Abstract
Poaceae, one of the largest flowering plant families in angiosperms, evolved distinct inflorescence and flower morphology diverging from eudicots and other monocots. However, the mechanism underlying the specification of flower morphology in grasses remains unclear. Here we show that floral zygomorphy along the lemma-palea axis in rice (Oryza sativa) is partially or indirectly determined by the CYCLOIDEA (CYC)-like homolog RETARDED PALEA1 (REP1), which regulates palea identity and development. The REP1 gene is only expressed in palea primordium during early flower development, but during later floral stages is radially dispersed in stamens and the vascular bundles of the lemma and palea. The development of palea is significantly retarded in the rep1 mutant and its palea has five vascular bundles, which is similar to the vascular pattern of the wild-type lemma. Furthermore, ectopic expression of REP1 caused the asymmetrical overdifferentiation of the palea cells, altering their floral asymmetry. This work therefore extends the function of the TCP gene family members in defining the diversification of floral morphology in grasses and suggests that a common conserved mechanism controlling floral zygomorphy by CYC-like genes exists in both eudicots and the grasses.
Morphological innovations are critical for the diversification of animals and plants to adapt to new environments and floral bilateral symmetry is an evolutionary adaptation that facilitates out-crossing by attracting pollinators (Cubas, 2004). Angiosperm flowers typically grow in patterns of monosymmetry (zygomorphy, bilateral symmetry), polysymmetry (actinomorphy, radial symmetry), or left-right asymmetry with one, several, or no symmetric plane (Endress, 1999, 2001). Phylogenetic and systematic studies confirm that zygomorphic flowers have evolved several times from radially symmetric ancestors (Endress, 2001; Cubas, 2004). Investigations in Antirrhinum majus (Veronicaceae), a model plant with bilaterally symmetrical flowers, revealed that the development of zygomorphic flowers requires the activity of CYCLOIDEA (CYC) and DICHOTOMA (DICH; Luo et al., 1996, 1999; Schwarz-Sommer et al., 2003), whose function is mediated through specific interaction with the MYB genes RADIALIS and DIVARICATA (Galego and Almeida, 2002; Corley et al., 2005). The CYC and DICH genes are members of the TCP gene family, coined from the conserved basic helix-loop-helix TCP domain found in TEOSINTE BRANCHED1 (TB1) in maize (Zea mays), CYC in A. majus, and the proliferating cell factor DNA-binding proteins of rice (Oryza sativa; Cubas et al., 1999a). This family has only been identified in angiosperms and shown to be involved in specifying plant morphology. Phylogenetic analysis indicates that continuous expansion of the TCP genes from genomic duplication events may have occurred in both higher and lower plants (Navaud et al., 2007). Several studies also show that repeated and independent recruitment of CYC-like genes might be a conserved evolution pathway in controlling zygomorphic flower development within eudicots (Coen and Nugent, 1994; Cubas et al., 1999a, 1999b; Hileman et al., 2003; Cubas, 2004; Citerne et al., 2006; Feng et al., 2006; Wang et al., 2008). However, evidence of TCP genes controlling floral zygomorphy outside eudicots is lacking.
Poaceae (grasses) is one of the largest flowering plant families in angiosperms, with about 10,000 species and 700 genera, including many economically important crops such as rice, barley (Hordeum vulgare), and maize (Clayton and Renvoize, 1986; Clark et al., 1995; Linder and Rudall, 2005). Evolutionary changes in the organization and structure of inflorescence and flower resulted in their distinct morphology in grasses diverging from those of higher eudicots and even other monocots (Grass Phylogeny Working Group, 2001; Kellogg, 2001; Rudall et al., 2005; Zanis, 2007). The grass inflorescence contains a number of spikelets, and each spikelet has several florets subtended by a pair of glumes. Each grass floret typically consists of three types of organs (i.e. a pistil, one or two whorls of three stamens, and two to three lodicules subtended by an inner bract or prophyll, called the palea, and the outer bract, called the lemma; Rudall and Bateman, 2004). Recent phylogenetic, genetic, and bioinformatics investigations have shed light on the molecular basis regulating the development of the inflorescence and spikelet in grasses (Grass Phylogeny Working Group, 2001; Kellogg, 2001; Bommert et al., 2005; Rudall et al., 2005; Zanis, 2007). Palea and lemma are unique structures found only in the Poaceae, where they are responsible for protecting the florets and kernels from pathogen and insect attack besides supplying carbohydrates to developing seeds (Abebe et al., 2004). Thus the establishment of the lemma/palea morphology might play a pivotal biological role in grass. Based on genetics analysis, some researchers refer to the palea and lemma as sepals or prophylls (Blaser, 1944; Clifford, 1987). Observations from palea or lemma defective mutants, dp1 and dp2 in rice, Osmads1 in rice, and leafy lemma and calcaroides in barley, imply nonequivalence of lemma and palea in the grass floret (Iwata and Omura, 1971a, 1971b; Pozzi et al., 2000; Prasad et al., 2005). However, the origin and the mechanism of lemma and palea development, and the underlying molecular mechanism defining grass flower morphological diversification, have long been controversial and still remain unclear (Schmidt and Ambrose, 1998; Irish, 2000; Ferrario et al., 2004).
