Over the past several years, our understanding of plant domestication has advanced substantially at the gene and genome levels. This is due largely to the rapid accumulation of genomic resources that provided genome-wide markers for population and molecular genetic analyses of crops and their wild relatives. A number of recent reviews captured some general aspects of these advances (Doebley et al., 2006; Ross-Ibarra et al., 2007; Vaughan et al., 2007; Burger et al., 2008). The primary goal of this article is to update the recent progress made in the grass family (Poaceae).
Grass domestication had a unique importance in the history of human civilization. Cereal crops, domesticated from wild grasses thousands of years ago, have provided food security for the human society. Of the modern cereals, maize (Zea mays), rice (Oryza sativa), and wheat (Triticum spp.), domesticated in different regions of the world 7,000 to 10,000 years ago, are the top three food crops with much higher annual production than others. Barley (Hordeum vulgare), arguably the fourth important cereal crop used as human food, animal feed, and brewing grains, is also among the earliest domesticated cereals. Sorghum ranks next in the annual cereal production, followed by oat (Avena sativa), rye (Secale cereale), and millets (http://faostat.fao.org/).
The number of studies published on cereal domestication appears to be correlated with the agronomic importance of the crops, with especially a wealth of data generated for maize, rice, wheat, and barley. The amount of genomic resources that became available over the past several years also seemed to have an impact on publications. The completion of rice genome sequencing (International Rice Genome Sequencing Project, 2005) has greatly accelerated the progress of studying rice domestication, resulting in a number of recent reviews on this subject (Kovach et al., 2007; Sang and Ge, 2007a, 2007b; Sweeney and McCouch, 2007; Vaughan et al., 2008).
Of the four top cereals, maize represents a rather unique case. It differs from barley, rice, and wheat by having undergone much more drastic morphological modifications during domestication. Considerable effort undertaken to investigate maize domestication has yielded classic literatures on the molecular basis of morphological evolution, which was thoroughly reviewed recently (Doebley, 2004; Doebley et al., 2006). For these reasons I will focus on updating recent progress made in barley, rice, and wheat, in which the studies of phenotypic transitions that allowed effective harvesting, such as reduction in shattering and improvement of threshing, will be the main theme of the article.
RICE
One of the recent advances in understanding rice domestication was the finding that the two major types of rice cultivars, rice subspecies indica and subspecies japonica, had distinct genomic backgrounds. Molecular clock dating with various markers suggested that the genomes of indica and japonica rice came from wild populations that diverged 0.4 to 0.2 million years ago, considerably preceding the time of rice domestication (Ma and Bennetzen, 2004; Vitte et al., 2004; Zhu and Ge, 2005). This lends support to the hypothesis of independent domestication of indica and japonica rice. The geographic locations for the domestication of indica and japonica cultivars were traced to south of the Himalayans and southern China, respectively (Londo et al., 2006).
Another line of advances was achieved through genetic analyses of morphological and physiological changes from the wild progenitors, Oryza nivara and Oryza rufipogon, to the cultivated rice. The number and chromosomal locations of quantitative trait loci (QTL) underlying important domestication traits have been estimated. These included the reduction in grain shattering and seed dormancy, synchronization of seed maturation, reduction in tiller number, increase in tiller erectness, increase in panicle branches, and the number of spikelets per panicle, and reduction in hull and pericarp coloration and awn length (Xiong et al., 1999; Cai and Morishima, 2002; Thomson et al., 2003; Li et al., 2006a; Onishi et al., 2007a). Of these changes often known as the domestication syndrome of cereals (Harlan, 1992), reduction in grain shattering is critical to effective harvesting and has been viewed as the hallmark of cereal domestication.
A major QTL, sh4, responsible for the reduction of grain shattering from the wild progenitors to cultivated rice was recently cloned (Li et al., 2006b). The gene encodes a transcription factor with a Myb3 DNA-binding domain. The mutation selected by early farmers to develop nonshattering cultivars was a single nucleotide substitution leading to an amino acid substitution from Lys to Asn in the DNA-binding domain. The substitution of the neutral for the positively charged amino acid, which weakened but did not knock out the gene function, caused the incomplete development and partial function of the abscission zone where a mature grain detaches from the pedicle. This disables the natural detachment of grains necessary for seed dispersal in the wild species, but allows manual separation of grains and pedicles at the abscission zone in cultivars. The fine tuning of the gene function is critical for maximizing the harvest efficiency because it prevents excessive grain loss during the first stage of harvest when tillers with mature panicles are cut and gathered. Equally important is the ability to subsequently remove and recover grains from the straws using simple threshing techniques such as beating against a wood tub.
