Abstract
Carbon is partitioned between export from the leaf and retention within the leaf, and this process is essential for all aspects of plant growth and development. In most plants, sucrose is loaded into the phloem of carbon-exporting leaves (sources), transported through the veins, and unloaded into carbon-importing tissues (sinks). We have taken a genetic approach to identify genes regulating carbon partitioning in maize (Zea mays). We identified a collection of mutants, called the tie-dyed (tdy) loci, that hyperaccumulate carbohydrates in regions of their leaves. To understand the molecular function of Tdy1, we cloned the gene. Tdy1 encodes a novel transmembrane protein present only in grasses, although two protein domains are conserved across angiosperms. We found that Tdy1 is expressed exclusively in phloem cells of both source and sink tissues, suggesting that Tdy1 may play a role in phloem loading and unloading processes. In addition, Tdy1 RNA accumulates in protophloem cells upon differentiation, suggesting that Tdy1 may function as soon as phloem cells become competent to transport assimilates. Monitoring the movement of a fluorescent, soluble dye showed that tdy1 leaves have retarded phloem loading. However, once the dye entered into the phloem, solute transport appeared equal in wild-type and tdy1 mutant plants, suggesting that tdy1 plants are not defective in phloem unloading. Therefore, even though Tdy1 RNA accumulates in source and sink tissues, we propose that TDY1 functions in carbon partitioning by promoting phloem loading. Possible roles for TDY1 are discussed.
Assimilates, RNA, and proteins are transported in the phloem tissue of veins (Lalonde et al., 2003a; van Bel, 2003; Lough and Lucas, 2006). Three distinct functional zones of the phloem, each with a different role in assimilate transport, have been described (van Bel, 2003). The collection phloem is actively involved in Suc loading into veins in leaves. The transport phloem functions after loading to distribute Suc through the vein system. The release phloem unloads Suc from veins into expanding cells in sink tissues. Suc movement within the phloem follows mass flow of solutes along a hydrostatic pressure gradient: high concentrations of nutrients and water enter the collection phloem and are conveyed through the transport phloem to the release phloem, where solutes and water are withdrawn, lowering hydrostatic pressure and maintaining the driving force (Lalonde et al., 2003a; van Bel, 2003).
The anatomy of a maize (Zea mays) leaf is intimately related to phloem loading and the transport of assimilates (Evert et al., 1978). Leaf blades have three different orders of longitudinal veins with distinct functions in carbohydrate distribution. Phloem loading occurs in the small and intermediate veins (Fritz et al., 1983), which intergrade into large veins for transport of Suc out of the leaves and into the stem (Russell and Evert, 1985; Fritz et al., 1989). The veins are surrounded by bundle sheath (BS) cells, which are encircled by mesophyll (M) cells (Esau, 1977). The path of Suc movement from the photosynthetic cells into the vein is as follows. Suc moves symplastically from its site of synthesis in the M cells (Lunn and Furbank, 1999), diffuses through plasmodesmata into the BS cells, and then enters into the vascular parenchyma (VP) cells (Russin et al., 1996). From the VP cells, Suc is exported to the apoplast by an unknown mechanism (Evert et al., 1978), prior to being imported into the phloem companion cells (CC) and/or sieve elements (SE). The pathway for carbon translocation in maize leaves was described 40 years ago (Hofstra and Nelson, 1969); however, we still know very little concerning the regulation of carbon partitioning.