Phylogenetic analyses, along with morphological studies, indicate that zygomorphy has evolved several times in monocotyledons, and enhancing or retarding organs within specific whorls renders the monocot bilateral symmetry flower (Rudall and Bateman, 2004). However, the definition of rice floral symmetry pattern and determination of its underlying molecular regulation mechanism still remain undefined. Recent genetic study of the rice palealess mutant suggests that the palea might be composed of two fused perianth parts, which, along with the lemma, would represent a modified trimerous calyx (Luo et al., 2005; Zanis, 2007). Moreover, recent analysis on Iberis amara, a Brassicaceae plant with monosymmetric flower, and a natural mutant with peloric flower demonstrated that IaTCP1, an ortholog of CYC, governs the formation of the monsymmetric corolla through suppressing adaxial petal cell growth (Busch and Zachgo, 2007). Therefore, identification of mutants with retarded or enhanced rice flower organs might help us to understand the molecular mechanism controlling rice flower organ development and floral zygomorphy. In this study, we first propose that the rice flower has bilateral symmetry along the rice lemma to palea (Le/Pa) axis and have characterized two palea defective mutants, retarded palea1-1 (rep1-1) and its allele rep1-2, whose palea growth is strongly retarded. Intriguingly, the palea identity of rep1 appears to have characteristics of the lemma with five vascular tissues present, suggesting a partial conversion to lemma identity and that REP1 may control rice floral zygomorphy by determination of palea identity and palea cell differentiation. Further molecular studies indicated that REP1 encodes a TCP gene, a homolog of the CYC gene in rice. The REP1 gene is only expressed in palea primordium during early flower development, but, at later floral stages, is radially dispersed in stamens, the vascular bundles of lemma, and palea. This work therefore provides unique insight into floral asymmetrical development in grasses.
RESULTS
Characterization of Floral Asymmetry in Rice
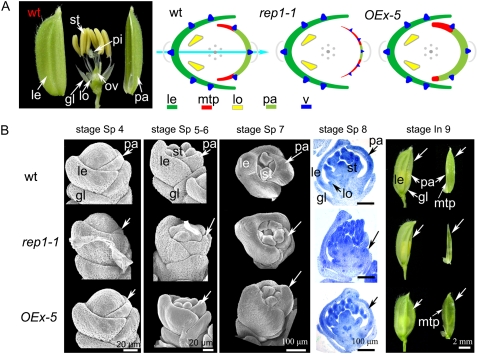
In this article, we propose that rice floret structure has bilateral symmetry along the Le/Pa axis, comprising lemma, palea, and lodicules in relation to their unique position and morphology (Fig. 1A). To be consistent with previous observations, we used the developmental stages defined by Ikeda et al. (2004) and the histological characterizations by Prasad et al. (2005). During floral development stages Sp 1 (formation of a pair of rudimentary glumes primordia) to Sp 7 (formation of carpel primordium), the floral primordia around the pistil primordium develops three obvious whorls: the outer whorl consisting of the lemma and palea, the second whorl consisting of two lodicules, and the third whorl consisting of six stamens (Fig. 1, A and B; Ikeda et al., 2004). Similar to the classic symmetry of floral organ arrangement in eudicots, rice stamens are symmetrically arranged in one whorl. However, the outer whorl and the second whorl in the rice floret are not radially symmetrical (i.e. the lemma and palea form interlocking structures, with the lemma larger than the palea; Fig. 1A; Ikeda et al., 2004), and the second whorl only consists of two lodicules biased to the lemma side (Fig. 1A; Supplemental Fig. S1G).
Figure 1.
Comparison of flower development in wild-type rice (9522 variety), rep1, and OEx-5 (REP1OEx line 5). A, Wild-type rice florets have six stamens, which grow actinomorphically in one whorl, surrounding one pistil in the center of the flower, whereas the outer and second whorl organs, including lodicules, lemma, and palea, grow zygomorphically along the lemma/lodicules to palea axis according to their organ identities and position. The flower diagrams show the relative position of different organs, with identities indicated by colors and shapes: lemma in dark green and curved; palea in pale-green; mtp in red and curved; lodicule in yellow and triangular; and vascular tissue as dark blue triangles. The floral zygomorphy along the Le/Pa axis, as defined by the different types of organs, is indicated by a sky-blue arrow. B, Compared to that of wild type, the rep1-1 palea development course is significantly retarded, while the mtp cells were overdifferentiated asymmetrically in OEx-5 (arrows). Abbreviations not defined in text: gl, empty glume; le, lemma; lo, lodicule; ov, ovary; pa, palea; pi, pistil; st, stamen; v, vascular bundle.
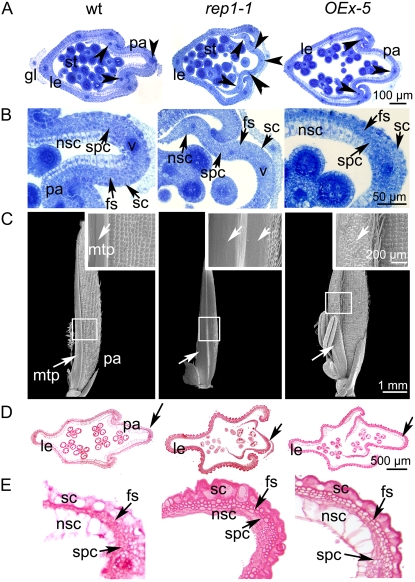
In addition to the asymmetric patterning of the rice floret, the outer whorl organs lemma and palea have distinctive cellular morphology (Prasad et al., 2005). Although the lemma and palea have very similar histology and both of them comprise silicified cells (sc), fibrous sclerenchyma (fs), spongy parenchymatous cells (spc), and nonsilicified cells (nsc; Fig. 2, A–C; Supplemental Fig. S1D), the palea has distinctive marginal tissue, which is absent in the lemma (Figs. 1A and 2C). This marginal tissue of the palea (mtp) differs from the rest of palea in a unique smooth epidermis, which lacks epicuticular, silicified thickening (Fig. 2C). Another striking difference between the lemma, palea, and lodicules is that they have different vascular tissue patterns (i.e. the lemma has five vascular bundles, whereas three exist in the palea; Fig. 2A, arrows; Ikeda et al., 2004; Prasad et al., 2005). While in lodicules, several vascular trachea elements are dispersed in the parenchyma cells (Supplemental Fig. S1G, arrowheads). As a result, coupled with the asymmetric position pattern, the rice floret presents bilateral symmetry along the Le/Pa axis according to the organ identities (Fig. 1A, sky-blue arrow), which is quite different from the floral symmetry defined in the core eudicots in relation to petal adaxial/abaxial position and morphology (Cubas, 2004).
