preferentially expressed in the shoots of young seedlings, while its transcript is barely detectable in the roots (Fig. 1A). LecRKA4.1 is also weakly expressed in the stems and flowers but strongly expressed in the leaves (Fig. 1A). Consistent with the RT-PCR results, a 1.2-kb fragment of LecRKA4.1 promoter directed GUS reporter expression in the shoots but not in the roots of young seedlings (Fig. 1, B and C). This indicates that the 1.2-kb promoter fragment likely contains all of necessary cis elements for proper expression of the LecRKA4.1 gene. Using three homozygous PLecRKA4.1:GUS lines, all with one T-DNA insertion, we found that the LecRKA4.1 promoter is very active in the vascular system and trichomes of the leaf (Fig. 1D), and it is also expressed in the guard cells (Fig. 1E). In addition, this promoter is active in anthers and stigmas but not in petals (Fig. 1F). We also found that the LecRKA4.1 promoter is active in germinating seeds, although its activity is absent in the protruding radicles (Fig. 1, G and H).
Figure 1.
Expression and protein subcellular localization patterns of LecRKA4.1. A, RT-PCR analysis of LecRKA4.1 expression in different organs. Shown are shoots (Sh) and roots (R) of 7-d-old seedlings, mature leaves (L), stems (St), and flowers (F). ACT2 was used as an internal control. RT was prepared as described elsewhere (Gao et al., 2008). B to H, Expression patterns of LecRKA4.1 as revealed by PLecRKA4.1:GUS. Shown are a 5-d-old seedling (B), an enlarged view of a root segment immediately from the root-hypocotyl junction (C), a mature leaf (D), an enlarged view of guard cells (E), an inflorescence with several flowers (F), and a germinating seed with and without seed coat (G and H). I, Transient expression of LecRKA4.1-GFP and LecRKA4.1351–682-GFP in leaf pavement cells. GFP images are shown at left. An mCherry version of RFP was used as a control (middle). The merged GFP and RFP images are shown at right. The arrows indicate the position of the nucleus. Bars = 20 μm.
The intronless LecRKA4.1 gene has a translated region of 2,049 bp, encoding a protein of 682 amino acids. The predicted protein has an N-terminal signal peptide (amino acids 1–27), legume lectin α and β domains (amino acids 29–278), and a transmembrane domain (amino acids 310–332) that is followed by a protein kinase domain (amino acids 367–641) and the C terminus. To confirm that LecRKA4.1 is a PM-localized protein, we transiently expressed LecRKA4.1-GFP fusion protein in leaf epidermal cells of a transgenic line that expresses an mCherry version of red fluorescent protein (RFP). Compared with RFP, which is localized to the cytoplasm and the nucleus, LecRKA4.1-GFP is exclusively localized to the PM (Fig. 1I). Because the N-terminal half of LecRKA4.1 contains the signal peptide, lectin domains, and a transmembrane domain, we constructed a GFP fusion with the C-terminal half of LecRKA4.1, LecRKA4.1351–682, which contains the predicted protein kinase domain. Transient expression in leaf epidermal cells showed that LecRKA4.1351–682-GFP was expressed in the nucleus and the cytoplasm (Fig. 1I). This pattern is similar to that of the RFP control, as shown in the merged RFP and GFP images. Therefore, we conclude that the transmembrane domain-containing N-terminal half of LecRKA4.1 is responsible for targeting LecRKA4.1 specifically to the PM.
A T-DNA Knockout Mutant of LecRKA4.1 Slightly Enhanced ABA Response in Seed Germination Inhibition
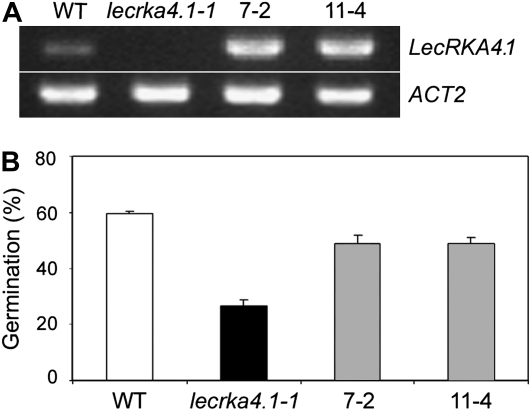
To demonstrate that the PM-localized LecRKA4.1 has a function in ABA response, we isolated and characterized a T-DNA line of LecRKA4.1 (SALK_ 070801), which is designated lecrka4.1-1. RT-PCR analysis showed that LecRKA4.1 transcript was absent in lecrka4.1-1 (Fig. 2A). Given that the T-DNA is located in the exon (1,434 bp from ATG), we believe that lecrka4.1-1 likely represents a null allele of LecRKA4.1. LecRKA4.1 is not expressed in the roots (Fig. 1A), and indeed, root growth inhibition response to ABA was not significantly altered in lecrka4.1-1 (data not shown). In addition, we did not observe any difference in leaf water loss in lecrka4.1-1 (data not shown), although the LecRKA4.1 promoter is active in the guard cells (Fig. 1E). Therefore, we focused our ABA phenotypic characterization on seed germination inhibition. After 3 d on the medium containing 0.6 μm ABA, the Columbia (Col) wild type had a germination percentage of 60%, but the germination percentage of lecrka4.1-1 was only 27% (Fig. 2B). This 55% reduction of seed germination percentage in lecrka4.1-1 indicates that the LecRKA4.1 mutant slightly increased the ABA sensitivity of seed germination inhibition. To substantiate that this enhancement is due to the loss of LecRKA4.1 function, a functional complementation test was performed. The full-length LecRKA4.1 cDNA under the control of a 2X35S cauliflower mosaic virus (CaMV) promoter was transformed into lecrka4.1-1, and two independently transformed lines, 7-2 and 11-4, had high levels of LecRKA4.1 transcript (Fig. 2A). In the absence of ABA, these two lines had almost 100% germination, similar to the wild type and lecrka4.1-1, and under 0.6 μm ABA, they had seed germination percentages close to that for the wild type compared with reduced germination for lecrka4.1-1 (Fig. 2B). These results suggest that LecRKA4.1 is a negative regulator of ABA response in seed germination.