Comparative sequence analyses further indicated that sh4 had a single origin and are now fixed in all rice cultivars (Li et al., 2006b; Lin et al., 2007; Onishi et al., 2007b). This finding was, however, somewhat surprising given the strong phylogenetic evidence for the independent domestications of indica and japonica rice. Two theoretical models were proposed to reconcile these apparently conflicting genetic and phylogenetic data (Sang and Ge, 2007a, 2007b).
The snowballing model considers that the earliest domesticated rice had fixed a set of critical domestication alleles such as sh4, which then spread into the populations of the wild progenitors, O. nivara and O. rufipogon, through introgression. One or both of the modern cultivars, indica and japonica, were derived from the hybrids that maintained the original set of domestication alleles and captured genomic background of wild populations best adapted to the local conditions. The core of the domestication alleles fixed in the founding cultivar thus acted to facilitate cultivar diversification as it rolled through the wild gene pool.
The combination model assumes that early rice cultivars were domesticated independently from wild populations with distinct genomic backgrounds at different locations. They initially fixed different sets of domestication alleles to make cultivation feasible and worthwhile. Subsequent crosses among these early cultivars followed by artificial selection for the best domestication alleles, including sh4, drove the fixation of the same set of alleles of different origins in nearly all modern cultivars.
Perhaps not in the strict sense, the snowballing model is more consistent with the conventional view of a single domestication, whereas the combination model leans toward the scenario of multiple domestications. In either case, introgression coupled with both artificial and natural selections played an important role in rice domestication. It served as an effective means to increase the genetic diversity of cultivars especially following the initial domestication bottleneck, and to produce cultivars adaptive to various climatic conditions in different geographic regions.
Other domestication-related genes cloned so far are not fixed in all rice cultivars. qSH1 is a homeobox gene responsible for the further reduction of grain shattering in some of the temperate japonica cultivars (Konishi et al., 2006). The causal mutation was a nucleotide substitution in the regulatory element located approximately 12 kb upstream of the coding region of qSH1, which substantially changed the level and pattern of gene expression and disrupted the development of the abscission zone between grains and pedicles.
For japonica cultivars with both qSH1 and sh4, the two genes contribute similar magnitude of phenotypic effect (Xiong et al., 1999; Cai and Morishima, 2002; Onishi et al., 2007a). The reasons why they have such different frequencies in rice cultivars can be speculated as follows. First, qSH1 was selected during the domestication of japonica rice, but was subsequently selected against and eliminated from most of the japonica cultivars after sh4 was introduced from cultivars of different origins, such as indica. The combination of qSH1 and sh4 could have made threshing too difficult and laborious at the time. Second, qSH1 was derived in the japonica cultivars that already had sh4 to further reduce shattering when the requirement for stronger threshing force was no longer a problem.
Another gene, Rc, encoding a bHLH protein that presumably regulates anthocyanin biosynthesis in the seed coat, was involved in the origin of white-seed cultivars from the red-seed wild progenitors (Sweeney et al., 2006). Two independent mutations in exon 6 of the gene could each be responsible for the loss of red pigment in the pericarps. A 14-bp deletion was found in nearly 98% of white rice and a nucleotide substitution resulting in a premature stop codon was found in the rest of white rice. Phylogenetic analyses indicated that the predominant 14-bp deletion originated in the japonica cultivar and spread into the indica cultivar through introgression (Sweeney et al., 2007). Although the reasons why white rice was favored over red rice thousands of years ago was not entirely clear, the selection for white seeds apparently was strong enough to spread the 14-bp deletion into the vast majority of modern cultivars.
BARLEY
Studies of barley domestication have progressed rapidly over the past few years (Pourkheirandish and Komatsuda, 2007). The phylogenetic relationships and population genetic structures of barley and its wild progenitor Hordeum spontaneum were analyzed using various types of molecular markers. Genes controlling key domestication traits were fine mapped or cloned. Similar to rice, the findings were brought on questions of whether barley was domesticated once or multiple times and to what extent introgression has influenced the domestication process.
Molecular phylogenetic analysis initially based on AFLP markers supported the hypothesis of a single origin of cultivated barley in the Fertile Crescent, and traced the location of barley domestication to the Israel-Jordan area (Badr et al., 2000). The conclusion, however, was soon called into question as the subsequent phylogenetic studies using different types of molecular markers favored the alternative hypotheses of multiple independent domestications. The analysis of chloroplast microsatellites suggested that there were additional centers of domestication west of the Fertile Crescent, including Ethiopia and western Mediterranean (Molina-Cano et al., 2005). A population genetic study based on sequences of seven nuclear loci also argued for the multiple origins of barley, and suggested that there were two centers of domestication, one in the Fertile Crescent and the other 1,500 to 3,000 km farther east (Morrell and Clegg, 2007). It further indicated that wild barley from the Fertile Crescent served as the primary genome donor for the modern cultivars in Europe and North America, while the wild populations from the eastern center contributed most of the genetic diversity of Asian cultivars.
