Abstract
In angiosperms, auxin phytohormones play a crucial regulatory role in fruit initiation. The expression of auxin biosynthesis genes in ovules and placenta results in uncoupling of tomato (Solanum lycopersicum) fruit development from fertilization with production of parthenocarpic fruits. We have identified two newly described genes, named Aucsia genes, which are differentially expressed in auxin-synthesis (DefH9-iaaM) parthenocarpic tomato flower buds. The two tomato Aucsia genes encode 53-amino-acid-long peptides. We show, by RNA interference-mediated gene suppression, that Aucsia genes are involved in both reproductive and vegetative plant development. Aucsia-silenced tomato plants exhibited auxin-related phenotypes such as parthenocarpic fruit development, leaf fusions, and reflexed leaves. Auxin-induced rhizogenesis in cotyledon explants and polar auxin transport in roots were reduced in Aucsia-silenced plants compared with wild-type plants. In addition, Aucsia-silenced plants showed an increased sensitivity to 1-naphthylphthalamic acid, an inhibitor of polar auxin transport. We further prove that total indole-3-acetic acid content was increased in preanthesis Aucsia-silenced flower buds. Thus, the data presented demonstrate that Aucsia genes encode a novel family of plant peptides that control fruit initiation and affect other auxin-related biological processes in tomato. Aucsia homologous genes are present in both chlorophytes and streptophytes, and the encoded peptides are distinguished by a 16-amino-acid-long (PYSGXSTLALVARXSA) AUCSIA motif, a lysine-rich carboxyl-terminal region, and a conserved tyrosine-based endocytic sorting motif.
In tomato (Solanum lycopersicum) and in many other angiosperms, the fruit originates from the ovary. At anthesis, the ovary is already formed; however, fruit set and growth usually take place only after pollination/fertilization. The current model of fruit set implies that ovary growth is blocked before pollination and that auxin is a key regulator of ovary growth derepression at fruit set (Goetz et al., 2007; Pandolfini et al., 2007). A large body of evidence supports the notion that also other phytohormones (e.g. gibberellin, cytokinin, brassinosteroid, ethylene, and abscisic acid) either affect and/or are involved in fruit initiation and/or development (Schwabe and Mills, 1981; Vriezen et al., 2008). Other factors, such as the influence of outer floral organs (Vivian-Smith et al., 2001) and environmental cues (e.g. temperature and light; George et al., 1984), are known to affect fruit initiation, and yet they are much less investigated.
Parthenocarpy (i.e. fruit set in the absence of pollination/fertilization) is currently obtained by treating tomato flowers with exogenous auxin or inhibitors of auxin transport or other phytohormones (Schwabe and Mills, 1981; Gillaspy et al., 1993; Serrani, 2007). By genetic manipulation, parthenocarpic fruit development has been conferred to tomato via alteration of auxin biology achieved by increasing auxin synthesis within the placenta/ovules (Ficcadenti et al., 1999), by increasing the auxin sensitivity in the fruit (Carmi et al., 2003), or by manipulating genes of the auxin or gibberellin signal transduction pathway (Wang et al., 2005; Goetz et al., 2006, 2007; Marti et al., 2007). For example, two members of the auxin signal transduction pathway, a tomato auxin/indole-3-acetic acid (AUX/IAA) gene, SlIAA9, and an Arabidopsis auxin response factor, AUXIN RESPONSE FACTOR8 (ARF8), have been shown to repress ovary growth before fertilization. Tomato fruit development can be uncoupled from fertilization also by silencing DELLA proteins, which are repressors of GA signaling (Marti et al., 2007). Experimental evidence indicates that GA-dependent degradation of DELLA proteins is stimulated by auxin (Fu and Harberd, 2003).
In angiosperms, auxin phytohormones control many different processes of plant growth and development (Woodward and Bartel, 2005; Leyser, 2006), their action being dependent on biosynthesis, transport, and sensitivity at the tissue and organ levels. Auxin (IAA) synthesis takes place mainly in apical meristems and young leaves (Woodward and Bartel, 2005), and IAA moves through the plant over both long and short distances (Woodward and Bartel, 2005; Leyser, 2006). The direction and flux of polar auxin transport are determined by the polarized localization of several transmembrane proteins, acting either as influx or efflux facilitators and/or transporters (Blakeslee et al., 2005; Leyser, 2006; Schulz and Kolukisaouglu, 2006; Yang et al., 2006; Blakeslee et al., 2007). Within the cell, changes in auxin movement and content are perceived by the SCF-TIR1 receptor complex (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007), triggering polyubiquitination and degradation of AUX/IAA transcriptional regulators (Woodward and Bartel, 2005; Leyser, 2006). AUX/IAAs are short-lived proteins that can form heterodimers with ARFs (Leyser, 2006). Degradation of AUX/IAA proteins usually derepresses the transcription of auxin-regulated genes (Woodward and Bartel, 2005; Leyser, 2006). Chemical and/or genetic alterations of polar auxin transport as well as mutations in genes of the auxin-signaling transduction pathway can lead to a variety of auxin-related developmental phenotypes (Leyser, 2006).
We show that a novel gene family, consisting of two genes encoding small peptides named AUCSIA (for auxin cum silencing action), regulates fruit initiation in tomato. Aucsia genes are ubiquitously expressed and yet preferentially expressed in flower buds before anthesis. We show that RNA interference (RNAi)-mediated suppression of Aucsia genes causes parthenocarpic fruit development and a 100-fold increase in total IAA content of preanthesis flower buds. Furthermore, Aucsia-silenced plants display other auxin-related phenotypes, such as alterations of leaf development, and exhibit reduced auxin-induced rhizogenesis, reduced polar auxin transport in roots, and increased sensitivity to 1-naphthylphthalamic acid (1-NPA), an inhibitor of polar auxin transport. Collectively, these data indicate that AUCSIA peptides play a role in auxin-regulated processes in tomato and, most likely, due to the high degree of sequence conservation, also in other angiosperms. The presence of Aucsia genes in both chlorophytes and streptophytes, the two major clades of green plants, indicates that Aucsia genes were present in green plants before the emergence of multicellular organisms and land colonization.
RESULTS
Tomato Aucsia Genes Are Highly Expressed in Flower Buds and Down-Regulated during Fruit Initiation
Comparative transcript analysis was carried out on preanthesis parthenocarpic and wild-type flower buds. Parthenocarpic lines used in this work have been obtained by genetically engineering tomato plants with either the DefH9-iaaM or the DefH9-RI-iaaM chimeric gene (DefH9-iaaM line 3 and DefH9-RI-iaaM line s5 described in Pandolfini et al., 2002). DefH9-iaaM and DefH9-RI-iaaM genes code for the same product (i.e. Trp monoxygenase) and are expressed under the control of the same promoter (from the DefH9 gene of Antirrhinum majus) but differ in the 5′ untranslated leader region. The two chimeric genes display in vitro different translational potential and in planta different levels of auxin (total IAA) in the flower buds: DefH9-RI-iaaM and DefH9-iaaM flower buds contain 10 and 50 times higher IAA than controls, respectively (Pandolfini et al., 2002). At preanthesis, DefH9-iaaM and DefH9-RI-iaaM flower buds are morphologically identical to wild-type buds, except for the ovaries, which are more enlarged than those of controls.
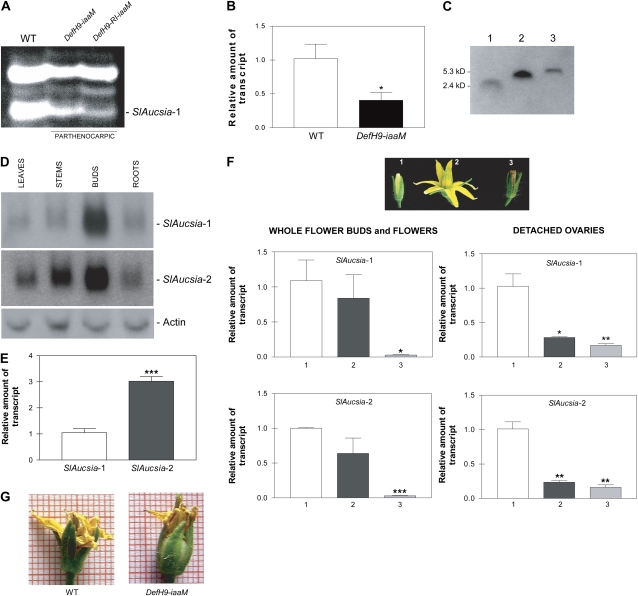
To identify differentially expressed genes, we performed a cDNA-amplified fragment length polymorphism (cDNA-AFLP) analysis using 32 different primer combinations (BstT/C + n – Mse + n, where n represents a selective nucleotide). Two hundred sixty-six cDNA-AFLP clones resulted in differentially expressed parthenocarpic flower buds (data not shown). One of these clones, a 129-bp-long cDNA fragment amplified with the BstCC-MseC primer combination (see cDNA sequence in Supplemental Fig. S1A), was identical to a portion of an EST (BG126668) and of a full-length cDNA (AK224828 and BW690880) of tomato. This gene, which we have named SlAucsia-1, was repressed in parthenocarpic flower buds (Fig. 1A). Quantitative reverse transcription (qRT)-PCR analysis showed that SlAucsia-1 mRNA was reduced by 60% in preanthesis parthenocarpic auxin-synthesis flower buds compared with wild-type buds (Fig. 1B).