Figure 2.
Observation of the cellular changes of wild type, rep1-1, and OEx-5. A, Flower transverse sections of wild-type, rep1-1, and OEx-5 transgenic plants at stage Sp 8. Note that five vascular bundles form in the rep1-1 palea (arrowheads) as opposed to three in wild type. B, Close-up of A, showing palea cell features. Note that four types of cell identity can be observed in the wild-type palea (i.e. sc, fs, spc, and nsc). Compared to that of wild type, the nsc cell size is smaller in the rep1-1 palea and bigger in the OEx-5 transgenic plant palea. C, SEM observation of the paleas of wild type, rep1-1, and OEx-5 at stage In 8. Insets are close-ups of C, marked in squares; note that the palea cell differentiation process is blocked in the rep1-1 mutant, while the mtp cells overdifferentiate in the OEx-5 transgenic plant. D, Flower transverse sections of wild-type, rep1-1, and OEx-5 transgenic plants around stage In 8. E, A close-up of D marked by arrows. Note that the palea cells seem smaller and underdifferentiated in rep1-1 compared to that of wild type, whereas cells differentiated fully in OEx-5. Abbreviations not defined in text: gl, empty glume; le, lemma; pa, palea; st, stamen; v, vascular bundle.
Identification and Isolation of the rep1 Mutants
To reveal the molecular mechanism regulating rice floret asymmetric development, we identified a mutant with a defect in palea morphology, rep1-1, from our rice mutant library, which is in the japonica subspecies 9522 background, created by using γ-ray radiation as previously reported (Li et al., 2006). The rep1-1 mutant was back-crossed with the wild-type plant 9522 variety three times and then used for genetic and phenotypic analysis. When the rep1-1 plants were pollinated with wild-type pollen, all F1 progeny displayed the wild-type phenotype, indicating that rep1-1 is a recessive mutant. F2 progeny tests yielded segregation of 387 normal and 133 mutant plants (χ2 [3:1] = 0.064; P > 0.5), indicating monofactorial recessive inheritance of the mutant characteristic.
Compared to the wild-type growth pattern, rep1-1 showed no obvious difference during vegetative growth (Supplemental Fig. S2, A and B). At the heading and flowering stage (In 9), the morphology of the rachis, primary, and secondary inflorescence branch also displayed normal morphology in the rep1-1 mutants (Supplemental Fig. S2, C–E), and the rep1-1 spikelet showed normal development of rudimentary and empty glumes, lemma, two lodicules, six stamens, and one pistil as seen in wild-type plants from stage Sp 4 to Sp 8 (Fig. 1B). The main defect of the rep1-1 floret was only observed at the palea position commencing at stage Sp 4 with the formation of palea primordium, as shown in Figure 1B. Compared to that of the wild-type plants, the rep1-1 palea primordium seemed smaller and the developmental process was dramatically delayed (Fig. 1B). By the stage In 9, the rep1-1 palea differed significantly from wild type, appearing undifferentiated and pale (Fig. 1B). This was further supported by longitudinal sections that showed abnormal development of the rep1-1 palea during the formation of lodicule and stamen primordia (stages Sp 5 and Sp 6; Supplemental Fig. S2, G and H). Another mutant with a palea defect, designated rep1-2, was obtained as a natural variant of ZF86 (indica; Supplemental Fig. S3A) and an allelism test indicated that rep1-2 was an allele of rep1-1. These morphological studies demonstrated that the palea development of rep1-2 is significantly retarded and that the rep1-2 mutant exhibits an identical phenotype to that of rep1-1 (Fig. 2, A, B, and D; Supplemental Fig. S3), despite the different genetic backgrounds.
The defect in the rep1 florets therefore appears to disrupt zygomorphic floral symmetry by suppressing palea development, although the growth pattern of both the stamens and lodicules in the rep1 mutant was not obviously affected (Figs. 1, A and B, and 2D; Supplemental Fig. S1, G and H). Genetic and molecular studies on the rep1 mutants therefore provide a valuable resource to help elucidate the molecular mechanism controlling palea development and the establishment of rice floral asymmetry along the Le/Pa axis.
Cloning and Annotation of the REP1 Gene
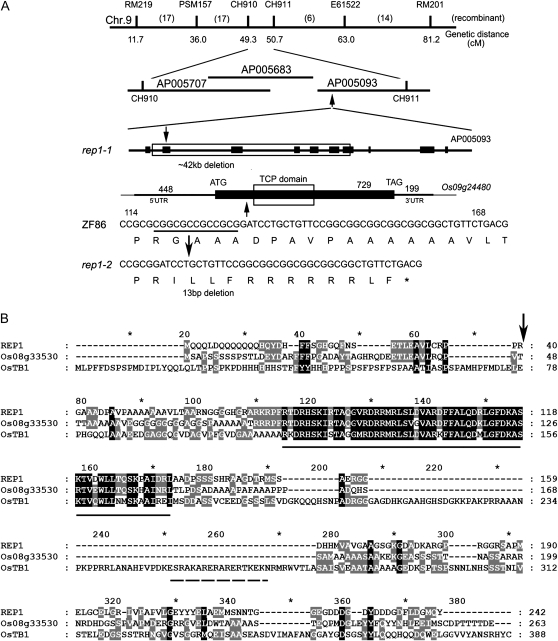
To identify the REP1 gene, we used a map-based cloning approach and localized the REP1 locus between two insertion-deletion (InDel) markers on chromosome 9, CH910 and CH911, which covered bacterial artificial chromosome (BAC) AP005707, AP005683, and AP005093 (Fig. 3A). By sequencing rep1-1 genomic DNA, we revealed that there was an approximate 42-kb deletion between the two InDel markers in the rep1-1 genome. Within this 42-kb region, there are six annotated genes in National Center for Biotechnology Information (NCBI) and The Institute for Genomic Research (TIGR; Fig. 3A), and we speculated that one of these, Os09g24480, encoding a predicted TCP domain protein, was the candidate gene related to the rep1-1 mutant. In agreement with this, a 13-bp deletion was also found in Os09g24480 of the rep1-2 genome (Fig. 3A), causing a frame shift and premature translational termination (Fig. 3, A and B, arrows). Furthermore, the Os09g24480 was confirmed to be REP1 by a functional complementation experiment using the DNA fragment carrying the wild-type genomic fragment, including a 2,968-bp promoter and a 726-bp gene sequence (REP1com; see “Materials and Methods” for detailed information), which was able to rescue the retarded palea phenotype of the rep1-1 plants (Supplemental Fig. S2F).