Despite a growing body of evidence supporting multiple origins of barley, the phylogenetic analysis alone could not rule out the possibility that barley was domesticated once in the Fertile Crescent and spread to other regions of the world where it hybridized with the wild species (Badr et al., 2000; Kilian et al., 2006b). Crosses between the early cultivars and wild populations followed by artificial selection for domestication traits and natural selection for better adaptation to local climates could have given rise to cultivars with highly divergent genomic backgrounds. This scenario is similar to the snowballing model of rice domestication.
Genetic analyses of domestication traits have recently yielded new insights into the process of barley domestication. The essential domestication transitions that have been subjected to extensive genetic studies in barley include the reduction in grain shattering (or acquisition of nonbrittle rachis), the separation of seeds from hulls (or appearance of naked seed), and the change from two- to six-rowed ears.
On barley ears, each spike serving as a seed dispersal unit consists of three spikelets, of which the two lateral ones are reduced with only awns left to assist the dispersal of the fully developed central spikelet in the wild species. This trait did not change during the initial domestication of the two-rowed barley. In the more advanced cultivar, six-rowed barley, the two lateral spikelets become fully developed so that the number of rows of grains is tripled. The gene, Vrs1, that controls the development of the lateral spikelets has been cloned (Komatsuda et al., 2007). In the two-rowed barley, Vrs1 encodes an HD-ZIP-containing transcription factor that is expressed specifically in the lateral-spikelet primordia and suppresses the development of the lateral rows. The loss-of-function mutation of Vrs1 allowed the further development of the lateral rows and gave rise to six-rowed barley. Three different loss-of-function mutations have been identified in the six-rowed barley, indicating the multiple independent origins of the trait (Komatsuda et al., 2007).
In wild barley, hulls are firmly adherent to the seed coats and fully protect seeds from biotic and abiotic stresses (Taketa et al., 2008). This trait is retained in a portion of barley cultivars known as covered barley, which makes it the only modern cereal crop with inseparable hulls and seeds. Covered barley is primarily used as animal feed or brewing grains. For the purpose of brewing, hulls serve as a filtration medium in the separation of fermentable extract during malt processing. The appearance of naked barley in which seeds are easily released from the hulls was a result of selection for direct human consumption.
Unlike rice where grains are recovered at first from straws through threshing and seeds are subsequently separated from hulls during the milling process, seeds of free-threshing barley and wheat can be directly removed from hulls that remain on the straws. This saved an intermediate step of removing grains from panicles, but required additional mutations that allowed easy release of seeds from hulls. The gene, Nud, targeted by this selection was recently cloned in barley (Taketa et al., 2008). In the covered barley, the gene encodes an ethylene response factor that regulates lipid biosynthesis in the seed coat, which produces adhesive lipid between seed coats and the hulls. A 17-kb deletion in the chromosome region containing Nud was identified in the naked barley. The comparison of flanking sequences indicated that the deletion mutation was selected only once, suggesting that the trait of naked seeds had a single origin.
Brittle rachis of barley, equivalent to grain shattering in rice, was found to be controlled primarily by two tightly linked loci, Btr1 and Btr2. The homozygous recessive genotype at one of the loci, btr1btr1/Btr2Btr2 or Btr1Btr1/btr2btr2, confers the nonbrittle phenotype. Cultivars from the western parts of the world have predominantly the btr1btr1/Btr2Btr2 genotype, while most of eastern cultivars have the Btr1Btr1/btr2btr2 genotype. Although neither locus has been cloned, phylogenetic analysis of DNA sequences tightly linked to these loci showed that eastern and western cultivars formed their own groups, indicating the independent origins of nonbrittle rachis from the eastern and western regions (Azhanguvel and Komatsuda, 2007). This corroborates the recent phylogenetic studies with multiple genes in supporting the independent domestications of barley. The double homozygous recessive genotype btr1btr1/btr2btr2, however, has not been found in any barley cultivars, probably due to the tight linkage of the two loci (Komatsuda et al., 2004).