Figure 1.
Expression of SlAucsia-1 and SlAucsia-2 genes in tomato. A, cDNA-AFLP analysis showing down-regulation of SlAucsia-1 in preanthesis flower buds from two parthenocarpic auxin-synthesis (iaaM) tomato lines (DefH9-iaaM line 3 and DefH9-RI-iaaM line s5; Pandolfini et al., 2002). B, qRT-PCR analysis of SlAucsia-1 mRNA steady-state level in preanthesis flower buds from wild-type (WT) and parthenocarpic auxin-synthesis (iaaM) tomato lines. The analysis was performed on four independent auxin-synthesis parthenocarpic lines. C, Identification of tomato AUCSIA-1 peptide using affinity-purified antibodies specific for the AUCSIA-1 21 C-terminal amino acids. Lane 1, Synthetic peptide consisting of the 21 C-terminal amino acids of the AUCSIA-1 peptide; lane 2, synthetic AUCSIA-1 peptide lacking the three N-terminal amino acids (i.e. MAP); lane 3, protein extract from tomato seedlings. D, Northern-blot analysis of SlAucsia-1 and SlAucsia-2 mRNA steady-state levels in different organs of tomato plants. E, SlAucsia-1 and SlAucsia-2 mRNA levels in preanthesis flower buds. The relative mRNA level of SlAucsia-2 was assessed by qRT-PCR compared with SlAucsia-1. F, Expression pattern of SlAucsia-1 and SlAucsia-2 mRNAs in different stages of tomato flower/fruit development. Top panel, Image 1, flower bud at 1 to 3 d before anthesis; image 2, open flower; image 3, flower at 4 to 5 d after anthesis. The left panels at bottom show relative mRNA levels of SlAucsia-1 and SlAucsia-2 assessed by qRT-PCR in open flowers and in flowers after fertilization compared with preanthesis flower buds. The right panels at bottom show qRT-PCR performed on ovaries collected at the same stages. G, Precocious ovary growth in a parthenocarpic (iaaM) auxin-synthesis tomato flower bud compared with a wild-type bud. Error bars in B, E, and F represent sd. * P < 0.05, ** P < 0.01, *** P < 0.001. [See online article for color version of this figure.]
Tomato has two Aucsia genes. SlAucsia-2 (AK224647) transcript is a homolog of SlAucsia-1: the sequences of SlAucsia-1 and SlAucsia-2 are 85% identical in their coding regions (Supplemental Fig. S1A). SlAucsia-1 and SlAucsia-2 mRNAs encode predicted peptides of 53 amino acids (Table I) with molecular masses of 5,596 and 5,662 D, respectively. Affinity-purified antibodies specific for the 21 C-terminal amino acids of tomato AUCSIA-1 detected a peptide with an apparent molecular mass of approximately 6 kD (Fig. 1C). SlAucsia-1 and SlAucsia-2 mRNAs were both expressed in stems, roots, and leaves and at a higher level in preanthesis flower buds (Fig. 1D). In all organs analyzed, SlAucsia-2 mRNA was more abundant than SlAucsia-1 mRNA (Fig. 1, D and E). For instance, in preanthesis flower buds (i.e. closed flower buds), the SlAucsia-2 mRNA level was three times higher than the SlAucsia-1 mRNA level (Fig. 1E). After pollination/fertilization (i.e. 4–5 d after anthesis), the levels of both SlAucsia-1 and SlAucsia-2 mRNAs decreased dramatically (97%) compared with their levels in preanthesis flower buds (Fig. 1F). Analysis of steady-state levels of SlAucsia-1 and SlAucsia-2 mRNAs in the ovaries collected from the same developmental stages (i.e. before anthesis, stage 1; open flowers, stage 2; after anthesis, stage 3; Fig. 1F) confirmed the data obtained from whole flower buds and showed that SlAucsia-1 and SlAucsia-2 mRNA levels were already decreased (approximately 75%) at the stage of open flower (Fig. 1F). These data suggest a possible involvement of Aucsia genes in controlling fruit initiation.
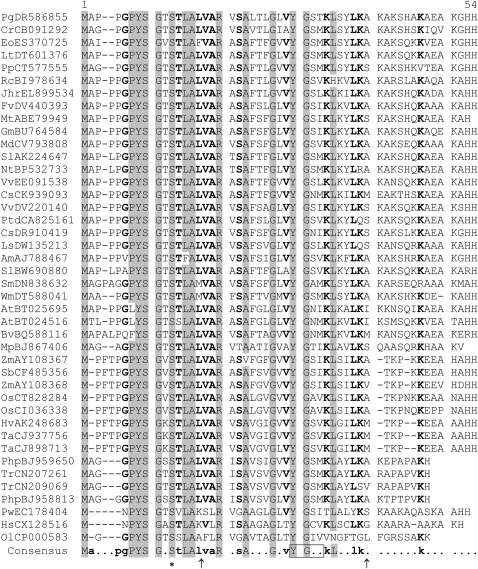
Table I.
Sequence comparison of putative AUCSIA peptides
Conserved residues are highlighted in gray (consensus level, 90%) and printed in bold (consensus level, 70%). Arrows indicate the positions of the two introns deduced from the analysis of the genomic sequences from Arabidopsis, Oryza sativa, Medicago truncatula, and Physcomitrella patens. The Ser residue candidate for alternate phosphorylation/O-GlcNAc glycosylation is indicated by an asterisk. The Tyr-based sorting motif (YXX[LMVIF]) is boxed. Pg, Picea glauca; Cr, Cycas rumphii; Eo, Elaeis oleifera; Lt, Liriodendron tulipifera; Pp, Pinus pinaster; Rc, Rosa chinensis; Jhr, Juglans hindsii × Juglans regia; Fv, Fragaria vesca; Mt, Medicago truncatula; Gm, Glycine max; Md, Malus × domestica; Sl, Solanum lycopersicum; Nt, Nicotiana tabacum; Vv, Vitis vinifera; Cs, Citrus sinensis; Ptd, Populus trichocarpa × Populus deltoides; Ls, Lactuca sativa; Am, Antirrhinum majus; Sm, Selaginella moellendorffii; Wm, Welwitschia mirabilis; At, Arabidopsis thaliana; Bv, Beta vulgaris; Mp, Marchantia polymorpha; Zm, Zea mays; Sb, Sorghum bicolor; Os, Oryza sativa; Hv, Hordeum vulgare; Ta, Triticum aestivum; Php, Physcomitrella patens; Tr, Tortula ruralis; Pw, Prototheca wickerhamii; Hs, Helicosporidium species; Ol, Ostreococcus lucimarinus. The alignment was created as described by Corpet (1988).
In normal fruit development, fertilization triggers the derepression of ovary growth, whereas DefH9-iaaM parthenocarpic plants show precocious ovary growth (Fig. 1G). On the basis of the down-regulation of SlAucsia-1 mRNA in preanthesis DefH9-iaaM and DefH9-RI-iaaM parthenocarpic flower buds and of the down-regulation of both SlAucsia-1 and SlAucsia-2 mRNAs early during fruit initiation in wild-type ovaries/flowers, we have been prompted to test whether silencing of Aucsia genes causes parthenocarpic fruit development.