Figure 2.
The lecrka4.1-1 knockout mutant slightly enhances the ABA response in seed germination inhibition. A, RT-PCR analysis of LecRKA4.1 mRNA expression in seedlings of the wild type (WT), lecrka4.1-1, and two transgenic lines (7-2 and 11-4) expressing the 2X35S:LecRKA4.1 transgene in lecrka4.1-1. ACT2 was used as an internal control. B, Functional complementation of lecrka4.1-1 by 2X35S:LecRKA4.1. The transgenic lines 7-2 and 11-4 showed similar seed germination percentages as the wild type in the presence of 0.6 μm ABA. Germination was scored at 3 d after the cold treatment. Data represent averages and sd of germination percentages for three replicates, each with about 40 to 50 seeds.
Mutants of LecRKA4.2 and LecRKA4.3 Slightly Enhanced ABA Response in Seed Germination Inhibition
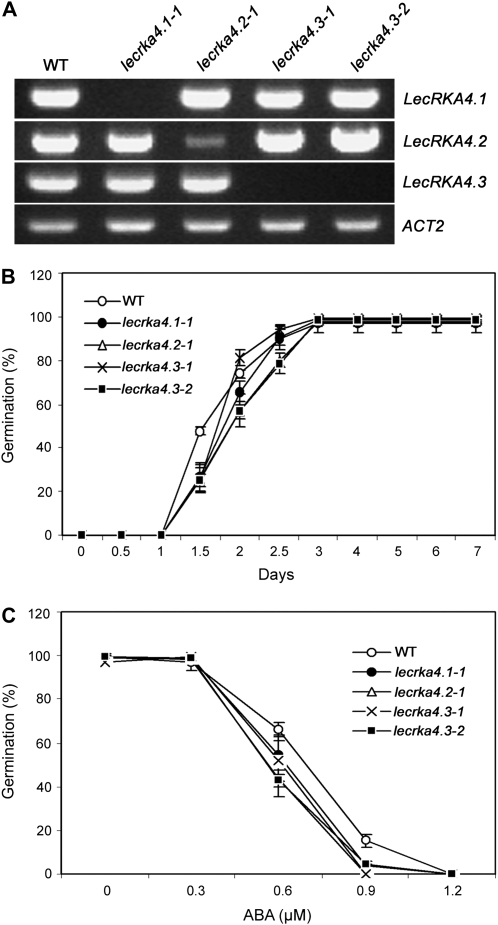
The weak phenotype of lecrka4.1-1 indicates that other members of the A4 subfamily of LecRKs could have redundant functions. Interestingly, two other members, At5g01550 (designated LecRKA4.2) and At5g01560 (LecRKA4.3), are immediately adjacent to LecRKA4.1 (At5g01540), and they share very high amino acid sequence identity (65%–74%). Therefore, we isolated and characterized their T-DNA insertional lines. In the Salk T-DNA database, there was one line for LecRKA4.2 (SALK_108000, designated lecrka4.2-1) and two lines for LecRKA4.3, SALK_003084 (designated lecrka4.3-1) and SALK_122197 (designated lecrka4.3-2). RT-PCR analysis showed that LecRKA4.2 transcript was lower than that in the wild type and that LecRKA4.3 was absent in the lecrka4.3-1 and lecrka4.3-2 alleles (Fig. 3A). As in the case of lecrka4.1-1, the T-DNA insertion in lecrka4.2-1, lecrka4.3-1, and lecrka4.3-2 did not interfere with the expression of other A4 subfamily genes.
Figure 3.
Mutants of LecRKA4.2 and LecRKA4.3 slightly enhance ABA response in seed germination inhibition. A, RT-PCR analysis of LecRKA4.1, LecRKA4.2, and LecRKA4.3 transcript levels in seedlings of the wild type (WT) and various mutants. ACT2 was used as an internal control. B and C, lecrka4.2 and lecrka4.3 mutants showed similar seed germination kinetic profiles with 0.3 μm ABA (B) and ABA dose-response patterns (C) as lecrka4.1-1. Germination for dose response was scored at 4 d after cold treatment. Data represent averages and sd of germination percentage for three replicates, each with about 40 to 50 seeds.
We then subjected the mutants to 0.3 μm ABA treatment. The germination kinetic profile showed that, similar to lecrka4.1-1, fewer lecrka4.2-1, lecrka4.3-1, and lecrka4.3-2 seeds germinated than wild-type seeds at 1.5 d after cold treatment, and after 3 d, they all germinated like wild-type seeds (Fig. 3B). At day 2, lecrka4.3-2 still germinated less, while lecrka4.3-1 had a similar germination percentage as the wild type. The reason for this allelic difference is unknown. A dose-response curve showed that after 4 d at both 0.6 and 0.9 μm ABA, the mutants for all three genes had lower germination rates than the wild type (Fig. 3C). These experiments indicate that the A4 subfamily of LecRKs encoded by the three genes clustered on chromosome 5 probably has a redundant function in ABA-mediated seed germination inhibition response.