Figure 3.
Map-based cloning and analysis of the REP1 gene. A, Location of REP1 locus on rice chromosome 9, showing gene annotation of rice AP005093 BAC clone in the TIGR Web site and the deletion information for the rep1 mutants. Note that six genes were deleted in the rep1-1 mutant. Furthermore, a 13-bp deletion was observed within Os09g24480 in the rep1-2 mutant, which caused premature termination of REP1 translation. Arrow shows the mutation site. B, Alignment of protein sequences of REP1, Os08g33530, and OsTB1 in the CYC/TB1 group. TCP domain is underlined with a straight line and R domain is underlined with a dashed line. Arrow indicates deletion position in the rep1-2 mutant. Note that both the REP1 and Os08g33530 proteins lack the R domain.
To further identify the coding sequences (CDS) for functional annotation of the REP1 gene, RACE-PCR was performed with the GeneRacer kit (Invitrogen) according to the user's manual. To our surprise, CDS of the REP1 gene does not match the sequence in the TIGR database in which the CDS length of LOC_Os09g24480 is predicted to be 777 bp, consisting of two exons (exon 1 runs from 1–715 bp and exon 2 from 1,518–1,579 bp of the genomic sequence). However, our RACE-PCR results show that the REP1 cDNA is 1,376 bp in length and does not contain any introns. The actual REP1 CDS runs from 1 to 729 bp with a 448-bp 5′-untranslated region (UTR) and a 199-bp 3′-UTR (Fig. 3; accession no. EU702407 [REP1]). The new 3′-terminal sequence was further confirmed using 3′-terminal specific sequences as primers for reverse transcription (RT)-PCR analysis (Fig. 4A).
Figure 4.
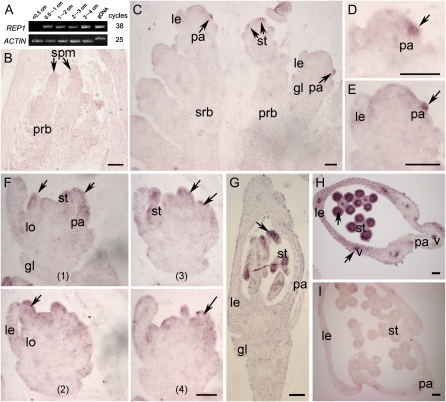
Expression pattern of the REP1 gene. A, Expression pattern of the REP1 gene detected by RT-PCR. B to G, Longitudinal and transverse sections through the main apex of wild-type flowers were hybridized with the REP1 antisense RNA probe labeled with digoxigenin-UTP. The transcript-specific hybridization signal is visualized in red. B, First and secondary rachis branch primordia emergence stage In 1 to In 4. C, Spikelet meristems around stage Sp 3 to Sp 5. D and E, Close-up of C, showing initiation of palea primordium around stage Sp 4. F, Serial sections showing lodicule and stamen primordia formation around stages Sp 5 and Sp 6. G, Longitudinal section of wild-type floret around stage Sp 8. H, Transverse section of wild-type flower during stage Sp 7 to Sp 8. I, Transverse section of wild-type flower with control REP1 sense RNA probes during stage Sp 7 to Sp 8. Abbreviations not defined in text: gDNA, genomic DNA; gl, empty glume; le, lemma; lo, lodicule; pa, palea; prb, primary rachis branches; spm, spikelet meristem; srb, secondary rachis branches; st, stamen; v, vascular bundle. Scale bar = 10 μm (B and C); 50 μm (D–I).
The REP1 protein was predicted to be a TCP family transcription factor in the NCBI and TIGR Web site, and our bioinformatics analysis and previous reports indicate that REP1 can be grouped into the CYC/TB1 subfamily with higher similarity to Os08g33530/OsTCP15, OsTB1, CYC, and DICH (Supplemental Fig. S4; Feng et al., 2006; Navaud et al., 2007; Yao et al., 2007). Therefore, functional analysis on REP1 will provide new insight toward elucidating whether there are TCP members defining the diversification of floral morphology in grasses.
Expression Pattern of REP1
As we described above, the rep1 mutations suppress palea development and disturb the establishment of the Le/Pa axis, but have minimal effect on rice vegetative growth and other flower organ development. To test the relationship between REP1 expression pattern and the rep1 phenotype, we first detected REP1 expression by RT-PCR using total RNA extracted from vegetative and reproductive organs. No REP1 transcript was detected in the vegetative organs such as root, stem, and leaf (Supplemental Fig. S5A) or in the inflorescence primordia from stage In 1 to In 4, when the inflorescence length was <0.5 cm (Fig. 4A). However, we observed the REP1 transcript at a low level in inflorescences of 0.5 to 4 cm in length when floral organs differentiate rapidly around stage In 7 (Fig. 4A). Interestingly, the REP1 transcript was detectable mainly in the stamens from the premeiosis stage to mature pollen stage (Supplemental Fig. S5B). In addition, the deletion in the rep1-2 mutant also appeared to produce a null allele of the REP1 gene because no REP1 signal could be detected in the rep1-2 inflorescence (Supplemental Fig. S5C). This could also explain why rep1-1 and rep1-2 display identical palea defects.