Several classes of genes that contribute to phloem loading of Suc have been identified. The genes directly responsible for Suc entry into the phloem are sucrose transporters (SUTs; Lalonde et al., 2004; Sauer, 2007). SUTs utilize the proton motive force generated by H+-ATPases to cotransport Suc and a H+ across a membrane (Bush, 1993; Gaxiola et al., 2007). Evidence demonstrating SUT function in phloem loading in vivo was provided by loss-of-function mutations that conditioned excess carbohydrate accumulation in leaves and reduced growth, delayed flowering, and decreased yield due to the reduced delivery of photoassimilates to growing sink tissues (Riesmeier et al., 1994; Kühn et al., 1996; Bürkle et al., 1998; Gottwald et al., 2000; Srivastava et al., 2008). Mutations in other phloem-expressed genes, such as a H+-ATPase (Zhao et al., 2000), K+ channel (Deeken et al., 2002), and aquaporin (Ma et al., 2004), which indirectly function in phloem loading of Suc, also led to carbon accumulation in leaves and reduced growth. In all of these cases, the mutant phenotype affects the entire leaf and does not result in a variegated phenotype. In contrast to phloem loading, phloem unloading in vegetative sink tissues often occurs symplastically and does not require Suc passage across a membrane (Giaquinta et al., 1983; Haupt et al., 2001; Lalonde et al., 2003a; Stadler et al., 2005).
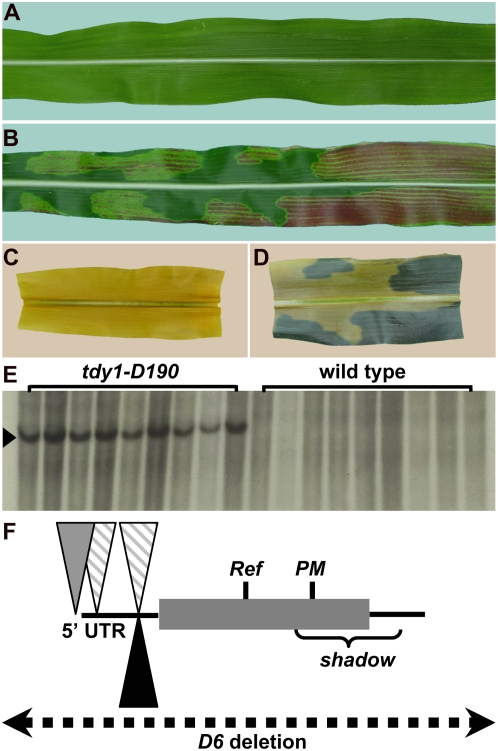
We have taken a genetic approach to identify genes regulating carbon partitioning in maize. We isolated mutants that develop chlorotic, carbon-hyperaccumulating regions in their leaves and designated this new class of mutants tie-dyed (tdy) due to their striking variegated appearance (Fig. 1, A and B; Braun et al., 2006; Baker and Braun, 2008). Our rationale was that by characterizing variegated mutants, in which some leaf regions contain excess carbohydrates while other regions appear similar to normal, the genes identified may function as regulators of carbon distribution rather than as transporters controlling sugar movement. The tdy mutants appear to be unique, as most other characterized variegated leaf mutants do not hyperaccumulate carbohydrates (Yu et al., 2007). The only mutant with a similar phenotype is sucrose export defective1 in maize, which we have shown functions independently of Tdy1 and Tdy2 (Russin et al., 1996; Provencher et al., 2001; Baker and Braun, 2008; Ma et al., 2008). From our previous analyses, the most salient features of the tdy phenotype related to carbon allocation are that (1) soluble sugars and starch hyperaccumulate in leaf regions (Fig. 1, C and D); (2) carbon accumulation is the earliest known defect and is proposed to be responsible for the chlorotic phenotype; (3) Tdy1 acts within the middle tissue layer of a leaf that contains the veins, BS, and interveinal M cells (Baker and Braun, 2007); (4) no blockages or plasmodesmatal structural perturbations are evident along the Suc symplastic transport path in tdy1 leaves, suggesting that carbon accumulation does not result from a physical impediment (Ma et al., 2008); and (5) the tdy1 phenotype is independent of starch metabolism, indicating that Tdy1 does not function in the photosynthetic cells in starch catabolism (Slewinski et al., 2008). Based on these and other data, we previously hypothesized that Tdy1 may function to promote Suc export from leaves by regulating SUT activity. Alternatively, we proposed that Tdy1 may function indirectly to affect Suc loading, for example, by functioning in a Suc-sensing signaling cascade controlling SUT abundance (Chiou and Bush, 1998; Vaughn et al., 2002; Ransom-Hodgkins et al., 2003).