lecrka4.1-1; LecRKA4.2 (RNAi) Lines Exhibited a Stronger ABA Response in Seed Germination Inhibition
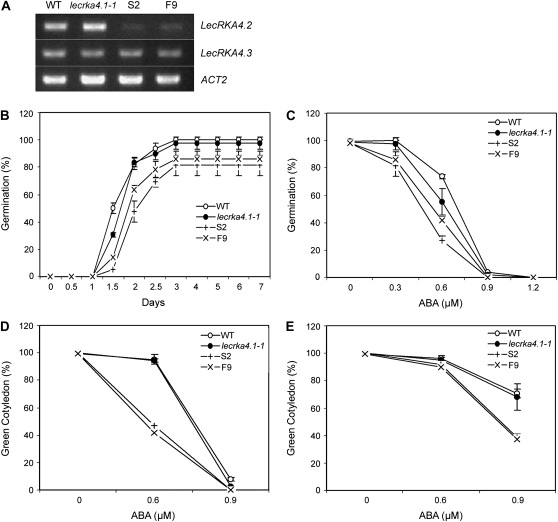
If LecRKA4.1, LecRKA4.2, and LecRKA4.3 have redundant functions as negative regulators of ABA response, we would expect to see a stronger phenotype in their double and triple mutants. However, their extremely tight genetic linkage prevented us from obtaining double or triple mutants by crossing with each other. Therefore, we decided to use RNA interference (RNAi) to suppress LecRKA4.2 and LecRKA4.3 gene expression in the lecrka4.1-1 background. Given their high sequence homology (77% identical for full-length cDNAs), we initially predicted that the LecRKA4.2 RNAi using either the partial fragment of LecRKA4.2 cDNA (1,130–1,760 bp) or the longer cDNA fragment (1–1,760 bp) would suppress both LecRKA4.2 and LecRKA4.3 expression. Two representative RNAi lines, S2 and F9, were chosen for further characterization. LecRKA4.2 mRNA expression was greatly suppressed in S2 and F9, but LecRKA4.3 expression was not altered (Fig. 4A).
Figure 4.
The lecrka4.1-1; LecRKA4.2 (RNAi) lines enhance the ABA response in seed germination inhibition. A, RT-PCR analysis of LecRKA4.2 and LecRKA4.3 mRNA expression in seedlings of the wild type (WT), lecrka4.1-1, and two LecRKA4.2 RNAi lines (S2 and F9) in the lecrka4.1-1 background. ACT2 was used as an internal control. B and C, S2 and F9 lines showed a stronger ABA seed germination inhibition response than lecrka4.1-1. Seed germination kinetic profiles (B) were determined using 0.3 μm ABA, and ABA dose response (C) was measured at 4 d after cold treatment. Each data point represents the average and sd of three replicates, each with about 40 to 50 seeds. D and E, S2 and F9 lines showed smaller green cotyledon percentages at 6 d (D) and 9 d (E) under ABA stress. Each point represents the average and sd of three replicates, each with about 40 to 50 seeds.
Seed germination kinetic profiles for the wild type, lecrka4.1-1, and the two RNAi lines, S2 and F9, at 0.3 μm ABA showed that after 1.5 d, while lecrka4.1-1 had a germination percentage about 50% lower than the wild type, S2 and F9 lines were even 50% to 80% lower than lecrka4.1-1 (Fig. 4B). After 2 d, although lecrka4.1-1 had a similar germination percentage as the wild type (approximately 82%), only 48% to 64% of seeds in S2 and F9 lines germinated. Furthermore, after 7 d, almost all of the wild-type and lecrka4.1-1 seeds germinated, but 14% to 19% of the seeds in S2 and F9 still did not germinate. On the other hand, an ABA dose-response curve showed that after 4 d, S2 and F9 lines had lower seed germination percentages than lecrka4.1-1 (Fig. 4C). We also examined the postgerminative seedling growth by scoring the cotyledon-greening phenotype, and the results showed that S2 and F9 had 50% fewer seedlings showing green cotyledons than lecrka4.1-1 after 6 d in the presence of 0.6 μm ABA (Fig. 4D) or after 9 d in the presence of 0.9 μm ABA (Fig. 4E). Taken together, these results show that suppression of LecRKA4.2 expression further enhances the ABA sensitivity observed in lecrka4.1-1.
To exclude the possibility that the enhanced response to ABA in lecrka4.1-1 and lecrka4.1-1; LecRKA4.2 (RNAi) lines is due to differences in seed germination or postgerminative seedling growth that are not related to ABA signaling, freshly harvested seeds of these mutant lines were allowed to germinate in the absence of ABA. The results showed that lecrka4.1-1 and the S2 and F9 lines exhibited a very similar germination kinetic profile (Supplemental Fig. S1). Furthermore, treatments with 1 and 10 μm GA3, which antagonizes the ABA inhibitory effect in seed germination, did not result in lower sensitivity to GA3 in lecrka4.1-1 and the S2 and F9 lines (Supplemental Fig. S2). These results support the idea that the observed seed germination response in the LecRKA4 subfamily gene knockout or RNAi lines is likely specific to ABA.
Expression of ABA-Responsive Genes Was Altered in lecrka4.1-1; LecRKA4.2 (RNAi) Lines
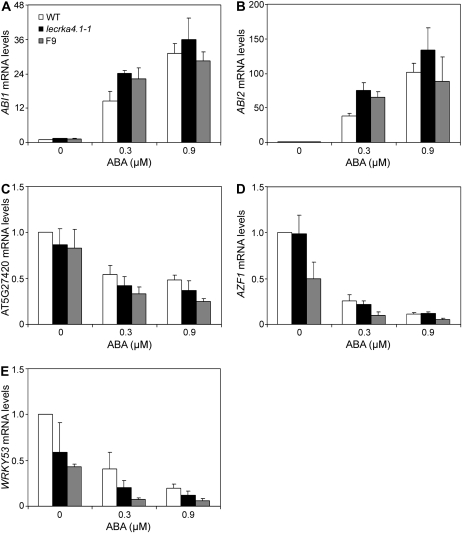
To test whether ABA-responsive genes are negatively regulated by the A4 subfamily LecRK, expression of ABI1 and ABI2 genes was quantified by real-time RT-PCR. Consistent with a previous report (Leung et al., 1997), transcripts of ABI1 and ABI2 were dramatically up-regulated by 0.3 and 0.9 μm ABA in the wild type (Fig. 5, A and B). In the presence of 0.3 μm ABA, the expression levels for ABI1 and ABI2 in both lecrka4.1-1 and the F9 RNAi line were slightly (60%–100%) higher than that in the wild type. However, there was no difference between lecrka4.1-1 and the F9 line, and at 0.9 μm ABA, the difference between the wild type and the mutant/RNAi line disappeared. Interestingly, a C3HC4-type zinc finger family protein gene (At5g27420), whose expression was not altered in the wild type by 1, 10, and 100 μm ABA (Xin et al., 2005), was slightly down-regulated by 0.3 and 0.9 μm ABA (Fig. 5C). Importantly, in the presence of 0.3 μm ABA, its expression level was slightly lower in lecrka4.1-1 than in the wild type, and F9 had an even lower expression level than lecrka4.1-1 (Fig. 5C). Although the treatment with 0.9 μm ABA did not further reduce the expression of At5g27420 in the wild type and lecrka4.1-1, the F9 line still displayed a similar difference compared with the wild type.