To precisely localize the expression pattern of REP1 in rice flowers, RNA in situ hybridization was performed on sections of wild-type inflorescences containing various stages of developing florets. Consistent with RT-RCR analysis, no obvious REP1 signal could be detected when the first and secondary rachis branch primordia emerged from stage In 1 to In 4 (Fig. 4B). The earliest expression of REP1 was observed asymmetrically in the palea primordia of the apical flowers on the primary and secondary branches at stage Sp 4, no signal was seen in the lodicules and lemma positions at this stage (Fig. 4, C–E). After the initiation of stamen primordia at stage Sp 6, the REP1 signal was clearly detected in stamens, palea, and lemma, but the signal was dispersed (Fig. 4F). Around stage Sp 8 during the development of pistil and stamen, the REP1 signal appeared stronger, radially in stamens and vascular bundles of lemma and palea (Fig. 4, G and H). Only background levels of signal were observed with the REP1 sense probe (Fig. 4I). The distinctive expression pattern of REP1 of initial asymmetrical expression and subsequent radial expression may therefore be critical in controlling rice palea development and floral morphology.
REP1 Determines Palea Primordium Identity and Enhances Cell Differentiation
To understand the role of REP1 on both palea development and the establishment of floral morphology at the cellular level in rice, the effects of REP1 on cell proliferation and differentiation during lemma and palea development were examined histologically. Compared to the cellular pattern of the wild-type floret described above, the rep1-1 lemma had no significant alternation and developed sc, fs, spc, and nsc (Supplemental Fig. S1, D and E); however, the rep1-1 palea cells seemed much smaller and less differentiated, with no clearly differentiated sc, fs, spc, and nsc (Fig. 2, A, B, D, and E). In addition, from the transverse sections we observed, five smaller vascular bundles in the rep1-1 palea (Fig. 2A, arrow), as seen in the lemma, suggesting that there may have been a partial developmental transformation from palea to lemma, occurs in the rep1-1 mutant (a similar observation was also seen in the rep1-2 mutant; see Supplemental Fig. S3B).
In the rep1-1 palea, even though the cell number was observed to be very close to that of wild type at stage Sp 8 (the innermost nsc no. was about 112 ± 2.5 in wild type and 110 ± 1.6 in rep1-1; n = 6), its cell size was obviously smaller than that of wild type (Fig. 2, A and B) and during later development, around stage In 8, the rep1 palea cells still remained less differentiated (Fig. 2, D and E; Supplemental Fig. S3, C and D). This result implies that loss of function of REP1 may dramatically affect the palea cell growth and expansion, especially in the epicuticula, the innermost cell layer, and vascular tissues (Fig. 2).
The alteration of palea cell morphology in the rep1 mutant was also observed by scanning electron microscopy (SEM) at stage In 8 (Fig. 2). Unlike that of wild type, the rep1-1 palea did not have the distinctive marginal tissue structure and most of the rep1-1 epicuticula grew smoothly without obvious silicified cells, imitating mtp-like cell growth patterns. Only a small part of epidermis differentiated to sc (Fig. 2C) and eventually the rep1-1 palea appeared bilaterally asymmetrical (Fig. 2, A, C, and D). These results also suggested that epicuticular thickening may be a necessary process for establishing palea identity.
Ectopic Expression of REP1 Enhances Palea Cell Differentiation and Disturbs Palea Internal Symmetry
To further test the cellular function of REP1, we constitutively expressed REP1 in wild-type rice under the control of the 35S promoter. In total, 63 transformants with near identical phenotypes were obtained, and one line, OEx-5 (REP1OEx line 5), was selected for phenotypic analysis; ectopic expression of REP1∷GFP signal obviously can be detected in this line in the lemma and palea organs (Supplemental Fig. S6C). In the complementation test, the REP1∷GFP fused construct with REP1 promoter was able to rescue the defects of rep1-1, suggesting that REP1∷GFP has biological function in rice. The vegetative growth of the OEx-5 appeared normal as wild type and the lemmas, lodicules, and inner whorls of the OEx-5 also had no visible differences compared with wild type (Supplemental Fig. S6). However, the development of palea of OEx-5 seemed abnormal (Supplemental Fig. S6B) and its mtp on one side of the palea displayed sc features (Fig. 2, C and D), and the palea cell size also appeared larger than that of wild type, especially in the mtp region (Fig. 2, B and E). Therefore, this result suggests that REP1 is indeed involved in enhancing palea cell differentiation and expansion. And, based on the fact that the OEx-5 palea showed asymmetrical differentiation of the mtp (Fig. 2C), we speculate that there may be an internal (IN) symmetry regulation pathway controlling palea bilateral development. Consistent with this hypothesis, recent work on isolation of the symmetric petals1 mutant in pea indicated that IN asymmetry for pea petal development was controlled by SYP1 (Wang et al., 2008).
DISCUSSION
The floral architecture in angiosperms falls into two major types, one with zygomorphic symmetry, such as in Antirrhinum, and the other with radial symmetry, as in Arabidopsis (Arabidopsis thaliana), petunia (Petunia hybrida), or tomato (Solanum lycopersicum; Coen and Nugent, 1994). Much progress has been made in understanding the genetic control of flower asymmetry in A. majus and other eudicots. Nevertheless, it is still of great interest to elucidate the genetic control of this developmental process in other plants, such as the grasses that have evolved unique flower and inflorescence structures.
REP1 Plays an Important Role in Establishing Palea Identity
Emerging work is beginning to shed some light on the genes that regulate the development of spikelet organs (for review, see Zanis, 2007); however, to date, there are very few examples of molecular characterization of palea development. In this study, we present REP1, which has a specific role in the regulation of palea development. In the rep1 mutant, palea growth is delayed and reduced, and cell differentiation, particularly in sc and nsc, is strongly affected (Fig. 2B). Furthermore, the rep1 palea has no obviously marginal tissue structure between the mtp and other parts of palea, with the rep1-1 epicuticula growing smoothly without obvious sc. In contrast, the mtp cells of the REP1 ectopic expression lines differentiated to sc-like cells (Fig. 2C). Asymmetrical expression of REP1 was seen in the palea primordium during early flower development and we propose that REP1 plays an important role in specifying rice palea development and, in turn, bilateral symmetry, while other aspects of floral development are unaffected.