Of the three traits discussed above, nonbrittle rachis appeared in the earliest barley cultivars approximately 10,000 years ago and is now fixed in all cultivars. Six-rowed ears and naked seeds appeared more than a millennium later, and are partially fixed in the modern cultivars. Six-rowed ears arose independently several times from various loss-of-function mutations of Vrs1. Naked seeds controlled by nud originated only once and is currently found in all two-rowed and six-rowed barley grown for direct human consumption (Taketa et al., 2008). Clearly, introgression between cultivars of either the same or different origins, coupled with strong artificial selection for free threshing, has allowed the spreading of nud.
WHEAT
Unlike in rice and barley where domestication occurred at the diploid level, the evolution of polyploidy genomes played an important role in wheat domestication (Dubcovsky and Dvorak, 2007). The diploid einkorn wheat (Triticum monococcum) and tetraploid emmer wheat (Triticum dicoccum) were domesticated independently in the Fertile Crescent approximately 10,000 years ago and served as important ancient crops (Salamini et al., 2002). Molecular phylogenetic studies revealed that einkorn wheat was domesticated from Triticum boeoticum, a wild diploid species with the assigned genome type of AA (Heun et al., 1997). Emmer wheat was domesticated from a wild tetraploid species, Triticum dicoccoides, with the AABB genome type that derived from the hybridization between an AA-genome species, Triticum urartu, and a close relative of the SS-genome species, Aegilops speltoides (Özkan et al., 2002; Kilian et al., 2006a). Hexaploid bread or common wheat, now the most widely grown modern cultivar, was derived from the hybridization between a tetraploid cultivar with the AABB genome and a wild diploid species, Aegilops tauschii, with the assigned genome type of DD (for phylogenetic evidence, see Huang et al., 2002; Petersen et al., 2006). Thus, bread wheat has the genome type of AABBDD.
Similar to rice and barley, selection for cultivars with nonshattering grains or nonbrittle rachis was a critical early step of wheat domestication. For tetraploid wheat of the AABB genome, nonbrittle rachis was controlled by recessive alleles at two loci, Br2 and Br3, located in the homologous regions of group 3 chromosomes, 3A and 3B, respectively (Watanabe et al., 2002). Thus, they are potentially the orthologous loci between the AA and BB genomes from the diploid parents. For hexaploid bread wheat, there was an additional brittle rachis locus, Br1, also mapped to the orthologous location of group 3 chromosome, 3D, of the DD genome (Nalam et al., 2006; Watanabe et al., 2006). Furthermore, comparative mapping showed that this chromosomal region of wheat might be orthologous to that of barley containing two tightly linked loci for brittle rachis, Btr1 and Btr2 (Nalam et al., 2006; Pourkheirandish and Komatsuda, 2007; but see Li and Gill, 2006). The region, however, is not orthologous to either of those harboring the rice shattering genes sh4 and qSH1.
Soon after the domestication of wheat with nonbrittle rachis, the free-threshing trait appeared in polyploid wheat. This led to the development of the two most commonly grown modern cultivars, the hexaploid bread wheat of the AABBDD genome and the tetraploid hard or durum wheat (Triticum durum), a descendent of the AABB-genome emmer wheat. The free-threshing condition was achieved through the appearance of softened and easily separable hulls that were tenacious and tightly enclosing in the wild species as well as in primitive cultivars such as einkorn and emmer wheat. Hulls of the free-threshing cultivars could open easily to release seeds under moderate mechanical force such as beating or grinding during harvest.
Genetic analysis between durum wheat and the wild progenitor of emmer wheat, T. dicoccoides, detected four QTL responsible for the origin of the free-threshing character (Simonetti et al., 1999). Of the four QTL, two with large effect, each accounting for approximately 25% of phenotypic variation, were mapped to the chromosome locations where two major free-threshing loci were previously identified. These were Tg on the short arm of chromosome 2B (group 2 chromosome of the BB genome) and Q on the long arm of chromosome 5A (group 5 chromosome of the AA genome). The free-threshing alleles, tg and Q, were partially recessive and partially dominant, respectively, at these two loci. Here the genotype of the free-threshing tetraploid wheat with the AABB genome is designated as tgtg2BQQ5A.
When tetraploid wheat with such a genotype was crossed with A. tauschii, the DD-genome parent of the bread wheat, the synthetic hexaploid was not free threshing due to the presence of the dominant allele at the Tg locus of the DD genome (Kerber and Rowland, 1974). Thus, an additional recessive mutation at the Tg locus of the DD genome was required for the development of the free-threshing condition in the hexaploid bread wheat, which gave rise to the genotype of tgtg2Btgtg2DQQ5A (Jantasuriyarat et al., 2004; Nalam et al., 2007).