Figure 5.
Real-time PCR analysis of gene expression in response to ABA. Four-day-old wild-type (WT), lecrka4.1-1, and F9 seedlings grown in liquid MS medium were treated for 12 h with 0, 0.3, and 0.9 μm ABA prepared in fresh MS medium. Expression of ABI1 (A), ABI2 (B), At5g27420 (C), and two genes, AZF1 (D) and WRKY53 (E), which were selected from the assembled protein-protein interaction network (Fig. 6), was quantified using real-time PCR analysis. The mRNA levels for each of these genes were normalized with ACT2, and relative mRNA levels in nontreated wild-type seedlings for each gene were set at 1. Average and se values for three biological repeats are shown. The wild type, lecrka4.1-1, and F9 are designated in all panels as shown in A.
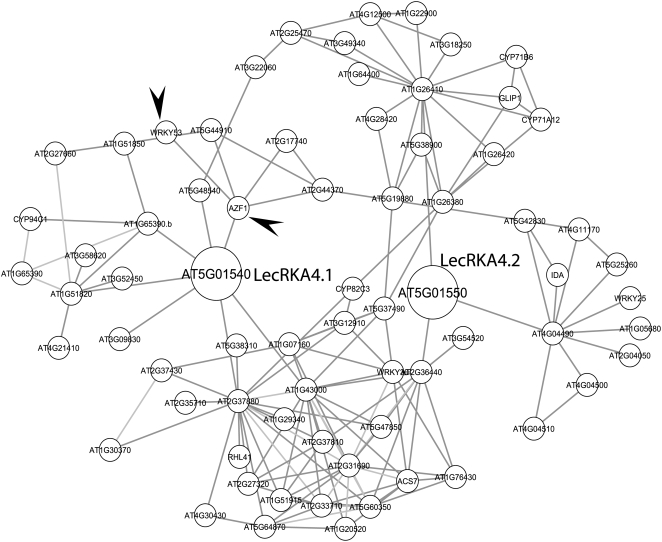
To further explore which genes are possibly regulated by the three members of A4 subfamily LecRK or their pathway, a protein-protein interaction network was assembled. The protein-protein interactions were based on both experimentally demonstrated or predicted physical interactions (Geisler-Lee et al., 2007) and functional interactions implicated by a large number of gene expression analyses (Ma et al., 2007). We found that LecRKA4.1 and LecRKA4.2, but not LecRKA4.3, were present in the network database, resulting in a total of 70 unique genes (nodes; Supplemental Table S1) that exhibit 149 physical or functional interactions (edges). Therefore, a network for LecRKA4.1 and LecRKA4.2 was assembled (Fig. 6). Functional categorical analysis of these 70 genes showed that genes involved in metabolism (in particular phosphate metabolism), protein fate (folding, modification, and destination), cellular transport, interaction with environments, and cell rescue, defense, and virulence were overly represented compared with the whole genome representation (Supplemental Table S1).
Figure 6.
A LecRKA4.1 and LecRKA4.2 gene interaction network. Three members of LecRKA4 genes were mapped to the protein-protein interaction database, with only LecRKA4.1 and LecRKA4.2 present in the database. This analysis revealed a total of 70 unique genes (Supplemental Table S1) that showed 149 interactions, and a network was then assembled based on these interactions. Arrowheads indicate the two genes (WRKY53 and AZF1) whose transcript levels were analyzed using real-time PCR (Fig. 5, D and E).
We then selected two transcription factor genes from the assembled network for quantitative analysis of their expression in lecrka4.1-1 and the F9 line. AZF1, although not yet updated in the database as ABA suppressed, was recently shown to be down-regulated by ABA (Sakamoto et al., 2004), and its close homolog, AZF2, was demonstrated to act as a transcription factor negatively regulating ABA response (Sakamoto et al., 2004). Consistently, we found that AZF1 mRNA level was reduced by about 75% and 90% by 0.3 and 0.9 μm ABA, respectively (Fig. 5D). Surprisingly, although AZF1 showed a direct functional interaction with LecRKA4.1 based on the assembled network (Fig. 6), there was no difference in its transcript levels between the wild type and lecrka4.1-1 at 0, 0.3, and 0.9 μm ABA (Fig. 5D). However, F9 had a 50% lower expression level than the wild type in the absence of ABA and a 65% lower level at 0.3 μm ABA. At 0.9 μm ABA, a similar trend was observed. In contrast to AZF1, WRKY53, another transcription factor gene that was down-regulated by ABA, as reported by the Genevestigator Web site (https://www.genevestigator.ethz.ch/gv/index.jsp) and in our experimental conditions (Fig. 5E), showed differential expression levels between the wild type, lecrka4.1-1, and F9 (Fig. 5E). Although there was no statistical difference between lecrka4.1-1 and the wild type in the no-ABA control, the 50% lower level difference in lecrka4.1-1 compared with the wild type at 0.3 μm ABA was statistically significant. Interestingly, as in the case of At5g27420 under the 0.3 μm ABA treatment (Fig. 5C), F9 showed a further reduction in WRKY53 expression compared with lecrka4.1-1 (Fig. 5E). In the control, there was an approximately 2-fold difference between F9 and the wild type, but at 0.3 μm ABA, they showed a 5-fold difference. At 0.9 μm ABA, there was still a 3-fold difference between F9 and the wild type.