Figure 1.
The cloning of Tdy1. A and B, Photographs of mature leaves. C and D, Photographs of segments from the same leaves, cleared and starch stained. A and C, Wild type. B and D, tdy1. E, Southern blot of tdy1-D190 mutants and wild-type siblings DNA probed with Mu1 (arrowhead). F, Schematic of mutations in different tdy1 alleles. Lines represent 5′ and 3′ untranslated regions (UTRs), the shaded box represents the Tdy1 protein-coding region, inverted triangles represent Mu insertions (striped, Mu1; gray, Mu8; black, Mu3), vertical lines indicate the locations of Ref and PM mutations, the bracket shows the region deleted in the shadow allele, and the dotted line indicates that the entire gene is deleted in the D6 allele. [See online article for color version of this figure.]
To gain insight into the function of Tdy1, we have cloned and characterized the Tdy1 gene. We found that Tdy1 encodes a novel predicted membrane-localized protein expressed exclusively in the phloem. In addition, we determined that Tdy1 RNA is expressed in source and sink tissues, suggesting that Tdy1 may function in both phloem loading and unloading processes. However, using a fluorescent dye to monitor solute transport, we observed a defect in phloem loading, but not unloading, in tdy1 plants. Therefore, these data suggest that Tdy1 functions in carbohydrate partitioning by promoting phloem loading. Potential TDY1 functions are discussed.
RESULTS
Cloning of Tdy1
To understand the molecular function of Tdy1, we cloned the gene using a Mutator (Mu) transposon-tagging approach (Chandler and Hardeman, 1992; Bennetzen et al., 1993). Mu-active lines were crossed by tdy1-Reference (tdy1-R) plants, and four new alleles were recovered with phenotypes indistinguishable from that of the tdy1-R allele. The new alleles were outcrossed to segregate away unlinked transposons. Southern-blot analysis was performed to detect cosegregation of a linked Mu transposon with a tdy1 mutant allele. A Mu1 element tightly linked to the tdy1-D190 allele was identified (no recombinants per 110 chromosomes, approximately 0.9 centimorgan; Fig. 1E). The DNA flanking the Mu1 element was cloned by constructing a subgenomic DNA λ library. Sequencing the DNA flanking the transposon identified an open reading frame with homology to expressed cDNAs in rice (Oryza sativa) and sorghum (Sorghum bicolor). We verified that the sequence cloned was the correct gene by analyzing independently derived alleles of tdy1. Two other alleles recovered from the transposon mutagenesis contained a Mu insertion at different sites in the same gene (tdy1-D24 and tdy1-D46; Fig. 1F; Supplemental Table S1). The tdy1-D6 allele was caused by a Mu element inserting into a nearby gene and resulting in an approximately 180-kb deletion encompassing the entire Tdy1 gene as well as several adjacent loci (Supplemental Table S2; Golubovskaya et al., 2006). Sequencing the tdy1-R allele identified a C-to-G substitution in the 112th amino acid that changed a Pro to Arg. Additionally, we found a 1-bp deletion in the tdy1-PM allele that caused a frameshift and premature termination. In total, we identified lesions in eight different tdy1 alleles, demonstrating that the gene we cloned corresponds to Tdy1.