Silencing of Tomato Aucsia Genes in Plants Transgenic for the rolC-hpAucsia Construct
In order to functionally characterize Aucsia genes, tomato plants were stably transformed with a hairpin (hp) construct driven by the rolC phloem-specific promoter to elicit RNAi of Aucsia transcripts. The hp construct driven by the rolC promoter has been used previously to confer systemic resistance to Plum pox virus without preventing local infections (Pandolfini et al., 2003). This observation indicates that RNAi elicited by the rolC hp construct is sufficiently robust to prevent the systemic spread of Plum pox virus via the phloem. The rolC-hpAucsia construct contains 200-nucleotide-long DNA fragments of the SlAucsia-1 gene placed in inverted orientation and separated by the spliceosomal intron of the rolA gene of Agrobacterium rhizogenes (Supplemental Fig. S1).
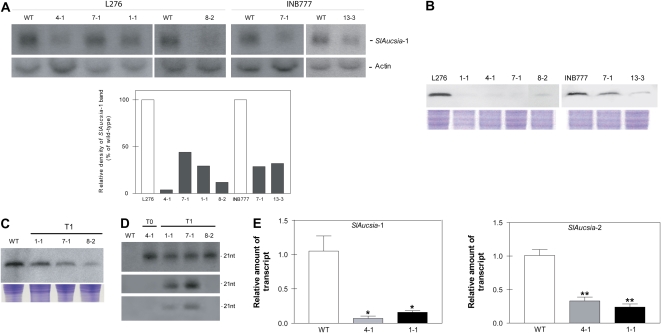
Young leaves from T0 tomato plants transgenic for the rolC-hpAucsia construct, raised in two different genetic backgrounds (L276 and INB777), were analyzed by northern blot. About 50% (13 of 25) of the transgenic plants showed reduced SlAucsia-1 mRNA steady-state levels in comparison with the wild type (data not shown). Six T0 Aucsia-silenced (Fig. 2A) independent lines (L276 1-1, 4-1, 7-1, and 8-2; INB777 7-1 and 13-3; Supplemental Fig. S2) were analyzed for AUCSIA peptide level. In all the T0 independent rolC-hpAucsia plants, AUCSIA peptide level was reduced in flower buds (Fig. 2B). In order to demonstrate that the silenced state is stably transmitted to the progeny, AUCSIA peptide level was evaluated in three T1 Aucsia-silenced transgenic lines (L276 1-1, 7-1, and 8-2). All T1 Aucsia-silenced plants showed reduced AUCSIA peptide levels (Fig. 2C).
Figure 2.
Molecular analysis of Aucsia-silenced plants. A (top), Northern blot analysis of SlAucsia-1 mRNA steady-state levels in leaves of six independent lines (belonging to two different genetic backgrounds, L276 and INB777) transgenic for the rolC-hpAucsia construct. A (bottom), Densitometric analysis performed on northern blot. The relative density of SlAucsia-1 bands was normalized against actin bands and reported as a percentage of the density of the corresponding wild-type (WT) bands. B, Western-blot analysis of protein extracted from flower buds of rolC-hpAucsia transgenic plants (T0) raised in two different genetic backgrounds (L276 and INB777). The four L276 transgenic plants analyzed are L276 1-1, 4-1, 7-1, and 8-2. The two INB777 transgenic plants analyzed are INB777 7-1 and 13- 3. C, Western-blot analysis of AUCSIA-1 peptide in three T1 independent L276 Aucsia-silenced lines: L276 1-1, 7-1, and 8-2. Top, AUCSIA-1 peptide analysis; bottom, Coomassie Brilliant Blue staining of total protein. D, Presence of siRNAs homologous to SlAucsia genes in leaves of T0 and T1 silenced lines (L276 genetic background). Top, siRNAs homologous to the SlAucsia-1 coding region; middle, siRNAs homologous to the 3′ UTR of SlAucsia-1 transcript; bottom, siRNAs homologous to the 3′ UTR of SlAucsia-2 transcript (see Supplemental Fig. S1A for a description of RNA probes). E, SlAucsia-1 and SlAucsia-2 steady-state mRNA levels in preanthesis flower buds from two Aucsia-silenced independent lines (L276 4-1 and 1-1). The relative mRNA levels were assessed by qRT-PCR in Aucsia-silenced plants compared with wild-type plants. Error bars in E represent sd. * P < 0.05, ** P < 0.01. [See online article for color version of this figure.]
The analysis of small interfering RNAs (siRNAs), the hallmark of RNAi, showed the presence of siRNAs homologous to SlAucsia genes in both T0 (L276 4-1) and T1 (L276 1-1, 7-1, and 8-2) plants using probes spanning the homologous coding region or the 3′ untranslated region (UTR) of either SlAucsia-1 or SlAucsia-2 mRNAs (Fig. 2D; Supplemental Fig. S1A). Thus, the rolC-hpAucsia construct elicited RNAi also against SlAucsia-2 transcript. This result is consistent with the high sequence homology of tomato Aucsia genes. Indeed, SlAucsia-1 and SlAucsia-2 transcripts are 85% identical over their coding regions, which include 16- to 20-nucleotide-long stretches of identical sequence. To provide a quantitative estimation of Aucsia gene silencing, qRT-PCR was performed on RNA extracted from flower buds of two lines (L276 4-1 and L276 1-1) that exhibited by northern-blot analysis a different degree of SlAucsia-1 silencing (Fig. 2A). The mRNA steady-state levels of SlAucsia-1 were about 90% and 80% reduced in L276 4-1 and L276 1-1, respectively, in comparison with wild-type flower buds (Fig. 2E). The mRNA steady-state levels of SlAucsia-2 were about 60% and 70% reduced in L276 4-1 and L276 1-1, respectively. Altogether, these data show that in rolC-hpAucsia transgenic tomato plants, both Aucsia genes are silenced and that the Aucsia-silenced state is transmitted to the progeny.
Aucsia-Silenced Tomato Plants Display Parthenocarpy
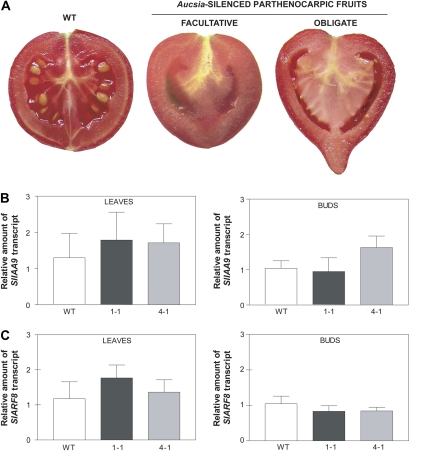
All six T0 Aucsia-silenced lines analyzed displayed parthenocarpic fruit development. Five of the six Aucsia-silenced lines showed facultative parthenocarpy; seedless fruits were generated from emasculated flowers (Fig. 3A; Table II), while pollinated flowers generated seeded fruits.
Figure 3.
Aucsia-silenced tomato plants are parthenocarpic. A, Wild-type (WT) pollinated fruit (left), seedless fruit obtained after flower emasculation of a facultative parthenocarpic Aucsia-silenced line (middle), and seedless fruit obtained from an obligate parthenocarpic Aucsia-silenced line (right). B and C, SlIAA9 (B) and SlARF8 (C) mRNA steady-state levels in leaves and in preanthesis flower buds from Aucsia-silenced L276 1-1 and 4-1 lines. The relative mRNA levels were assessed by qRT-PCR in Aucsia-silenced plants compared with wild-type plants. [See online article for color version of this figure.]
Table II.
Parthenocarpic fruit development in Aucsia-silenced plants
Number and percentage of fruits set per number of emasculated flowers and average weight of fruits obtained from selfed and emasculated flowers belonging to six independent T0 Aucsia-silenced lines obtained in two different genetic backgrounds (L276 and INB777). The weight values reported are means ± sd (wild type, n = 18–20 selfed fruits sampled from four plants; Aucsia silenced, n = 7–13 selfed fruits sampled from one to two plants). Statistically significant differences between fruit weights of Aucsia-silenced and wild-type plants were calculated using Student's t test. Significant differences are as follows: * P < 0.05, ** P < 0.01, *** P < 0.001.
| Line | Fruit Set
|
Average Weight
|
||
|---|---|---|---|---|
| No. of Fruit/No. of Emasculated Flowers | % | Emasculated | Selfed | |
| g | ||||
| L276 1-1 | 4/9 | 44 | 13.49 ± 0.85*** | 38.98 ± 2.70*** |
| L276 4-1a | 7/7 | 100 | 35.47 ± 12.03** | 38.43 ± 21.10* |
| L276 7-1 | 6/11 | 54 | 28.77 ± 4.25*** | 34.29 ± 2.04*** |
| L276 8-2 | 10/15 | 67 | 40.14 ± 2.93*** | 48.38 ± 3.13* |
| L276 wild type | 0/15 | 0 | 81.29 ± 4.39 | |
| INB777 7-1 | 20/25 | 80 | 18.27 ± 1.61*** | 48.72 ± 5.76** |
| INB777 13-3 | 14/20 | 70 | 18.47 ± 1.36*** | 61.04 ± 5.55* |
| INB777 wild type | 0/20 | 0 | 77.40 ± 4.24 | |
Obligate parthenocarpic line.