DISCUSSION
In this work, we provide genetic evidence that LecRKA4.1 and the other two most closely related LecRKs (LecRKA4.2 and LecRKA4.3) act as negative regulators of ABA response in seed germination. First, we show that single knockout mutants for each of these three genes exhibit subtle enhancements in ABA-inhibited seed germination. Second, down-regulation of LecRKA4.2 by RNAi further enhanced the response in lecrka4.1-1. Third, this enhancement is not due to the developmental delay in lecrka4.1-1 and lecrka4.1-1; LecRKA4.2 (RNAi) lines or to their altered sensitivity to GA3. Fourth, gene expression studies, coupled with gene network assembly, suggest that LecRKA4.1 and LecRKA4.2 control the expression of some ABA-responsive genes. Therefore, our results demonstrate that the A4 subfamily of LecRKs acts redundantly in the negative regulation of ABA response.
The identification of the PM-associated ABA signaling components, including ABA receptors, has been a significant challenge. Some of the components, such as RPK1 (Hong et al., 1997; Osakabe et al., 2005), are targeted for their possible functions in ABA response because of earlier observations that they are transcriptionally regulated by ABA in the wild type. However, the genes that are not transcriptionally affected by ABA in the wild type may not give investigators a clue to their possible function in ABA response and thus may potentially be overlooked. For example, LecRKA4.1, reported in this paper, was not regulated by 1, 10, and 100 μm ABA at the transcriptional level in the wild type in several DNA microarray experiments (Xin et al., 2005), nor was its promoter activity differentially regulated by low concentrations (0.3 and 0.9 μm) of ABA (Supplemental Fig. S3). Therefore, it would be difficult to assume its role in the control of ABA response. However, using the rop10-1 background and only in the presence of a low dose of ABA, this gene was revealed to be slightly up-regulated in rop10-1 (Xin et al., 2005). Clearly, transcriptome analysis involving various genetic backgrounds and different ABA doses will lead to the identification of novel components, in particular those associated with the PM, in the ABA signaling network.
However, it is difficult to elucidate how the A4 subfamily of LecRKs links to ROP10 small GTPase in ABA signaling. The observation that LecRKA4.1 was up-regulated by a low dose (1 μm) but not at higher doses (10 and 100 μm) of ABA in rop10-1 seedlings indicates that LecRKA4.1 might participate in the ROP10-gated, low-dose ABA response pathway (Xin et al., 2005). In this report, we show that the lecrka4.1-1; LecRKA4.2 (RNAi) lines enhanced the sensitivity to low doses (0.3–0.9 μm) of ABA in the seed germination response and the cotyledon-greening process, but we were unable to test this hypothesis in the true leaves because we failed to observe any difference in leaf water loss between the lecrka4.1-1; LecRKA4.2 (RNAi) lines and the wild type. It is possible that LecRKA4.3 and/or the residual LecRKA4.2 mRNA in these lines might still be functional to mask the low-dose ABA response in the true leaves. A further complication is that if LecRKA4.1 transcript is higher in rop10-1 (Xin et al., 2005), one would expect that increased LecRKA4.1 expression would be necessary for the enhanced ABA response in rop10-1 (Zheng et al., 2002). However, loss of LecRKA4.1 function (and that of two other LecRK genes) resulted in a similar phenotype as rop10-1. The facts that rop10-1 and lecrka4.1-1 are in different ecotypic backgrounds and they have weak phenotypes prevented us from testing whether they act in the same or different pathways. Although we did not find any apparent difference in ROP10 transcript level in the wild type and a lecrka4.1-1; LecRKA4.2 (RNAi) line treated by 0.3 and 0.9 μm ABA (data not shown), it is still possible that LecRKA4.1, LecRKA4.2, and LecRKA4.3 might function to regulate the ROP10 GTPase activity. If this is the case, this type of regulation is possibly subjected to the transcriptional control of the LecRK genes by ROP10 signaling. Given the complexity of the ABA response, in particular a need for plants to distinguish low versus high ABA concentrations and/or transient versus sustained ABA action, this kind of regulatory loop, if demonstrated, will be an important mechanism for plants to cope with their dynamic environments during growth and development.
Nevertheless, the identification of three members of the A4 subfamily of LecRKs as redundant, negative regulators of ABA response will provide novel insights into the complex ABA signaling networks. Recently, plastidic and nuclear ABA receptors have been found (Razem et al., 2006; Shen et al., 2006), together with the PM-localized RPK1, as an important ABA signaling component (Osakabe et al., 2005). One common feature of these components is that their mutants exhibit only weak or limited phenotypes (Osakabe et al., 2005; Razem et al., 2006; Shen et al., 2006). Similarly, we only observed the weak seed germination phenotype for various LecRKA4 single mutants and the lecrka4.1-1; LecRKA4.2 (RNAi) lines. This indicates that ABA perception and response likely involve distinct receptors and/or signaling branches. Therefore, the identification of various components involved in ABA perception and signaling remains an important step before we have a full picture of ABA sensing and signaling.
The role of the legume lectin-like extracellular domain in ABA signaling is intriguing. Lectins are known to bind to carbohydrates, and some of them have been shown to act as defense proteins or in plant development (Herve et al., 1996; Navarro-Gochicoa et al., 2003; Andre et al., 2005; Chen et al., 2006; Wan et al., 2008). In Arabidopsis, there are a total of 42 LecRKs, and most of them have not been functionally characterized yet (Barre et al., 2002). If oligosaccharides turn out to be the ligands for the A4 subfamily of LecRKs, how does that link to ABA response? Given that alterations in the biosynthesis of some polysaccharides, such as pectin and hemicellulose, lead to altered sugar responses (Li et al., 2007; Mouille et al., 2007; Gao et al., 2008) and that the cross talk of sugar and ABA responses exists in plants (Finkelstein and Gibson, 2002), one might speculate that oligosacchrides might exert an effect in sugar signaling that then indirectly affects the ABA response. However, the observed ABA-inhibited seed germination phenotype in the LecRKA4 mutants or transgenic lines is less likely due to sugar signaling, as we did not observe any altered sugar response (data not shown). Another possibility is the ABA signaling-defense response link. The network analysis indicates that LecRKA4.1 and LecRKA4.2 have interactions with some genes, such as WRKY53 (Murray et al., 2007) and WRKY25 (Zheng et al., 2007), which are involved in defense response. The involvement of ABA signaling in the defense response has been reported (de Torres-Zabala et al., 2007), and a function for LecRK in disease resistance was reported in rice (Oryza sativa; Chen et al., 2006). Therefore, it is also possible that the A4 subfamily of LecRKs may act through ABA signaling to modulate disease response, or that in response to oligosaccharides that are associated with disease response, LecRKA4 members indirectly exert their effect in the ABA response. However, the possibility that these LecRKs might directly bind ABA cannot be excluded. Clearly, testing the binding of the A4 subfamily of LecRKs by ABA at the physiologically relevant concentrations will be one future direction.