Plant organogenesis mainly compromises cell proliferation, expansion, and differentiation (Li et al., 2005). TCP-like genes are thought to act at the cellular level to specify plant morphology by regulating cell proliferation and differentiation (Gaudin et al., 2000; Li et al., 2005; Busch and Zachgo, 2007; Broholm et al., 2008). For instance, it has been reported that CYC represses the expression of cyclin D3b in the dorsal stamen of A. majus (Gaudin et al., 2000). In I. amara, IaTCP1 is expressed strongly in small cells of the adaxial petals, whereas an S-phase cell cycle marker gene, IaH4, showed low level expression (Busch and Zachgo, 2007), implying that IaTCP1 controls petal development through suppressing cell division. Furthermore, correlating with its specific expression pattern, CIN, a class II TCP gene, controls cell differentiation in petals directly or indirectly through effects on cell proliferation (Nath et al., 2003; Crawford et al., 2004). The observation of the smaller palea cell size in the rep1 mutant implies that REP1 may have a similar role to other TCP proteins in regulating the cell expansion and differentiation. Whether this observation excludes a role for REP1 in regulating the cell division, as in other reported TCP proteins, requires further experiments with cell proliferation markers. Currently, the rice MADS-box gene OsMADS1 is the only cloned gene implicated in lemma/palea development (Jeon et al., 2000; Prasad et al., 2001), which controls epidermal cell fate and proliferation, and internal cell differentiation during lemma development, while regulating internal cell layer differentiation in the palea (Prasad et al., 2005). It will therefore be interesting to test the relationship between OsMADS1 and REP1 during palea development in rice.
Because rice has the ability to self-pollinate in the exquisite flower structure enclosed by outer whorl organs, we hypothesize that the palea mtp is an essential tissue for locking the lemma and palea, and the establishment of the rice Le/Pa axis is possibly a crucial adaptation favoring seed propagation. Abnormal development and growth of lemma, palea, or lodicules would alter zygomorphic morphology of the rice spikelet. From this point of view, REP1 is partly or indirectly involved in the establishment of asymmetry through its impact on the formation of palea. However, the fact that overexpression of REP1 failed to transform lemma into palea indicates that palea was not simply derived from lemma by recruiting REP1. In other words, there are additional factors involved in the development of palea and establishment of floral asymmetry. Further experiments to identify genes interacting with REP1 will facilitate our understanding about rice floral zygomorphy.
REP1 Is a TCP Family Member with a Distinct Expression Pattern
In this article, we isolated the rice REP1 gene using a map-based cloning strategy, which encodes a putative protein belonging to a plant-specific TCP transcription factor family. These TCP family members have previously only been identified in angiosperms and have been shown to be essential in specifying plant morphology (Navaud et al., 2007). Among them, TB1 in maize (Doebley et al., 1997), OsTB1 in rice (Takeda et al., 2003), and Arabidopsis AtTCP12/BRC1 and AtTCP18/BRC2 proteins (Aguilar-Martinez et al., 2007) are associated with controlling the growth and development of axillary shoots. Other TCP family members, including CYC and DICH in A. majus, LCYC in Linaria vulgaris, McCYC and McDICH in Mohavea confertiflora, LjCYC2 in Lotus japonicus, LegCYC1B in Sinningia speciosa, IaTCP1 in I. amara, and LOBED STANDARD1 and KEELED WINGS in pea (Pisum sativa), seem to be involved in an evolutionary conserved pathway for controlling zygomorphic floral development within eudicot species (Luo et al., 1996, 1999; Cubas et al., 1999b; Hileman et al., 2003; Citerne et al., 2006; Feng et al., 2006; Busch and Zachgo, 2007; Wang et al., 2008). Therefore, a key question is whether there are TCP genes that control the diversification of floral asymmetry in grasses. Through genetic and molecular studies, we have addressed this point by elucidating at least the partial role of REP1 in controlling rice flower bilateral symmetry by regulating palea development. Thus, we propose that REP1 is the putative ortholog of CYC in rice, which has evolved independently in the rice genome.
During rice floret development, the REP1 gene is asymmetrically expressed in palea primordium during early flower development, but during later floral stages is radially dispersed in stamens, the vascular bundles of lemma, and palea. This expression pattern shows significant divergence from other CYC-like genes in eudicots, which are expressed in the dorsal regions of floral meristems, where they affect petal and stamen growth and asymmetry (Luo et al., 1996, 1999; Cubas et al., 1999b; Hileman et al., 2003; Feng et al., 2006; Busch and Zachgo, 2007; Broholm et al., 2008; Wang et al., 2008). It has been speculated that evolution of CYC-like genes, coupled with changes in their expression patterns and interactions with the target genes, may control zygomorphic flower development in flowering plants (Cubas, 2004). For instance, IaTCP1, the putative CYC ortholog from I. amara, lacks asymmetric early expression, but displays very strong differential expression in the adaxial corolla at later floral stages, which correlates strongly with the unequal petal growth (Busch and Zachgo, 2007). Recent studies on CYC-like genes further demonstrated that GhCYC2, a CYC-like TCP transcription factor in Gerbera, controls the Gerbera complex inflorescence structure by a gradient of expression, from highly expressed in the marginal, bilaterally symmetrical ray flowers, to absent in the central disc flowers (Broholm et al., 2008). Therefore, the REP1 expression pattern may have been altered during the evolution of grass floret morphology such that palea identity and development is at least partially determined by asymmetrical REP1 expression in the palea organs during early developmental stages and the symmetrical expression of REP1 during later flower development. Furthermore, REP1 may also have additional biological functions in stamens, as well as the vascular bundles of lemma and palea. While no obvious developmental defects were observed in the stamen and lemma of the rep1 mutants, this may be a reflection of functional redundancy of other genes with REP1 due to gene duplication events of TCP domain family members, or alternatively may mean that REP1 has no critical role in stamen and lemma development. Our phylogenetic analysis revealed 22 putative TCP domain-containing members in the rice genome (Yao et al., 2007), supporting the hypothesis that lineage-specific expansions of the TCP family members happened during rice genome evolution (Navaud et al., 2007). Therefore, investigations on the REP1-like proteins in the relative grass family will help us to elucidate whether the conserved pathway exists in controlling other grass floral zygomorphy.