The facultative parthenocarpic fruits were similar in shape to wild-type pollinated fruits (Fig. 3A), and no evident reduction in the number of seeds per fruit was noted in fruits generated from nonemasculated flowers. One plant, L276 4-1, displayed obligate parthenocarpy; also, nonemasculated flowers produced seedless fruits (Fig. 3A). These seedless fruits were umbonate (Fig. 3A). This alteration has been described in tomato fruits as a consequence of excessive auxin treatments and is indicative of hyperauxiny (Pandolfini et al., 2002). The obligate parthenocarpic plant also did not produce any seeds when used as male or female parent in crosses with wild-type flowers. The percentage of fruit set of Aucsia-silenced emasculated flowers ranged from 44% to 100% among the six independent T0 transgenic lines analyzed, belonging to two different genetic backgrounds (L276 and INB777; Table II). The average weight of either emasculated or pollinated Aucsia-silenced fruit was lower than that of pollinated wild-type fruit (Table II). The parthenocarpic trait displayed by Aucsia-silenced tomato plants resembles that caused by altered auxin biology. Parthenocarpy has been obtained previously by altering auxin signaling via down-regulation of an AUX/IAA gene, SlIAA9 (Wang et al., 2005), and via ARF8 gene loss of function (Goetz et al., 2006). Steady-state levels of SlIAA9 mRNA and SlARF8 mRNA analyzed in leaves and in preanthesis flower buds of Aucsia-silenced plants were not altered compared with those of wild-type plants (Fig. 3, B and C). These results indicate that Aucsia gene function is not directly linked to SlIAA9 and SlARF8 mRNA levels.
Aucsia-Silenced Tomato Plants Display Auxin-Related Alterations of Leaf Development
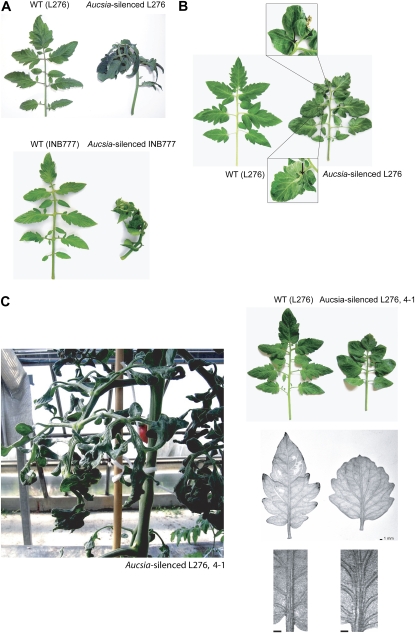
Tomato has nonpinnate, compound leaves consisting of a terminal leaflet, three pairs of major lateral leaflets, and, in between, a variable number of smaller leaflets (Fig. 4A). The leaves of all six Aucsia-silenced tomato lines analyzed were reflexed (Fig. 4A) and very often displayed leaflet fusions (Fig. 4B). The Aucsia-silenced plant displaying obligate parthenocarpy (i.e. L276 4-1) showed the most severe leaf alterations (Fig. 4C). In line L276 4-1, the leaves were frequently barely lobed and the leaf vasculature was drastically altered due to an increased number of parallel strands in the midvein (Fig. 4C). In adult L276 4-1 plants, leaves were dark green, extensively wrinkled, and showed abnormal development of the lamina (Fig. 4C, right). The height and growth habits, the number and diameter of internodes, flowering time, as well as apical dominance did not appear to be influenced in the Aucsia-silenced plants. Alterations of leaf morphogenesis exhibited by Aucsia-silenced plants suggest that these plants might be defective in auxin action and/or distribution. Leaf fusion and alteration in vascular patterning observed in Aucsia-silenced plants were also described in parthenocarpic SlIAA9-inhibited plants (Wang et al., 2005).
Figure 4.
Leaf development is altered in Aucsia-silenced tomato plants. A, Leaves of Aucsia-silenced plants are reflexed. Downcurling of the leaves is shown in the L276 (top) and INB777 (bottom) backgrounds. B, Examples of leaf fusions displayed by Aucsia-silenced plants. C (left), Leaf alterations displayed by an obligate parthenocarpic L276 4-1 plant; top right, reduced lobation of the leaves; bottom right, alteration of leaf vasculature. [See online article for color version of this figure.]
Aucsia-Silenced Tomato Plants Show Reduced Rhizogenesis in Response to Exogenous Auxin
To investigate whether Aucsia silencing alters auxin-induced rhizogenesis, root formation was evaluated in cotyledon explants in the presence of increasing concentrations of auxin.
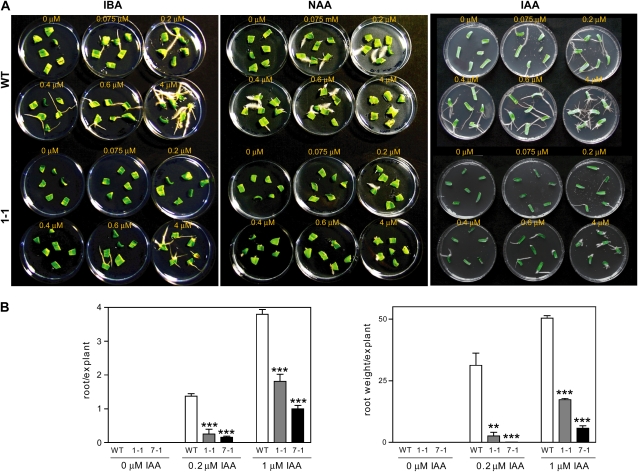
In wild-type tomato cotyledon explants, root formation was observed starting from 0.075 μm of the synthetic auxin naphthalene acetic acid (NAA) or the natural auxins indole-3-butyric acid (IBA) and IAA (Fig. 5A). In Aucsia-silenced cotyledon explants, root formation was promoted at higher auxin concentration, 0.2 and 0.6 μm NAA/IAA and IBA, respectively (Fig. 5A). To provide a quantitative evaluation of adventitious root formation capacity, both root number per explant and root weight per explant were evaluated using the natural auxin IAA. Cotyledon explants from two facultative parthenocarpic lines, L276 7-1 and L276 1-1, showing either intermediate (55%) or low (44%) fruit set efficiencies in emasculated flowers (Table II), were tested. The number of roots produced by Aucsia-silenced cotyledon explants of the two lines was on average 15% and 36% of the number of roots formed by wild-type cotyledon explants at 0.2 and 1.0 μm IAA, respectively (Fig. 5B). Root growth evaluated as root weight per explant was also significantly reduced in Aucsia-silenced lines compared with wild-type plants (Fig. 5B). These data indicate that the Aucsia-silenced seedlings are impaired in auxin-induced adventitious root formation. Aucsia-silenced seedlings were also defective in lateral root formation (Supplemental Fig. S3). The number of lateral roots of L276 1-1 and 7-1 seedlings was approximately 50% reduced (Supplemental Fig. S3).
Figure 5.
Effects of exogenous auxin on root formation in Aucsia-silenced cotyledon explants. A, Cotyledon explants were treated with increasing concentrations (0.075, 0.2, 0.4, 0.6, and 4 μm) of IBA, NAA, and IAA. B, Root number per explant (left) and root weight per explant (right) were evaluated in two Aucsia-silenced lines, L276 1-1 and 7-1, compared with the wild type (WT). Two IAA concentrations, 0.2 and 1 μm, were used in the quantitative assay. Error bars represent sd. ** P < 0.01, *** P < 0.001. [See online article for color version of this figure.]