Another interesting question that needs to be addressed concerns the pathways controlled by these LecRKs. Consistent with their weak ABA response phenotypes, the mutants and transgenic lines exhibit slight differences in ABI1/ABI2 expression from their wild types, indicating that these LecRKs may not control a major ABA response pathway but only modulate a minor branch of the pathway. Although more dramatic differences might be observed if three members of the A4 subfamily of LecRK genes are knocked out, currently this possibility cannot be tested because they physically cluster on the same chromosomal location. Furthermore, the assembled network and gene expression studies indicate that LecRKA4.1 and LecRKA4.2 may have overlapping and distinct response pathways. For example, while the mutation in LecRKA4.1 increased ABI1 and ABI2 expression, LecRKA4.2 (RNAi) did not further enhance their expression (Fig. 5, A and B). Similarly, it is less likely that AZF1 was predominantly affected by LecRKA4.1, but a reduction in LecRKA4.2 expression could enhance the ABA suppression of AZF1 expression (Fig. 5D). Nevertheless, down-regulation of the genes At5g27420 (encoding a zinc finger family protein) and WRKY53 (a transcription factor) was enhanced by the loss of LecRKA4.1 and LecRKA4.2 function (Fig. 5, C and E). Regardless of whether or not these LecRKs modulate a minor ABA signaling branch through overlapping or distinct pathways, it is important to first identify their downstream factors in ABA signaling in the future. Therefore, genetic and biochemical analyses of those components on the assembled network will provide further insights into the complex ABA signaling networks.
MATERIALS AND METHODS
Plant Materials, T-DNA Mutant Isolation, and Growth Conditions
The lecrka4.1-1 (SALK_ 070801), lecrka4.2-1 (SALK_108000), lecrka4.3-1 (SALK_003084), and lecrka4.3-2 (SALK_122197) mutants of Arabidopsis (Arabidopsis thaliana) were isolated from T-DNA insertion lines maintained in the Arabidopsis Biological Resource Center at The Ohio State University. For screening homozygous mutants, the following primers were used: XZP136 (sense, 5′-CATCCTCGCAGATTGAGATACAG-3′) and XZP60 (antisense, 5′-TCGACGAAACATATCCTACCAACT-3′) for lecrka4.1-1; XZP211 (sense, 5′-GTTTTCAAAGGGGAAGAACATACG-3′) and XZP212 (antisense, 5′-GGGTTGGAGATAGCGTGAAGGTG-3′) for lecrka4.2-1; and XZP213 (sense, 5′-CTCTGTCTTCTTTGCTTCTTCATCTCG-3′) and XZP214 (antisense, 5′-CGGCTATAAAGAAGGTCCCAGAG-3′) for lecrka4.3-1 and lecrka4.3-2. Seeds of the Arabidopsis wild type (Col), T-DNA insertion mutants, and transgenic plants were cold treated at 4°C for 3 to 4 d and then germinated and grown in the greenhouse for harvesting seeds, or in an incubator at 22°C with a 16-h-light/8-h-dark cycle for seed germination assay.
ABA and GA Response Assays
Seed germination was performed on half-strength Murashige and Skoog (MS) medium supplemented with various ABA concentrations. Petri plates were incubated at 22°C in the light after cold treatment (4°C for 3–4 d). Germination was scored every 12 h after incubation in the light, with the criterion being set as the complete protrusion of the radicle. For cotyledon-greening characterization, seedlings were observed from 5 to 9 d after cold treatment. For the seed germination assay with GA3, seeds were plated on half-strength MS medium supplemented with various GA3 concentrations. After 3 d of cold treatment, germination was scored every 2 h after incubation in the light, with the same germination criterion used above.
Regular and Real-Time RT-PCR Analyses
For regular PCR analysis of LecRK gene expression, 7-d-old young seedlings were used for total RNA using TRIzol (Invitrogen). A total of 5 μg of RNA was reverse transcribed in a 20-μL reaction using SuperScript III reverse transcriptase and oligo(dT)12–18 primer (Invitrogen) according to the instructions provided by the vender. PCR was performed using Taq DNA polymerase, with ACT2 as the internal control, using the primers ACT2S and ACT2A described previously (Xin et al., 2005). Gene-specific primer pairs XZP196 (sense, 5′-CTCTGCAGATGCATCCTCGCAGATTGAGATAC-3′) and XZP197 (antisense, 5′-GTGCTACTGACTGATACGAGAAGTCG-3′); XZP206 (sense, 5′-TTCTTAATTAACCATGGATGCTCGTGCTCTTCTTGCT-3′) and XZP205 (antisense, 5′-AAGGATCCATTTAAATGAATCTCTCCTCTCGCGTGCAAT-3′); and XZP213 (sense, 5′-CTCTGTCTTCTTTGCTTCTTCATCTCG-3′) and XZP232 (antisense, 5′-CTCCGCTTCTTCTCGGTTTACTG-3′) were used for the amplification of LecRKA4.1, LecRKA4.2, and LecRKA4.3, respectively. These experiments were repeated once.