The recent molecular cloning of Q showed that it was a gene belonging to the AP2 family of transcription factors (Faris et al., 2003; Simons et al., 2006). The Q allele had a higher level of transcription than the wild-type allele, q, in spikes, leaves, and roots. The coding region of Q differs from that of q by an amino acid substitution, which was responsible for an increased abundance of homodimer formation of Q protein when tested in yeast. This mutation in the coding region, together with regulatory mutations potentially including a substitution at the microRNA-binding site (Chuck et al., 2007), led to the gain-of-function mutation of Q that confers the free-threshing phenotype. In addition to free threshing, Q had pleiotropic effect on several other domestication-related traits, such as plant height, rachis fragility, and spike shape and emergence time, resulting in tougher rachis and higher yield.
Comparative sequence analyses indicated that Q had a single origin in wheat (Simons et al., 2006). One hypothesis concerning the evolution of free-threshing wheat is that the free-threshing condition was developed in the AABB-genome wheat with the genotype of tgtg2BQQ5A, which then served as the tetraploid parent of hexaploid bread wheat. In this scenario, an additional recessive mutation at Tg of the DD genome was sufficient (Faris et al., 2006). This would be similar to the development of tough rachis in the hexaploid wheat, where a recessive allele of Br1 presumably orthologous to Br2 and Br3 of the tetraploid parent was selected following the allopolyploid formation, though it is unclear why tg was not found in the AA genome of the free-threshing polyploid wheat. Alternatively, the tetraploid parent was only partially free threshing, with the genotype of tgtg2Bqq at the time of hybridization. Q was selected in the hexaploid wheat at first and then spread into tetraploid wheat through introgression.
ONE GENE FOR ONE TRAIT
These recent findings raise a general question of how many genes and mutations were required for a critical domestication transition. In the cases where the causal mutations have been identified, a single mutation controls primarily nonshattering in all rice cultivars, free threshing or naked seeds in barley, and naked grains of maize (tga1; Wang et al., 2005). One gene with different mutations accounts for the origin of white seeds of rice and six-rowed ears of barley. In maize, tb1 that encodes a TCP-family transcription factor was primarily responsible for the reduction of lateral branches of the wild progenitor teosinte (Doebley et al., 1997). While the causal mutation(s) have been located in the intergenic region approximately 58 to 69 kb upstream of the tb1 coding region, it was not entirely clear whether a single mutation or multiple mutations of independent origins were involved in this domestication transition (Wang et al., 1999; Clark et al., 2004, 2006).
The next category of examples includes nonbrittle rachis in wheat and barley, where the major QTL have been narrowly located on chromosomes but not yet cloned. The QTL primarily responsible for nonbrittle rachis in wheat were mapped to the orthologous chromosomal positions between the different diploid genomes in the tetraploid and hexaploid wheat, implying that orthologous genes could have been the targets of selection. In barley, it was proposed based on genetic analyses that two tightly linked loci, btr1 and btr2, controlled nonbrittle rachis. However, the linkage has never been broken up in experimental crosses and the double-recessive genotype has never been found in nature. This seems to warn the possibility that btr1 and btr2 could be different mutations of the same gene. One can entertain the hypothesis that the allele with the same mutation cannot form homodimers and confer the nonbrittle phenotype, but alleles with different mutations are able to form heterodimers that fulfill the same function of the protein as in the wild progenitor.
It is intriguing that in six (sh4, Rc, Nud, Vsr1, tb1, and tga1) of the seven cases where an important domestication transition has been characterized at the gene level, one gene controls primarily one trait. Of six cases where causal mutations are precisely identified, three (sh4, Nud, and tga1) fit the scenario of one mutation for one trait. Even for nonbrittle rachis of barley and wheat where the major QTL have not been cloned, there is a good chance that each transition was controlled by the same or orthologous genes.
One exception to the scenario of one gene primarily for one trait was free threshing of wheat, which was controlled by two major loci of similar magnitude of phenotypic effect. Interestingly, however, the two loci are quite different in other aspects of their effects. Tg is dominant over Q for the hulled or non-free-threshing condition, and Q improves many other domestication-related traits in addition to free threshing. It is thus plausible that tg, conferring a reasonable degree of free threshing, was selected at first during wheat domestication and Q was then selected and quickly driven to fixation in both tetraploid and hexaploid cultivars because it was such a beneficial allele with dominant effect on numerous traits.
Thus, it is reasonable to conclude based on these findings that in most cases a single gene played a pivotal role in moving the population over the trajectory of a key domestication transition. QTL of smaller effect or modifier genes played relatively minor but necessary roles in the optimization of a domestication trait. This observation seems remarkable given that there are multiple regulators in a developmental pathway that could be potentially targeted by domestication selection.