Aucsia-silenced line L276 7-1 was also tested for root gravitropic response and for sensitivity to IAA-induced inhibition of primary root growth. Aucsia-silenced L276 7-1 plants did not exhibit any alteration in gravitropic root curvature or root growth in the presence of inhibitory IAA concentrations (0.1 and 0.5 μm; data not shown).
Aucsia Gene Silencing Impairs Auxin Transport in the Root and Increases Sensitivity to 1-NPA
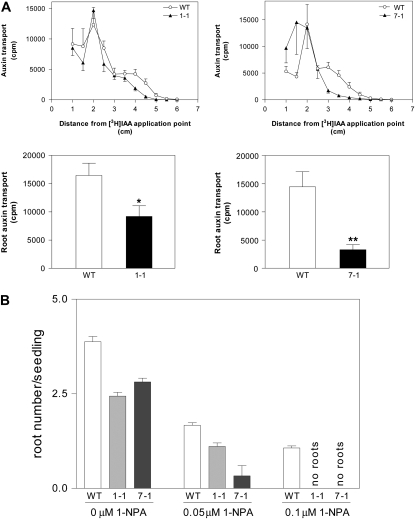
The phenotypes exhibited by Aucsia-silenced plants are suggestive of defects in either polar auxin transport or auxin signaling. With the aim of testing whether polar auxin transport is impaired in Aucsia-silenced plants, [3H]IAA transport was monitored in Aucsia-silenced and wild-type seedlings. Auxin transport was measured at 5 h after application of [3H]IAA to the shoot apex. A decreased amount of [3H]IAA was detected in Aucsia-silenced roots starting at 3 to 3.5 cm from the [3H]IAA application point (Fig. 6A, top row). The total [3H]IAA measured in the roots (segments located between 3 and 6 cm from the [3H]IAA application point) of Aucsia-silenced tomato L276 7-1 and L276 1-1 seedlings was 25% and 55%, respectively, of the [3H]IAA transported to the roots of wild-type seedlings (Fig. 6A, bottom row).
Figure 6.
Polar auxin transport and 1-NPA sensitivity. A, Inhibition of auxin transport in Aucsia-silenced seedlings. In the top row, [3H]IAA was applied between the cotyledons and after 5 h the seedlings were cut in 5-mm segments for radioactivity measurements. In the bottom row, total radiolabeled [3H]IAA was transported to roots of wild-type (WT) and Aucsia-silenced tomato seedlings. The values are sums of the radioactivity measured in the segments located between 3 and 6 cm from the [3H]IAA application point. B, 1-NPA sensitivity of tomato seedlings. The number of adventitious roots per seedling in Aucsia-silenced (L276 1-1 and 7-1) lines and in wild-type plants treated with either 0.05 or 0.1 μm 1-NPA or without 1-NPA is shown. Error bars represent sd. * P < 0.05, ** P < 0.01.
Since the decrease in auxin transport was evident only in the root, auxin transport was also evaluated in internodes of young plants and total IAA content was quantified in the flower at the preanthesis stage of development. No alteration of auxin transport has been detected in young internodes of lines L276 7-1 and L-276 1-1 in comparison with the wild type (data not shown). On the other hand, the flowers of Aucsia-silenced plants at preanthesis contain about 100 times more total IAA compared with wild-type plants (Table III).
Table III.
Total IAA content in preanthesis flower buds of Aucsia-silenced plants
The values (means ± se; n = 2) are expressed in nanomoles per gram of tissue (fresh weight).
| Line | Total IAA Content |
|---|---|
| L276 1-1 | 113.6 ± 0.99 |
| L276 7-1 | 261.0 ± 1.41 |
| L276 wild type | 1.9 ± 0.33 |
Aucsia-silenced plants were also tested for their sensitivity to the polar auxin transport inhibitor 1-NPA. Aucsia-silenced and wild-type seedlings were completely deprived of their roots and cultivated in medium supplemented or not with inhibitory concentrations of 1-NPA (0, 0.05, and 0.1 μm; Fig. 6B). In the absence of 1-NPA, derooted seedlings of both wild-type and Aucsia-silenced lines produced adventitious roots, but in Aucsia-silenced plants, the root number per seedling was lower compared with that in wild-type plants. These data confirm that Aucsia-silenced lines possess an intrinsic impaired capability to form adventitious roots. At 0.05 μm 1-NPA, root number per seedling (Fig. 6B) was significantly reduced to about 43% in the wild type and to about 41% in L276 1-1 compared with untreated wild-type and L276 1-1 seedlings, respectively. L276 7-1 showed a stronger sensitivity to 0.05 μm 1-NPA; the number of adventitious roots was reduced to 12% with respect to untreated seedlings. At 0.1 μm 1-NPA, adventitious root formation took place in wild-type seedlings, while it was completely abolished in the two Aucsia-silenced lines (Fig. 6B). Thus, Aucsia-silenced tomato plants showed an increased sensitivity to the inhibitory effect of 1-NPA on rhizogenesis.
Aucsia Is a Green Plant Gene Family
Aucsia genes are present in both chlorophytes and streptophytes (Table I). Thus, Aucsia represents an ancient gene family predating land plant evolution. Aucsia genes of both vascular (e.g. Arabidopsis [Arabidopsis thaliana], At3g01130 and At5g15320; Oryza, AL606631; Medicago, AC148293) and nonvascular land plants (e.g. Physcomitrella, ASYA88961.b1 and BHYX476795.y1; http://moss.nibb.ac.jp) are always split, at conserved nucleotide positions, by two introns that divide their coding sequences, with a central exon encoding 22 amino acids flanked by 5′ and 3′ exons encoding 14 to 16 and 9 to 15 amino acids, respectively (Table I). Thus, the structure of Aucsia genes and their copy number appear to be quite stable during land plant evolution. We did find Aucsia sequelogues in Prototheca wickerhamii, Helicosporidium species, and Ostreococcus lucimarinus (Table I), but we were unable to find an Aucsia sequelogue in the genome of Chlamydomonas reinhardtii. It is worth noting that the Aucsia gene of Ostreococcus species (i.e. O. lucimarinus and O. tauri), which are unicellular green algae, is present in a single copy per genome and does not contain any intron.
The sequence alignment of the predicted AUCSIA peptides from several green plant species highlights conserved features (Table I). A stretch of 16 amino acids, PYSGXSTLALVARXSA, containing 14 conserved amino acids, represents the AUCSIA motif.
The first Ser residue of the AUCSIA motif is a likely candidate for alternate phosphorylation/O-GlcNAc glycosylation (www.cbs.dtu.dk/services/YinOYang/).
AUCSIA peptides contain a Lys-rich C-terminal region usually ending with a His residue. Lys residues are often a target for ubiquitination (Dreher and Callis, 2007). Polyubiquitination targets proteins for degradation, while monoubiquitination is involved in protein localization and endocytosis (Bonifacino and Traub, 2003; Dreher and Callis, 2007). AUCSIA peptides also have a conserved Tyr-based (YXX[LMVIF]) sorting motif involved in endocytosis (Bonifacino and Dell'Angelica, 1999; Bonifacino and Traub, 2003).
DISCUSSION
Parthenocarpy obtained by increasing auxin synthesis in ovule/placenta (DefH9-iaaM gene; Rotino et al., 1997) is characterized as other auxin-related parthenocarpy (e.g. SlIAA9 down-regulation; Wang et al., 2005) by a precocious growth of the ovary suggesting that the repression of ovary growth is relieved before anthesis. Therefore, it is reasonable to assume that in auxin-synthesis (iaaM) parthenocarpic flower buds, the genetic program for fruit development has already been switched on well before anthesis. Genes that are differentially or heterocronically expressed in iaaM-parthenocarpic flower buds can be candidate genes playing a role in fruit initiation.
This work has functionally characterized what to our knowledge are two new tomato genes, named Aucsia, which are highly expressed at preanthesis and drastically down-regulated at anthesis during normal fruit development. In auxin-synthesis (iaaM) parthenocarpic plants, Aucsia genes are precociously down-regulated before anthesis. Silencing of SlAucsia genes causes parthenocarpic development of the fruit. Aucsia-silenced tomato plants are characterized by facultative parthenocarpy, producing seedless fruits when flowers are emasculated. However, Aucsia-silenced plants displayed, albeit rarely, obligate parthenocarpy (i.e. seedless fruit is produced also from pollinated flowers). Thus, Aucsia genes are involved in restricting the carpel from developing fruit before pollination.