For real-time PCR analysis of gene expression, seeds of lecrka4.1-1, its wide type (Col), and a lecrka4.1-1; LecRKA4.2 (RNAi) line (F9) were incubated in 50 mL of liquid MS medium, pH 5.7, at 4°C for 3 d and then allowed to germinate and grow at 22°C for 4 d under a 16-h-day/8-h-night regime with continuous shaking at 150 rpm. After three rinses with freshly prepared MS liquid medium, young seedlings were treated with 0, 0.3, and 0.9 μm ABA that was dissolved with dimethyl sulfoxide (with the final concentration in the medium of 0.1%, v/v) and prepared in the MS medium. After 12 h of treatment, the seedlings were immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation. Three biological replicates were performed at different times for all genotype/treatment combinations. RNA RT was described above, and real-time quantitative PCR analysis was performed using the QuantiTect SYBR Green PCR kit (Qiagen). Real-time PCR was carried out in the MasterCycler II (Cypheid) according to the manufacturer's protocol. The primers for ACT2 were designed previously (Zheng et al., 2002). Gene-specific primer pairs were as follows: for ABI1 (At4g26080), XZP331 (sense, 5′-AGAGTGTGCCTTTGTATGGTTTTA-3′) and XZP332 (antisense, 5′-CATCCTCTCTCTACAATAGTTCGCT-3′); for ABI2 (At5g57050), XZP333 (sense, 5′-GATGGAAGATTCTGTCTCAACGATT-3′) and XZP334 (antisense, 5′-GTTTCTCCTTCACTATCTCCTCCG-3′); for At5g27420, XZP177 (sense, 5′-CAGAGACATGACCCGTATGC-3′) and XZP178 (antisense, 5′-GCGCCAGTACAGTGACGTAT-3′); for AZF1, XZP281 (sense, 5′-AGCGGTTCCGTTGTTATTA-3′) and XZP282 (antisense, 5′-ACGCAAACGACTTGAAACAG-3′); and for WRKY53, XZP286 (sense, 5′-AATCTCCGGCATCGATAAAC-3′) and XZP288 (antisense, 5′-GCCTCTCTCTGGGCTTATTC-3′).
Construction of 2X35S:LecRKA4.1
The LecRKA4.1 full-length cDNA was amplified using primers XZP147 (sense, 5′-ACTGCAGATGGGCACACAAAGATCCATG-3′) and XZP148 (antisense, 5′-GCAGATCTCTACTGACTGATACGAGAAGTC-3′), with the underlined bases indicating the introduced restriction enzyme sites for cloning. The PCR product was digested by PstI and BglII and then cloned into GZ42, which is based on pCAMBIA1301 (B4), containing a hygromycin resistance selection marker and a 2X35S promoter of CaMV.
Construction of PLecRKA4.1:GUS
The LecRKA4.1 promoter fragment containing a 1.2-kb region upstream of the ATG codon and 20 bp of coding DNA downstream of ATG was amplified with primers XZP199 (sense, 5′-GGCGTCGACCTCAGTCCCATCTTTCTC-3′) and XZP200 (antisense, 5′-CATGGATCCTTGTGTGCCCATGGGTG-3′) using TaKaRa ExTaq (TaKaRa Bio). Underlined bases show the restriction enzyme sites for cloning. The amplified DNA fragment was digested by SalI and NcoI and then cloned into SalI and NcoI sites of the binary vector pCAMBIA1301 (B4) that contains GUS and the CaMV 35S terminator. The resulting plasmid, XZ22, was then transformed into Col using the floral dip method (Clough and Bent, 1998). Independently transformed lines were used for GUS activity assay as described elsewhere (Dan et al., 2007).
Construction and Expression of LecRKA4.1-GFP, LecRKA4.1351–682-GFP, and RFP
LecRKA4.1 full-length cDNA was amplified with XZP147 (sense, as described above) and XZP198 (antisense, 5′-CATCTAGACTGACTGATACGAGAAGTCGAAGAAAC-3′), then digested by PstI and XbaI and cloned into the pCAMBIA3301-based binary vector GZ6 that contains GFP and the CaMV 35S promoter and terminator, resulting in the 35S:LecRKA4.1-GFP plasmid, designated XZ16. A cDNA fragment (1,051–1,339 bp) of LecRKA4.1 was amplified with XZP196 (sense, as described above) and XZP193 (antisense, 5′-CTAAGCTTCCGTTGGGGATGTAATCG-3′) and then digested by PstI and HindIII. This fragment was cloned into XZ16 (digested by PstI and HindIII), giving rise to the LecRKA4.1351–682-GFP plasmid, designated XZ25. XZ16 and XZ25 were bombarded into young leaves of 35S:mCherry (RFP) lines, according to the instructions with the Bio-Rad Biolistic PDS-1000/He Particle Delivery System. The 35S:mCherry plasmid was constructed by replacing GFP in GZ6 (described above) with a cDNA fragment encoding the mCherry version of RFP (Shaner et al., 2004), which was PCR amplified from the pRSET-B mCherry plasmid kindly provided by Dr. Bo Liu (University of California, Davis). Two primers were used: the sense primer ZZP63 (5′-TATCTAGAATGGTGAGCAAGGGCGAGGA-3′) and the antisense primer ZZP64 (5′-GGGTTACTTGTACAGCTCGTCCATG-3′). Fluorescence was visualized under a Bio-Rad Radiance 2000 confocal laser scanning device (Carl Zeiss Microimaging) and analyzed using ImageJ 1.37 software (http://rsb.info.nih.gov/ij/), as described elsewhere (Yang et al., 2007).
Production of lecrka4.1-1; LecRKA4.2 (RNAi) Transgenic Plants
The LecRKA4.2 cDNA fragment (1–1,760 bp) was amplified with primers XZP206 (sense, 5′-TTCTTAATTAACCATGGATGCTCGTGCTCTTCTTGCT-3′) and XZP205 (antisense, 5′-AAGGATCCATTTAAATGAATCTCTCCTCTCGCGTGCAAT-3′) using TaKaRa ExTaq (TaKaRa Bio). The amplified fragment was digested by SwaI and NcoI and then cloned into the pFGC5941 double-stranded RNA vector. Subsequently, this amplified fragment was digested by BamHI and PacI and cloned into the above resulting plasmid, giving rise to the RNAi plasmid, designated XZ20. Another RNAi plasmid, XZ21, was similarly obtained by amplifying the LecRKA4.2 cDNA fragment (1,130–1,760 bp) with primers XZP204 (sense, 5′-AATCTAGACCATGGTCCGAGGAAACTTATCGTCACC-3′) and XZP205 (antisense, as described above). This fragment was then digested by SwaI-NcoI and BamHI-XbaI and sequentially cloned into pFGC5941. XZ20 and XZ21 were then transformed into the lecrka4.1-1 knockout mutant using the floral dip method (Clough and Bent, 1998). Two representative lines, S2 and F9, respectively, from XZ21- and XZ20-transfromed populations, were chosen for ABA response assays.