In summary, we describe rice floral zygomorphy along the Le/Pa axis in this study and, through characterization of the palea-specific regulator, REP1, provide new insights into the morphological establishment of grass flowers, and propose that this rice TCP domain gene might have evolved to regulate palea development and rice floral zygomorphy. Further molecular study on rice relatives, especially in basal grass with symmetric sepals, or asymmetric androecium, would facilitate understanding of the link between REP1-like protein expression and grass flower asymmetric structure.
MATERIALS AND METHODS
Plant Materials
Plant growth conditions were as previously described (Chu et al., 2006); the 9522 cultivar was used as a wild-type strain for observation of phenotypes and for RNA in situ analysis.
Microscopy and Image Processing
Inflorescences at successive developmental stages were collected from wild-type and the mutant plants over 3 to 5 d from August to September. The materials were fixed in formaldehyde-acetic acid, then sectioned and viewed with a Zeiss light microscope. SEM was performed as we described previously (Li et al., 2006). Fresh tissues of wild-type and mutant plants were photographed using a Nikon E995 digital camera (Nikon). Images were processed using Photoshop 7.0 software (Adobe).
Map-Based Cloning of REP1
The REP1 locus was first mapped to a region closely linked to a sequence-tagged site marker E61552 on the long arm of chromosome 9 by using 10 F2 plants of rep1-1 and Guang-lu-ai 4 (spp. indica). Then, by using 738 F2 plants, the REP1 locus was narrowed to a region between two InDel markers, CH910 and CH911. Mutations in rep1-1 and rep1-2 were determined by PCR amplification and sequence analysis.
Isolation of REP1 cDNA and Sequence Analysis
To obtain the full-length cDNA of REP1, total RNA was isolated using TRIzol reagent (Invitrogen) from wild-type inflorescence as previously described (Li et al., 2006) and RACE-PCR was performed with the GeneRacer kit (Invitrogen) as described in the user's manual. PCR amplification of the REP1 cDNA was carried out using 2 μL of RACE product and rice (Oryza sativa) REP1-specific primers 5′-RACE-A1 and 3′-RACE-S1 with Phusion hot start high-fidelity polymerase (Novagen). Internal reverse primer 5′-RACE-A2 was designed 20 nucleotides upstream to the putative start codon to amplify the 5′-UTR. Similarly, forward primer 3′-RACE-S2 was designed based on the sequence between 165 and 183 nucleotides upstream of the putative stop codon to determine the 3′-UTR. The ClustalX tool was used to do multiple sequence alignments and a phylogenetic tree was constructed with the aligned TCP protein sequences using the MEGA (version 3.0) software as we described before (Yao et al., 2007).
RT-PCR and in Situ Hybridization
Gene-specific primers REP1-RTF and REP1-RTR were used for RT-PCR analysis with 38 to 40 cycles following protocol as we describe previously (Li et al., 2006). REP1-specific probe was generated by inserting the REP1 cDNA fragment into pMD18-T (TaKaRa; gene-specific primers REP1IF and REP1IR), and this DNA fragment, digested with EcoRI and HindIII, was subcloned into pBluescript SK(+) and sequenced for confirmation. RNA hybridization and immunological detection of the hybridized probes were performed as previously described (Chu et al., 2006).
Molecular Cloning and Agrobacterium-Mediated Rice Transformation
Primers REP1-CF1 and REP1-CR1 were used for complementary clone construction. The 3,694-bp genomic DNA fragment including 2,968-bp promoter and 726-bp gene sequence was ligated into pDONR201 entry vector and then pGWB4 destination vector (Invitrogen; Os09g24480pro2968bp∷Os09g24480gDNA726 bp∷sGFP, named REP1com in this article) for rice transformation in rep1-1 background. For ectopic expression of REP1, a 726-bp gene cDNA was amplified with primers REP1-AF and REP1-CR1, and this REP1 fragment was ligated into pDONR201 entry vector and pGWB5 vector (35S∷Os09g24480gDNA726 bp∷sGFP, named REP1OEx in this article) for transformation in 9522 cultivar background. For Agrobacterium-mediated rice transformation, constructs were transformed into Agrobacterium strain EHA105, and transformation of rice followed the protocol described by Nishimura (2006). For the complementary experiment, 15 transformants were obtained and line 2 (Com-2), showing similar phenotypes to the other transgenic plants, was used to do phenotype analysis. For the ectopic expression experiment, a total of 63 transformants were obtained, which showed similar phenotypes and line 5 (OEx-5) was used for phenotypic analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU702407.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Lemma and lodicule cellular phenotypes of wild-type, rep1-1, and OEx-5 (REP1OEx line 5) plants.
Supplemental Figure S2. Phenotypes of the rep1-1 mutants.
Supplemental Figure S3. Phenotypes of the rep1-2 mutants.
Supplemental Figure S4. Phylogenetic tree visualizing the orthology among REP1 and TCP proteins.
Supplemental Figure S5. Expression patterns of the REP1 gene by RT-PCR.
Supplemental Figure S6. Phenotypes of the OEx-5 transgenic plants.
Supplemental Text S1. Primers list.
Supplementary Material
Acknowledgments
We thank B. Han (Rice Genome Resource Center) for providing the BAC clone. We gratefully acknowledge M.J. Cheng and Z.J. Luo for mutation screening and generating F2 populations for mapping, C.X. Ying for assistance with in situ hybridization, X.S. Gao, J.Q. Li, and Z.P. Zhang for SEM analyses, L. Zhang for assistance with the illustrations, and Ning Jiang for helpful suggestions and critical reading of the manuscript.