The most straightforward explanation of the Aucsia-silenced parthenocarpic phenotype is the elevated amount of auxin in young flower buds that can substitute the signal(s) derived from fertilization in triggering fruit set. In this regard, Aucsia silencing-induced parthenocarpy is very similar to iaaM parthenocarpy. The IAA content of iaaM- and Aucsia-silenced parthenocarpic flower buds was approximately 10 to 60 (Pandolfini et al., 2002) and 100 times higher than that in wild-type buds, respectively. Phenotypical fruit modification, such as a remarkable accentuation of the umbone displayed by an obligate parthenocarpic Aucsia-silenced line, is also indicative of a higher auxin content (Pandolfini et al., 2002). At maturity, both obligate and facultative parthenocarpic fruits obtained by Aucsia gene silencing are reduced in size and weight compared with wild-type fruits. The smaller size of Aucsia-silenced fruits could be related either to the involvement of Aucsia genes in later stages of fruit development or alternatively to other factors controlling final fruit development in addition to or in place of Aucsia genes. It is worthwhile to note that the size and weight of DefH9-iaaM parthenocarpic fruits are usually similar to those of wild-type fruits (Rotino et al., 2008).
Besides fruit initiation, Aucsia genes are involved in vegetative development as well. Aucsia-silenced tomatoes are altered in leaf morphogenesis, displaying leaf fusions and curled leaves. Mutations affecting auxin biology can cause perturbation of leaf venation and abnormally shaped leaves (Leyser, 2006). Leaf fusion and alteration in vascular patterning were also described in SlIAA9-inhibited parthenocarpic plants where these phenotypes were enhanced by 1-NPA treatment (Wang et al., 2005). However, SlIAA9 down-regulated and Aucsia-silenced parthenocarpic plants differ in their response to exogenous auxin. In fact, in SlIAA9 down-regulated plants, auxin promotes root formation at lower concentrations than in wild-type plants (Wang et al., 2005), while root formation in Aucsia-silenced cotyledon explants occurs at higher auxin concentrations than in the wild type.
The finding that root formation in Aucsia-silenced plants is resistant to exogenous auxins (IBA, IAA, and NAA) suggests either a reduced responsiveness to the phytohormones or a reduced capacity of auxin cellular transport. The lower sensitivity of Aucsia-silenced explants to IBA with respect to IAA and NAA could be dependent on differences in cellular transport machineries. NAA influx is independent of influx carriers, while NAA efflux is carrier mediated (Woodward and Bartel, 2005; Leyser, 2006). Both IBA influx and efflux are carrier mediated, probably via IBA-specific carriers (Rashotte et al., 2003).
Auxin transport is curtailed in tomato Aucsia-silenced roots but apparently not in the stems. However, Aucsia-silenced plants show an increased sensitivity to the inhibitory effect of 1-NPA on adventitious root formation. NPA is a polar auxin transport inhibitor that affects endocytic recycling of some ABC-PGP auxin efflux carriers (Noh et al., 2001; Murphy et al., 2002; Geisler et al., 2003; Terasaka et al., 2005; Geisler and Murphy, 2006). Overall, these results could be interpreted as relating to the need for the presence of one or more interacting partners of AUCSIA proteins whose distribution could be tissue/organ dependent.
The mechanisms leading to parthenocarpic fruit development via silencing of Aucsia genes cannot be explained by mechanisms involving either auxin hypersensitivity or down-regulation of IAA9/ARF8. In fact, in flower buds of Aucsia-silenced plants, SlIAA9 and SlARF8 mRNA levels do not change with respect to the wild type. This means that Aucsia-related parthenocarpy is not linked to the down-regulation of IAA9 and ARF8 genes at the transcript level.
Our data indicate that Aucsia silencing-induced parthenocarpy is strictly connected to auxin accumulation in reproductive organs at early stages of flower/fruit development. Several direct and indirect mechanisms could be responsible for the increase in auxin content in the flower buds: an increased auxin synthesis, a decreased auxin degradation, an alteration of auxin transport within the ovary and between the ovary and other floral organs, or probably via the synergistic action of more than one of these mechanisms.
Although AUCSIA's mechanism of action is unknown at present, AUCSIA peptides, due to their minimal molecular mass, are likely regulatory subunits of protein complexes. Reversible and alternative posttranslational modifications of AUCSIA peptides via phosphorylation/O-GlcNAc glycosylation of the first Ser residue of the AUCSIA motif (PYSGXSTLALVARXSA) might be a mechanism used to control AUCSIA function. In this respect, O-GlcNAc glycosylation often targets proteins to multiprotein complexes (Huber and Hardin, 2004), while phosphorylation is also used as a signal for protein disassembly and/or degradation (Dreher and Callis, 2007).
Two O-GlcNAc transferases, SPINDLY and SECRET AGENT, which modify Ser or Thr residues, are present in the Arabidopsis genome (Hartweck et al., 2002, 2006). Arabidopsis spindly mutants show parthenocarpic fruit development and other alterations of growth and development indicative of altered gibberellin and cytokinin responses (Jacobsen and Olszewski, 1993; Greenboim-Wainberg et al., 2005). Regulation of fruit development includes the action and cross talk of different phytohormones (Serrani et al., 2008). A role for Aucsia in the interaction between auxin and other phytohormones involved in fruit growth and development, such as gibberellins, cytokinins, and ethylene, can well be hypothesized.
AUCSIA peptides contain also a conserved Tyr-based sorting motif involved in endocytosis (Bonifacino and Dell'Angelica, 1999). Notably, endocytic recycling of membrane complexes necessary for auxin movement (Kleine-Vehn et al., 2006; Dhonukshe et al., 2007; Zazimalova et al., 2007) is a way to control polar auxin transport.
The presence of Aucsia genes in O. lucimarinus, P. wickerhamii, and Helicosporidium species indicates that an ancestral Aucsia gene was present in green plants before the divergence of streptophytes and chlorophytes. Green algae contain IAA but apparently lack the IAA-specific transport systems (Cooke et al., 2002). At high concentration, IAA is toxic to eukaryotic cells (de Melo et al., 2004). Consequently, the unicellular green algal ancestor of land plants might have possessed a mechanism for IAA efflux and/or detoxification. The primordial Aucsia gene might have been involved in the process of auxin detoxification.
In conclusion, this work has identified a novel gene family affecting auxin biology and involved in tomato fruit initiation. Aucsia genes may allow us to develop a cis-genesis method that could be included in a general strategy that employs parthenocarpy to improve fruit quality and productivity in crop plants (Pandolfini et al., 2009). The identification of AUCSIA's molecular partners will be crucial to unraveling the mechanisms of action of AUCSIA peptides in auxin-regulated plant development.
MATERIALS AND METHODS
cDNA-AFLP Analysis
The cDNA-AFLP analysis was performed according to Vos et al. (1995) with some modifications (Breyne and Zabeau, 2001). Starting from 1 μg of mRNA, first-strand cDNA from tomato (Solanum lycopersicum) was synthesized with SuperScript II (Invitrogen) using oligo(dT)25 biotinylated primer, and then the double-stranded cDNA was obtained following the manufacturer's instructions (Invitrogen). Double-stranded cDNA was digested with the “rare-cutter” BstYI enzyme; 3′ ends of cDNA were captured from the solution with magnetic streptavidin-coated beads, while the other fragments were washed away. Second digestion was performed with the “frequent-cutter” MseI. Double-digested fragments were ligated to adapters. The product of the ligation reaction was subjected to a nonselective PCR using primers complementary to the adapters, with nonselective nucleotides on the 3′ ends. Afterward, a selective PCR was performed on a 1:20 dilution of the first PCR mixture, using primers complementary to the anchors but modified on their 3′ ends with selective nucleotides. The BstYI primer was fluorescein labeled on its 5′ end. PCR product was size fractionated on a 6% polyacrylamide gel at 150 W for about 2 h. The size resolution of this gel is from 50 to 500 bases. After running, the gel was introduced in a scanner apparatus (Genomyx) and the image of the gel was elaborated with Acquire software (Beckman Coulter). A virtual grid was created by the software and superimposed on the gel image, allowing the excision of differentially expressed cDNA-AFLP bands. cDNAs were eluted from acrylamide gels in 100 μL of sterile distilled water. A 5-μL aliquot of eluted cDNAs was reamplified by PCR using the nonlabeled primers identical to those adopted for selective PCR. The PCR products were purified and subsequently sequenced. All sequences were analyzed for homology using BLASTN and/or BLASTX (Altschul et al., 1997) exploiting the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) databases.
rolC-hpAucsia Construct Design and Tomato Transformation
The rolC promoter, from the rolC gene of Agrobacterium rhizogenes strain A4, was fused to two 200-nucleotide-long DNA fragments placed in inverted orientation and separated by an 87-nucleotide-long DNA fragment containing the 85-nucleotide-long intron of the rolA gene of A. rhizogenes. The 200-nucleotide-long sequence of the SlAucsia-1 gene includes 21 nucleotides upstream of the ATG initiation codon, 159 nucleotides spanning the coding region, and the first 20 nucleotides of the 3′ UTR. A 251-nucleotide DNA fragment containing the termination and polyadenylation signals from the rolC gene was placed downstream of the SlAucsia-1 antisense arm (Supplemental Fig. S1B). The resulting chimeric gene was subcloned in the T-DNA region of a derivative of pBin19 binary vector and then used to transform Agrobacterium tumefaciens strain LBA4404. Transgenic tomato plants of L276 and INB777 lines were obtained according to Ficcadenti et al. (1999).