Network Assembly
We assembled a protein-protein interaction network using the network assembly methods as adopted previously (Chuang et al., 2007; Bromberg et al., 2008). Briefly, our network database includes the known protein interaction databases, which contain the data from yeast two-hybrid experiments, predicted interactions via orthology and cocitation (Geisler-Lee et al., 2007; http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi), curation of the literature, and the recently reported interaction database derived from analysis of large-scale DNA microarray analyses (Ma et al., 2007). Three LecRKA4 genes were mapped to their corresponding proteins in the network database. LecRKA4.3 was not present in the assembled network database; thus, the resulting interactions were used to build the LecRKA4.1 and LecRKA4.2 interaction network.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g01540 (LecRKA4.1), At5g01550 (LecRKA4.2), and At5g01560 (LecRKA4.3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Seed dormancy behaviors of lecrka4.1-1, S2, and F9.
Supplemental Figure S2. Seed germination patterns of lecrka4.1-1, S2, and F9 are similar to those of the wild type in response to GA3.
Supplemental Figure S3. Promoter activity of LecRKA4.1 in response to ABA doses.
Supplemental Table S1. List of 70 genes in the network and their functional categories.
Supplementary Material
Acknowledgments
We are grateful to Dr. Bo Liu (University of California, Davis) for providing the mCherry version of RFP, with permission for use from Dr. Roger Y. Tsien (University of California, San Diego). We thank Dr. Matthew Geisler (Southern Illinois University) for his advice on the Arabidopsis protein-protein interaction database. We also thank the Arabidopsis Biological Resource Center at The Ohio State University for providing various T-DNA lines. We appreciate the constructive and helpful comments on the manuscript from the editor and the reviewers.
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 2004–35304–14911 to Z.-L.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhi-Liang Zheng (zhiliang.zheng@lehman.cuny.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andre S, Siebert HC, Nishiguchi M, Tazaki K, Gabius HJ (2005) Evidence for lectin activity of a plant receptor-like protein kinase by application of neoglycoproteins and bioinformatic algorithms. Biochim Biophys Acta 1725 222–232 [DOI] [PubMed] [Google Scholar]
- Barre A, Herve C, Lescure B, Rouge P (2002) Lectin receptor kinases in plants. Crit Rev Plant Sci 21 379–399 [Google Scholar]
- Bromberg KD, Ma'ayan A, Neves SR, Iyengar R (2008) Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science 320 903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46 794–804 [DOI] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T (2007) Network-based classification of breast cancer metastasis. Mol Syst Biol 3 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ (2002) GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA 99 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan H, Yang G, Zheng ZL (2007) A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol Biol 63 221–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI (2002) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5 26–32 [DOI] [PubMed] [Google Scholar]
- Gao P, Xin Z, Zheng ZL (2008) The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS ONE 3 e1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zeng Q, Guo J, Cheng J, Ellis BE, Chen JG (2007) Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. Plant J 52 1001–1013 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O'Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve C, Dabos P, Galaud JP, Rouge P, Lescure B (1996) Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J Mol Biol 258 778–788 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6 470–479 [DOI] [PubMed] [Google Scholar]
- Hong SW, Jon JH, Kwak JM, Nam HG (1997) Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol 113 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth CJ, Parkes KE, Snell CR, Mullineaux PM, Reynolds CA (2008) Criteria for confirming sequence periodicity identified by Fourier transform analysis: application to GCR2, a candidate plant GPCR? Biophys Chem 133 28–35 [DOI] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH (2001) Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev 15 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW (2007) Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 19 2500–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L (2007) A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315 1712–1716 [DOI] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ (2007) An Arabidopsis gene network based on the graphical Gaussian model. Genome Res 17 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hematy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50 605–614 [DOI] [PubMed] [Google Scholar]
- Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20 1431–1438 [DOI] [PubMed] [Google Scholar]
- Navarro-Gochicoa MT, Camut S, Timmers AC, Niebel A, Herve C, Boutet E, Bono JJ, Imberty A, Cullimore JV (2003) Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula: structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol 133 1893–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Razem FA, El-Kereamy A, Abrams SR, Hill RD (2006) The RNA-binding protein FCA is an abscisic acid receptor. Nature 439 290–294 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22 1567–1572 [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, et al (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443 823–826 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang S, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Patel A, Mathieu M, Kim SY, Xu D, Stacey G (2008) A lectin receptor-like kinase is required for pollen development in Arabidopsis. Plant Mol Biol 67 469–482 [DOI] [PubMed] [Google Scholar]
- Wang XF, Zhang DP (2008) Abscisic acid receptors: multiple signal-perception sites. Ann Bot (Lond) 101 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072 [DOI] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frey NF, Leung J (2008) An update on abscisic acid signaling in plants and more. Molecular Plant 1 198–217 [DOI] [PubMed] [Google Scholar]
- Xie Z, Ruas P, Shen QJ (2005) Regulatory networks of the phytohormone abscisic acid. Vitam Horm 72 235–269 [DOI] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL (2005) Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol 139 1350–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Yoshida S, Asami T, Kuchitsu K (2003) Visualization of abscisic acid-perception sites on the plasma membrane of stomatal guard cells. Plant J 35 129–139 [DOI] [PubMed] [Google Scholar]
- Yang G, Gao P, Zhang H, Huang S, Zheng ZL (2007) A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE 2 e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Xin Z (2008) Plasma membrane-associated signaling proteins in the abscisic acid response network. In A Hemantaranjan, ed, Advances in Plant Physiology, Vol 10. Scientific Publishers, Jodhpur, India, pp 17–29
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.