Equally intriguing is the question of how distant among the evolutionary lineages the targets of domestication selection are conserved. Comparative mapping previously suggested that the conservation might be widespread for key domestication traits in the grass family (Paterson et al., 1995). The recent findings from fine mapping and cloning of domestication QTL of cereals offer an opportunity to examine this hypothesis at a finer genomic scale. The evidence so far seems to indicate that the conservation has a more stringent phylogenetic constraint than previously thought. The best example is the comparison of genetic basis of the nonshattering trait among rice, barley, and wheat. While the QTL responsible for nonshattering were mapped to the orthologous chromosomal regions in barley and wheat that belong to the same tribe of the subfamily Pooideae, these regions are not orthologous to those containing shattering genes, sh4 and qSH1, in rice that belongs to a closely related but different subfamily Ehrhartoideae (Kellogg, 2001). Even for barley and wheat, the free-threshing trait is controlled by different genes, nud for barley and Q for wheat.
Why is the phenomenon of one gene primarily controlling a domestication transition common in individual crops but not between crops especially as they become more distantly related to each other? First of all, it has been well established that genes involved in the important domestication transitions are regulatory genes whose mutations can generate substantial phenotypic modifications that serve as suitable targets for strong artificial selection in the key steps of crop evolution (Doebley and Lukens, 1998; Doebley et al., 2006). Taking one step further from this theory, one can imagine that knocking out or drastically altering the function of a regulatory gene without severely negative pleiotropic effect may not be an easy genetic modification to engineer. It is thus not surprising to find that repeated selection experiments performed independently by early farmers ended up at the same target genes. Examples include the origins of white rice and six-rowed barley, where the same phenotypic modification in each case was accomplished through independent selection for the loss-of-function mutations of the same gene. This demonstrates that there was the limited number of suitable targets in the developmental pathway for the artificial selection that aimed at developing the most desirable phenotype for cultivation. Quite conceivably, this constrain is relaxed as the developmental pathways become increasingly divergent between more distantly related crop species.
Furthermore, strong artificial selection coupled with introgression could drive the fixation of the most beneficial gene for a key domestication transition of a crop (Fig. 1). Even for cultivars with different origins and partial reproductive isolation, gene flow could spread domestication genes across the entire gene pool of a crop and provide opportunities for replacing less favorable genes with the most beneficial ones, especially when there was negative epistasis between them. This eventually led to the fixation of a gene of large phenotypic effect for a domestication trait, such as sh4 for nonshattering rice and nud for naked barley. This mechanism, however, does not work between crops that are reproductively isolated.
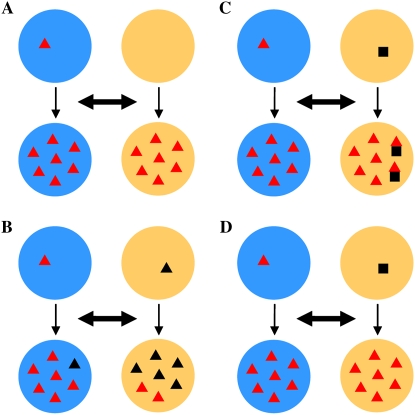
Figure 1.
Domestication genes and domestication processes. Circles with blue and orange backgrounds represent independently domesticated cultivars of a crop. Double arrowheads indicate gene flow between them during the process of domestication. A, Red triangle in the top blue circle represents the single origin of a domestication allele. The beneficial allele is subsequently fixed in this cultivar and spreads and also becomes fixed in the other cultivar. Examples for this scenario include nud in barley and sh4 in rice if qSH1 was derived in some japonica cultivars after the fixation of sh4 (see text for explanation). B, Black triangle in the top orange circle represents the independent origin of another allele at the same locus as the red triangle. Two alleles (red and black) have the similar function and coexist in the modern cultivars. Examples for this scenario include the loss-of-function alleles of Vrs1 in barley and Sc in rice. C, Alleles at two different loci, represented by a red triangle and a black square, are selected during the independent domestications. The red allele is more beneficial than the black allele and there is negative epistasis between them. Artificial selection fixes the red gene in both cultivars with only a small portion of the orange cultivar still retaining the black gene. The example may include the coexistence of sh4 and qSH1 in some japonica cultivars in the case that qSH1 originated in japonica before the introgression of sh4. D, In the case of two domestication loci being initially selected, if the red gene is more beneficial than the black gene and/or there is a negative epistasis between them, the black gene is eliminated from the cultivars and the red gene ends up being the only domestication gene fixed in the modern cultivars. This is a more generalized case for scenario A assuming that there had been another gene targeted by artificial selection at the initial stage of domesticating the orange cultivar.