Plant Material, Growth Conditions, and Phenotypic Analysis
Tomato plants were grown in a greenhouse under a 10-h/14-h light/dark cycle at 24°C and 18°C, respectively. For cDNA-AFLP analysis (Vos et al., 1995), iaaM-parthenocarpic tomato plants transgenic for the auxin-synthesizing gene (Pandolfini et al., 2002) and wild-type plants were used. Tomato seeds were first surface sterilized and then sown in Magenta boxes containing Murashige and Skoog (MS) culture medium (50% MS basal salt mixture including 0.5 mg L−1 thiamine, 0.25 mg L−1 nicotinic acid, 0.5 mg L−1 pyridoxine, 1.5% [w/v] Suc, and 0.8% [w/v] agar, pH 5.9). Selection plates were prepared by adding to MS medium kanamycin at the final concentration of 50 mg L−1. The seedlings were grown under sterile conditions in a climatic chamber at a constant 25°C during a 10-h/14-h light/dark cycle, with an average irradiance of 120 μmol m−2 s−1 of photosynthetically active radiation. For lateral root number measurement, Aucsia-silenced seedlings were grown for 15 d on MS agar. For auxin sensitivity analysis, cotyledon explants (four to six cotyledons per dish) from 21-d-old tomato Aucsia-silenced and wild-type seedlings were transferred for 10 d on MS medium supplemented with IBA, NAA, and IAA (0, 0.075, 0.2, 0.4, 0.6, and 4 μm). For quantitative evaluation of IAA sensitivity, cotyledon explants were grown on MS medium containing 0, 0.2, and 1 μm IAA. The number and weight of regenerated roots were collected from each cotyledon after 15 d. For determination of venation patterning, leaves were fixed in 14% acetic acid/84% ethanol, dehydrated with ethanol, and finally clarified with a solution of chloral hydrate. For 1-NPA treatment, Aucsia-silenced and wild-type seeds were previously germinated on MS medium. Rooted seedlings were cut at 0.5 cm from the base of the stem and transferred to Magenta boxes (three to four per box) containing MS medium supplemented with 0.05 or 0.1 μm 1-NPA. The number of regenerated roots was measured after 15 d of cultivation. Statistical analysis was conducted using Student's t test.
Transgenic State of Aucsia-Silenced Tomato Plants
Genomic DNA was extracted from 0.5 to 1 g of frozen leaves using the Nucleon PhytoPure system (Amersham Biosciences) according to the manufacturer's instructions. A total of 15 μg of genomic DNA was digested with HindIII, electrophoresed on a 0.7% agarose gel, and transferred to a nylon membrane (Hybond-N+; Amersham Biosciences). The membrane was hybridized with 100 ng of fluorescein-labeled probe prepared using the random prime labeling module (Amersham Biosciences). After overnight hybridization at 60°C, filters were first washed in 1× SSC/0.1% SDS at 60°C and then in 0.5× SSC/0.1% SDS at 60°C. Blots were detected using the alkaline phosphatase-based chemiluminescent detection assay (Amersham Biosciences). Probe for the analysis, corresponding to a portion of the rolC promoter, was obtained by amplification with the following forward (F) and reverse (R) primers: F, 5′-TGAGATTCCATAGACCACAAACCACC-3′; R, 5′-GTTAACAAAGTAGGAAACAGGT-3′.
Auxin Transport Analysis
For auxin transport measurement on intact plantlets, 3 d after germination wild-type and Aucsia-silenced seedlings, showing similar root growth, were placed vertically on slanted agar plates. A total of 12 pmol of radiolabeled auxin [3H]IAA (Amersham Biosciences; specific activity of 962 GBq mmol−1) was applied between the cotyledons. After 5 h, seedlings were sectioned into 5-mm segments, starting just below the cotyledons. Each section was placed in scintillation fluid (Perkin-Elmer) overnight under shaking before being analyzed in a Beckman scintillation counter (Beckman Instruments). Statistical analysis was conducted using Student's t test.
For polar auxin transport analysis in internodes of 4-week-old tomato plants, the protocol from Al-Hammadi et al. (2003) was adopted, using 100 nCi mL−1 [3H]IAA in the agar donor block. After 2, 5, and 8 h of incubation, the receiver blocks were removed, placed in liquid scintillation cocktail, and shaken before counting.
Total IAA Analysis
The total auxin content, which includes both free and conjugated IAA, was quantified in wild-type and Aucsia-silenced lines as described by Pandolfini et al. (2002). Preanthesis flower buds for each line were collected from two plants at the same developmental stage.
qRT-PCR Analysis
RNA was isolated using the RNeasy mini kit (Qiagen) starting from 100 mg of frozen tissues and treated with RQ1 DNase (Promega). mRNA samples were checked for DNA contamination by performing PCR on the actin gene. Comparative PCR analysis was carried out using first-strand cDNA obtained with oligo(dT) primer and the ImProm-II Reverse Transcription System (Promega). cDNA was amplified using SYBR Green qPCR Supermix-UDG (Invitrogen) on the ABI Prism 7000 Sequence Detection System (Applied Biosystems). qRT-PCR was performed using the following cycling conditions: 2 min at 50°C, 2 min at 95°C, 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s, and finally 72°C for 2 min. All quantitations were normalized to the actin gene as an endogenous control. For each determination of mRNA levels, three cDNA samples derived from three independent RNA extractions were analyzed. Relative quantitation of transcript levels was carried out following Livak and Schmittgen (2001). Statistical analysis was conducted using Student's t test. For tomato qRT-PCR analysis, forward (F) and reverse (R) primers used for amplification of the target genes were as follows: for SlAucsia-1 (F, 5′-GCTCTTTGCTGATAGAATAAGAT-3′; R, 5′-TAATATTTGATCGTGGAGAACCC-3′); for SlAucsia-2 (F, 5′-TAACTGTTCAATTTTGTGTTCACG-3′; R, 5′-GATGTAACAAGAAGATTATTATGC-3′); for SlIAA9 (F, 5′-TGGCCACCCATTCGATCTTTT-3′; R, 5′-TTCTGAGGTCCACTTTCCTC-3′); for SlARF8 (F, 5′-GGCAGCTTGTATTTGTTGACAGGGA-3′; R, 5′-TTTCTGCACATCCTCGGGTGAAAG-3′); and for actin (F, 5′-CCCGTTCAGCAGTGGTGGT-3′; R, 5′-TACGAGGGTTATGCTTTGCC-3′).