This work was supported by the National Basic Research Program of China (grant no. 2009CB941500), National “863” High-Tech Project (grant no. 2006AA10A102), National Natural Science Foundation of China (grant nos. 30725022, 30600031, and 90717109), and Shanghai Leading Academic Discipline Project (grant no. B205).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Da-Bing Zhang (zhangdb@sjtu.edu.cn).
The online version of this article contains Web-only data.
References
- Abebe T, Skadsen RW, Kaeppler HF (2004) Cloning and identification of highly expressed genes in barley lemma and palea. Crop Sci 44 942–950 [Google Scholar]
- Aguilar-Martinez JA, Poza-Carrion C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H (1944) Studies in the morphology of the Cyperaceae. II. The prophyll. Am J Bot 31 53–64 [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46 69–78 [DOI] [PubMed] [Google Scholar]
- Broholm SK, Tahtiharju S, Laitinen RA, Albert VA, Teeri TH, Elomaa P (2008) A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA 105 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Zachgo S (2007) Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci USA 104 16714–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Qian Q, Liang WQ, Yin C, Tan H, Yao X, Yuan Z, Yang J, Huang H, Luo D, et al (2006) The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 142 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citerne HL, Pennington RT, Cronk QC (2006) An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci USA 103 12017–12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L, Zhang W, Wendel J (1995) A phylogeny of the Grass family (Poaceae) based on ndhF sequence data. Syst Bot 20 436–460 [Google Scholar]
- Clayton W, Renvoize S (1986) Genera Graminum. HMSO, London, pp 1–389
- Clifford H (1987) Spikelet and Floral Morphology: Grass Systematics and Evolution. Smithsonian Institution, Washington, DC, pp 21–30
- Coen ES, Nugent JM (1994) Evolution of flowers and inflorescences. Dev Suppl 107–116
- Corley SB, Carpenter R, Copsey L, Coen E (2005) Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc Natl Acad Sci USA 102 5068–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford BC, Nath U, Carpenter R, Coen ES (2004) CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P (2004) Floral zygomorphy, the recurring evolution of a successful trait. Bioessays 26 1175–1184 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999. a) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18 215–222 [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E (1999. b) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401 157–161 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Endress PK (1999) Symmetry in flowers: diversity and evolution. Int J Plant Sci 160 S3–S23 [DOI] [PubMed] [Google Scholar]
- Endress PK (2001) Evolution of floral symmetry. Curr Opin Plant Biol 4 86–91 [DOI] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario S, Immink RG, Angenent GC (2004) Conservation and diversity in flower land. Curr Opin Plant Biol 7 84–91 [DOI] [PubMed] [Google Scholar]
- Galego L, Almeida J (2002) Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev 16 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA, Coen E, Doonan JH (2000) The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol 122 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88 373–457 [Google Scholar]
- Hileman LC, Kramer EM, Baum DA (2003) Differential regulation of symmetry genes and the evolution of floral morphologies. Proc Natl Acad Sci USA 100 12814–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54 147–156 [Google Scholar]
- Irish VF (2000) Variations on a theme: flower development and evolution. Genome Biol. 1 reviews1015.1–reviews1015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata N, Omura T (1971. a) Linkage analysis by reciprocal translocation method in rice plants (Oryza sativa L.). I. Linkage groups corresponding to the chromosome 1, 2, 3 and 4. Jpn J Breed 21 19–28 [Google Scholar]
- Iwata N, Omura T (1971. b) Linkage analysis by reciprocal translocation method in rice plants (Oryza sativa L.). II. Linkage groups corresponding to the chromosome 5, 6, 8, 9, 10 and 11. Sci Bull Fac Agr Kyushu Univ 25 137–153 [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, et al (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al (2006) The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. Plant Cell 18 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder H, Rudall P (2005) Evolutionary history of Poales. Annu Rev Ecol Evol Systemat 36: 107–124
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383 794–799 [DOI] [PubMed] [Google Scholar]
- Luo Q, Zhou K, Zhao X, Zeng Q, Xia H, Zhai W, Xu J, Wu X, Yang H, Zhu L (2005) Identification and fine mapping of a mutant gene for palealess spikelet in rice. Planta 221 222–230 [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O, Dabos P, Carnus E, Tremousaygue D, Herve C (2007) TCP transcription factors predate the emergence of land plants. J Mol Evol 65 23–33 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Aichi I, Matsuoka M (2006) A protocol for Agrobacterium-mediated transformation in rice. Nat Protocols 1 2796–2802 [DOI] [PubMed] [Google Scholar]
- Pozzi C, Faccioli P, Terzi V, Stanca AM, Cerioli S, Castiglioni P, Fink R, Capone R, Muller KJ, Bossinger G, et al (2000) Genetics of mutations affecting the development of a barley floral bract. Genetics 154 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211 281–290 [DOI] [PubMed] [Google Scholar]
- Rudall PJ, Bateman RM (2004) Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytol 162 25–44 [Google Scholar]
- Rudall PJ, Stuppy W, Jennifer C, Kellogg EA, Briggs BG (2005) Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am J Bot 92 1432–1443 [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Ambrose BA (1998) The blooming of grass flower development. Curr Opin Plant Biol 1 60–67 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Davies B, Hudson A (2003) An everlasting pioneer: the story of Antirrhinum research. Nat Rev Genet 4 657–666 [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33 513–520 [DOI] [PubMed] [Google Scholar]
- Wang Z, Luo Y, Li X, Wang L, Xu S, Yang J, Weng L, Sato S, Tabata S, Ambrose M, et al (2008) Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc Natl Acad Sci USA 105 10414–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Ma H, Wang J, Zhang DB (2007) Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J Integr Plant Biol 49 885–897 [Google Scholar]
- Zanis MJ (2007) Grass spikelet genetics and duplicate gene comparisons. Int J Plant Sci 168 93–110 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.