From the literature reviewed above, one gene primarily responsible for a critical domestication transition emerges as a common phenomenon. This could be attributed to strong artificial selection on the limited number of suitable genes for phenotypic changes that essentially turned a wild species into a crop. Introgression among cultivars of even different origins greatly facilitated the spread of the most beneficial genes. Although further generalization would have to rely on cloning additional domestication genes from a larger number of crops, what has been learned so far seems to imply that a drastic phenotypic modification for improving an existing crop or domesticating a new crop can potentially be achieved by selecting or inducing mutations of one gene. However, it may not be easy to come across or engineer such a mutation because the number of target genes can be small.
Acknowledgments
I thank Toby Kellogg, Claire Lorts, and anonymous reviewers for helpful comments on the previous version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tao Sang (sang@msu.edu).
References
- Azhanguvel P, Komatsuda T (2007) A phylogenetic analysis based on nucleotide sequence of a marker linked to the brittle rachis locus indicates a diphyletic origin of barley. Ann Bot (Lond) 100 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr A, Müller K, Schäfer-Pregl R, Rabey HEL, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17 499–510 [DOI] [PubMed] [Google Scholar]
- Burger JC, Champan MA, Burke JM (2008) Molecular insights into the evolution of crop plants. Am J Bot 95 113–122 [DOI] [PubMed] [Google Scholar]
- Cai HW, Morishima H (2002) QTL clusters reflect character associations in wild and cultivated rice. Theor Appl Genet 104 1217–1228 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S (2007) The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spkelet1. Nat Genet 39 1517–1521 [DOI] [PubMed] [Google Scholar]
- Clark RM, Linton E, Messing J, Doebley JF (2004) Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc Natl Acad Sci USA 101 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley JF (2006) A distant upstream enhance at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38 594–597 [DOI] [PubMed] [Google Scholar]
- Doebley JF (2004) The genetics of maize evolution. Annu Rev Genet 38 37–59 [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127 1309–1321 [DOI] [PubMed] [Google Scholar]
- Doebley JF, Lukens L (1998) Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley JF, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386 485–488 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploidy wheat under domestication. Science 316 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris JD, Feller P, Brooks SA, Gill BS (2003) A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris JD, Simons KJ, Zhang Z, Gill BS (2006) The wheat super domestication gene Q. Wheat Inf Serv 100 129–148 [Google Scholar]
- Harlan JR (1992) Crops and Man, Ed 2. American Society of Agronomy and Crop Science Society of America, Madison, WI
- Heun M, Schafer-Pregl R, Klawan D, Castagna R, Accerbi M, Borhi B, Salamini F (1997) Site of einkorn wheat domestication identified by DNA fingerprinting. Science 278 1312–1314 [Google Scholar]
- Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploidy wheat. Proc Natl Acad Sci USA 99 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Jantasuriyarat C, Vales MI, Watson CJW, Riera-Lizarazu O (2004) Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness. Theor Appl Genet 108 261–273 [DOI] [PubMed] [Google Scholar]
- Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber RE, Rowland GG (1974) Origin of the threshing character in hexaploid wheat. Can J Genet Cytol 16 145–154 [Google Scholar]
- Kilian B, Özkan H, Deusch O, Effgen S, Brandolini A, Kohl J, Martin W, Salamini F (2006. a) Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol 24 217–227 [DOI] [PubMed] [Google Scholar]
- Kilian B, Özkan H, Kohl J, von Haeseler A, Barale F, Deusch O, Brandolini A, Yucel C, Martin W, Salamini F (2006. b) Haplotype structure at seven barley genes: relevance to gene pool bottlenecks, phylogeny of ear type and site of barley domestication. Mol Genet Genomics 276 230–241 [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Maxim P, Senthil N, Mano Y (2004) High-density AFLP map of nonbrittle rachis 1 (btr1) and (btr2) genes in barley (Hordeum vulgare L.). Theor Appl Genet 109 986–995 [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicher T, Tagiri A, et al (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312 1392–1396 [DOI] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23 578–587 [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T (2006. a) Global dissemination of a single mutation conferring white pericarp in rice: genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170 185–194 [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T (2006. b) Rice domestication by reducing shattering. Science 311 1936–1939 [DOI] [PubMed] [Google Scholar]
- Li W, Gill BS (2006) Multiple genetic pathways for seed shattering in the grasses. Funct Integr Genomics 6 300–309 [DOI] [PubMed] [Google Scholar]
- Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, Zhang W, Wang X, Xie D, Sun C (2007) Origin of seed shattering in rice (Oryza sativa L.). Planta 226 11–20 [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Cano JL, Russell JR, Moralejo MA, Escacena JL, Arias G, Powell W (2005) Chloroplast DNA microsatellite analysis supports a polyphyletic origin for barley. Theor Appl Genet 110 613–619 [DOI] [PubMed] [Google Scholar]
- Morrell PL, Clegg MT (2007) Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA 104 3289–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam VJ, Vales MI, Watson CJW, Johnson EB, Riera-Lizarazu O (2007) Map-based analysis of genetic loci on chromosome 2D that attract glume tenacity and threshability, components of the free-threshing habit in common wheat (Triticum aestivum L). Theor Appl Genet 116 135–145 [DOI] [PubMed] [Google Scholar]
- Nalam VJ, Vales MI, Watson CJW, Kianian SF, Riera-Lizarazu O (2006) Map-based analysis of genes affecting the brittle rachis character in tetraploid wheat (Triticum turgidum L.). Theor Appl Genet 112 373–381 [DOI] [PubMed] [Google Scholar]
- Onishi K, Horiuchi Y, Ishigoh-Oka N, Takagi K, Ichikawa N, Maruoka M, Sano Y (2007. a) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed Sci 57 7–16 [Google Scholar]
- Onishi K, Takagi K, Kontani M, Tanaka T, Sano Y (2007. b) Different patterns of genealogical relationships found in the two major QTL causing reduction of seed shattering during rice domestication. Genome 50 757–766 [DOI] [PubMed] [Google Scholar]
- Özkan H, Brandolina A, Schäfer-Pregl R, Salamini F (2002) AFLP analysis of a collection of tetraploid wheat indicates the of emmer and hard wheat domestication in southeast Turkey. Mol Biol Evol 19 1797–1801 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lin YR, Li Z, Schertz KF, Doebley JF, Pinson SRM, Liu SC, Stansel JW, Irvine JE (1995) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269 1714–1718 [DOI] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Yde M, Berthelsen K (2006) Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol Phylogenet Evol 39 70–82 [DOI] [PubMed] [Google Scholar]
- Pourkheirandish M, Komatsuda T (2007) The importance of barley genetics and domestication in a global perspective. Ann Bot (Lond) 100 999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA 104 8641–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamini F, Ozkan H, Brandolini A, Schafer-Pregl R, Marin W (2002) Genetics and geography of wild cereal domestication in the Near East. Nat Rev Genet 3 420–441 [DOI] [PubMed] [Google Scholar]
- Sang T, Ge S (2007. a) The puzzle of rice domestication. J Integr Plant Biol 49 760–768 [Google Scholar]
- Sang T, Ge S (2007. b) Genetics and phylogenetics of rice domestication. Curr Opin Genet Dev 17 533–538 [DOI] [PubMed] [Google Scholar]
- Simonetti MC, Bellomo MP, Laghetti G, Perrino P, Simeone R, Blanco A (1999) Quantitative trait loci influencing free-threshing habit in tetraploid wheats. Genet Resour Crop Evol 46 267–271 [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, McCouch SR (2007) The complex history of the domestication of rice. Ann Bot (Lond) 100 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR (2007) Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3 e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch SR (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, et al (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, Tai TH, McClung AM, Lai X, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and Oryza sativa cultivar jefferson. Theor Appl Genet 107 479–493 [DOI] [PubMed] [Google Scholar]
- Vaughan DA, Balazs E, Heslop-Harrison JS (2007) From crop domestication to super-domestication. Ann Bot (Lond) 100 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA, Lu BR, Tomooka N (2008) The evolving story of rice evolution. Plant Sci 174 394–408 [Google Scholar]
- Vitte C, Ishii T, Lamy F, Brar D, Panaud O (2004) Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.). Mol Gen Genet 272 504–511 [DOI] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li BL, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF (2005) The origin of the naked grains of maize. Nature 436 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley JF (1999) The limits of selection during maize domestication. Nature 398 236–239 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Fujii Y, Kato N, Ban T, Martinek P (2006) Microsatellite mapping of the genes for brittle rachis on homoeologous group 3 chromosomes in tetraploid and hexaploid wheats. J Appl Genet 47 93–98 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Sugiyama K, Yamagishi Y, Sakata Y (2002) Comparative telosomic mapping of homoeologous genes for brittle rachis in tetraploid and hexaploid wheats. Hereditas 137 180–185 [Google Scholar]
- Xiong L, Liu K, Dai K, Xu C, Zhang Q (1999) Identification of genetic factors controlling domestication-related traits for rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor Appl Genet 98 243–251 [Google Scholar]
- Zhu Q, Ge S (2005) Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol 167 249–265 [DOI] [PubMed] [Google Scholar]