Northern-Blot Analysis of Aucsia-Silenced Plants
Total RNA (15 μg) isolated with Trizol (Invitrogen) was separated on 1% agarose-formaldehyde denaturing gels. The DNA probes were labeled with [32P]dCTP using Ready to Go DNA labeling beads (−dCTP; Amersham Biosciences). Unincorporated nucleotides were removed with Probe G-50 microcolumns (Amersham Biosciences). The Hybond-N+ membranes (Amersham Biosciences) were hybridized overnight at 42°C in ULTRAhyb buffer (Ambion). Labeled probe (106 cpm mL−1) was added to the hybridization buffer. The membranes were washed twice in 2× SSC containing 0.1% SDS for 5 min and twice in 0.1× SSC containing 0.1% SDS for 15 min at 42°C. For SlAucsia-1, SlAucsia-2, and actin mRNA analysis, the DNA probes were obtained by PCR with the same primers adopted for qRT-PCR. Densitometric analysis of band intensity was determined with a GS-710 densitometer (Bio-Rad) using the software Quantity One (Bio-Rad).
siRNA Analysis of Aucsia-Silenced Plants
siRNA analysis was performed following the protocol described previously (Hamilton and Baulcombe, 1999). Small RNAs (40 μg per lane) were separated on a 15% polyacrylamide-7 m urea gel and electroblotted for 1 h at 100 V in 0.5× Tris-borate/EDTA on a Hybond-N+ membrane (Amersham Biosciences). Radiolabeled [32P]single-stranded RNA probes were obtained by the Riboprobe in vitro transcription system (Promega). The membrane was hybridized overnight at 40°C after the addition of hydrolyzed probe. The membrane was rinsed in 2× SSC/0.2% SDS at 50°C. Oligoribonucleotides of 23 and 21 nucleotides were used as molecular markers.
Affinity Purification of Anti-AUCSIA Antibodies
Twenty-one-amino-acid-long synthetic peptide, corresponding to the final C-terminal portion of tomato AUCSIA-1 peptide (KLKYLKAKAKSQKKAEAKARH) conjugated to an ovalbumin carrier, was used as immunogen in rabbit in order to obtain polyclonal antiserum (Primm). Anti-AUCSIA antibodies were purified from serum using 21-amino-acid-long synthetic peptide immobilized on AminoLink Plus Coupling Gel (Pierce). Antigen-antibody interaction was dissociated using the ImmunoPure gentle Ag/Ab elution buffer (Pierce).
Western-Blot Analysis
Total proteins were extracted in 30 mm Tris-HCl, pH 8.2, buffer containing 50 mm KCl, 0.5% Tween 20, 0.1% polyvinylpolypyrrolidone, 1 mm EDTA, 0.04% β-mercaptoethanol, and 0.1% plant protease inhibitor cocktail (Sigma). A total of 50 μg of proteins was loaded on 15% Tris-Tricine SDS-polyacrylamide gels and transferred to Immobilon polyvinylidene difluoride membranes (Millipore). Blots were first incubated with 1 μg mL−1 affinity-purified anti-AUCSIA antibodies. The antigen-antibody complexes were developed using the chemiluminescent system based on CDP-Star substrate (Amersham Biosciences).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK224828, BW690880 (SlAucsia-1 cDNA), and AK224647 (SlAucsia-2 cDNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequences of the cDNA fragments isolated with cDNA-AFLP analysis corresponding to SlAucsia-1, SlAucsia transcripts, and the rolC-hpAucsia construct.
Supplemental Figure S2. Southern-blot analysis of tomato Aucsia-silenced plants transgenic for the rolC-hpAucsia construct.
Supplemental Figure S3. Lateral root formation in Aucsia-silenced lines (L276 1-1 and 7-1).
Supplementary Material
Acknowledgments
We thank V. Rizzatti, M.G. Tacconi, R. Pedretti, and F. Cavallanti for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Angelo Spena (angelo.spena@univr.it).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Al-Hammadi AS, Sreelakshmi Y, Negi S, Siddiqi I, Sharma R (2003) The polycotyledon mutant of tomato shows enhanced polar auxin transport. Plant Physiol 133 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Peer WA, Murphy AS (2005) Auxin transport. Curr Opin Plant Biol 8 494–500 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Dell'Angelica EC (1999) Molecular basis for the recognition of tyrosine-based sorting signals. J Cell Biol 145 923–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72 395–447 [DOI] [PubMed] [Google Scholar]
- Breyne P, Zabeau M (2001) Genome-wide expression analysis of plant cell cycle modulated genes. Curr Opin Plant Biol 4 136–142 [DOI] [PubMed] [Google Scholar]
- Carmi N, Salts Y, Dedicova B, Shabtai S, Barg R (2003) Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217 726–735 [DOI] [PubMed] [Google Scholar]
- Cooke TJ, Poli D, Sztein AE, Cohen JD (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49 319–338 [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo MP, de Lima TM, Pithon-Curi TC, Curi R (2004) The mechanisms of indole-acetic acid cytotoxicity. Toxicol Lett 148 103–111 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17 520–527 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficcadenti N, Sestili S, Pandolfini T, Cirillo C, Rotino GL, Spena A (1999) Genetic engineering of parthenocarpic fruit development in tomato. Mol Breed 5 463–470 [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743 [DOI] [PubMed] [Google Scholar]
- Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, Dudler R, et al (2003) TWISTED DWARF-1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporter AtPGP1 and AtPGP19. Mol Biol Cell 14 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Murphy AS (2006) The ABC of auxin transport: the role of P-glycoproteins in plant development. FEBS Lett 580 1094–1102 [DOI] [PubMed] [Google Scholar]
- George W, Scott J, Splittstoesser W (1984) Parthenocarpy in tomato. Hortic Rev (Am Soc Hortic Sci) 6 65–84 [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Hooper LC, Johnson SD, Rodrigues JC, Vivian-Smith A, Koltunow AM (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunov AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Genger RK, Grey WM, Olszewski NE (2006) SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heynh. J Exp Bot 57 865–875 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE (2002) Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Hardin SC (2004) Numerous post translational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr Opin Plant Biol 7 318–322 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe R, Swarup P, Bennet M, Friml J (2006) Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN-1. Plant Cell 18 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2006) Dynamic integration of auxin transport and signalling. Curr Biol 16 424–433 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Marti C, Orzáez D, Ellul P, Moreno V, Carbonell J, Granell A (2007) Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J 52 865–876 [DOI] [PubMed] [Google Scholar]
- Murphy AS, Hoogner KR, Peer WA, Taiz L (2002) Identification, purification and molecular cloning of N-1-naphtylphtalamic acid-binding plasma membrane-associated aminopeptidase from Arabidopsis. Plant Physiol 128 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T, Molesini B, Avesani L, Spena A, Polverari A (2003) Expression of self-complementary hairpin RNA under the control of the rolC promoter confers systemic disease resistance to plum pox virus without preventing local infection. BMC Biotechnol 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T, Molesini B, Spena A (2007) Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci 12 327–329 [DOI] [PubMed] [Google Scholar]
- Pandolfini T, Molesini B, Spena A (2009) Parthenocarpy in crop plants. In L Ostergaard, ed, Fruit Development and Seed Dispersal. Wiley Blackwell, Oxford (in press)
- Pandolfini T, Rotino GL, Camerini S, Defez R, Spena A (2002) Optimisation of transgene action at the post-transcriptional level: high quality parthenocarpic fruits in industrial tomatoes. BMC Biotechnol 2 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Poupart JC, Waddell S, Muday GK (2003) Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol 133 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotino GL, Pandolfini T, Lo Scalzo R, Sabatini A, Fibiani M, Spena A (2008) Field trials of genetically modified tomato: fruit quality and productivity. In VR Preedy, RR Watson, eds, Tomato and Tomato Products. Science Publishers, Enfield, NH, pp 47–66
- Rotino GL, Perri E, Zottini M, Sommer H, Spena A (1997) Genetic engineering of parthenocarpic plants. Nat Biotechnol 15 1398–1401 [DOI] [PubMed] [Google Scholar]
- Schulz B, Kolukisaouglu HU (2006) Genomics of plant ABC transporters: the alphabet of photosynthetic life forms or just holes in membranes? FEBS Lett 580 1010–1016 [DOI] [PubMed] [Google Scholar]
- Schwabe WW, Mills JJ (1981) Hormones and parthenocarpic fruit set: a literature survey. Hortic Abstr 51 661–699 [Google Scholar]
- Serrani JC (2007) Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv micro-tom of tomato. J Plant Growth Regul 26 211–221 [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, García-Martínez JL (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J 56 922–934 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645 [DOI] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, et al (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A, Luo M, Chaudhury A, Koltunow A (2001) Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development 128 2321–2331 [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177 60–76 [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latchè A, Pech JC, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16 1123–1127 [DOI] [PubMed] [Google Scholar]
- Zazimalova E, Krecek P, Skupa P, Hoyerova K, Petrasek J (2007) Polar transport of the plant hormone auxin: the role of PIN-FORMED (PIN) proteins. Cell Mol Life Sci 64 1621–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.