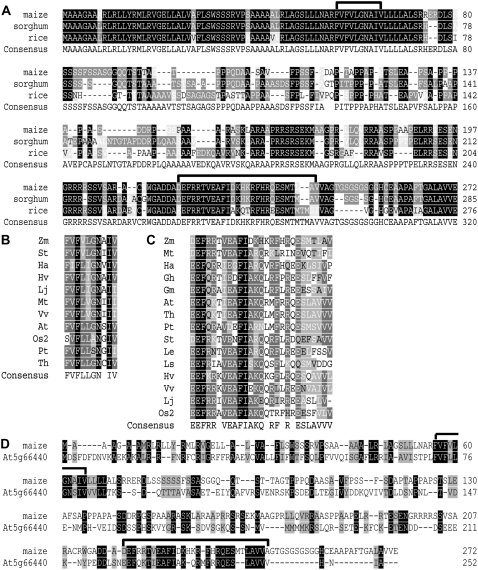
BLAST analyses of nucleotide and deduced amino acid sequences determined that Tdy1 encodes a novel protein of 272 amino acids. Orthologous proteins having the highest sequence similarity were identified in sorghum and rice (Fig. 2A). These genes showed homology throughout the protein-coding region and mapped to syntenic genomic positions (Gale and Devos, 1998). The N- and C-terminal regions of the protein were most highly conserved, with the middle portion of the protein showing the least conservation. TDY1 is predicted to be a membrane-localized protein (Fig. 3A). The N terminus contains two stretches of hydrophobic amino acids, the first predicted to be a signal peptide and the second a transmembrane domain. The middle and C-terminal regions of TDY1 are predicted to be localized in the cytoplasm. The middle portion of TDY1 contains 10 interspersed Pro residues that are conserved in the grasses and may be important for protein folding. Interestingly, the second conserved Pro is mutated to an Arg residue in the tdy1-R allele, suggesting that the mutant protein may not fold correctly. BLAST analyses also detected two domains conserved in TDY1 and proteins of unknown function from other plants (Fig. 2, B and C). Domain I is 10 amino acids long, located within the predicted transmembrane domain, and highly conserved in both monocots and dicots. Domain II is 27 amino acids in length, located near the C terminus of the protein, and also well conserved across angiosperms. We did not identify TDY1-like sequences in animal or prokaryotic genomes.
Figure 2.
TDY1 protein alignment. A, Alignment of maize, sorghum, and rice TDY1 orthologs. Domains I and II are indicated by black brackets above the sequences. Maize (FJ376984), sorghum (Sb09g020660.1), rice (BAF18345). B, Alignment of domain I. Zm (Zea mays), St (Solanum tuberosum; DN939221), Ha (Helianthus annuus; DY923064), Hv (Hordeum vulgare; BM816550), Lj (Lotus japonicus; BAF98220), Mt (Medicago truncatula; AJ500673), Vv (Vitis vinifera; CAN77202), At (Arabidopsis thaliana; NP_201445), Os2 (Oryza sativa related sequence2; EAY75070), Pt (Populus trichocarpa; ABK92540), Th (Thellungiella halophila; ABB45849). C, Alignment of domain II. Abbreviations and sequences are the same as in B, except Gh (Gossypium hirsutum; AAC33276), Gm (Glycine max; BE807829), Le (Lycopersicon esculentum [now Solanum lycopersicum]; BP894688), Ls (Lactuca sativa; BU011277). D, Alignment of maize TDY1 with the closest related Arabidopsis protein (At5g66440, NP_201445). Black brackets indicate domains I and II.
Figure 3.
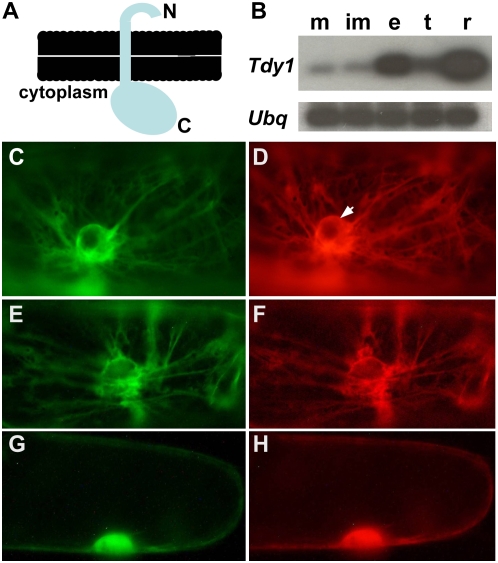
RT-PCR of Tdy1 RNA in maize tissues and TDY1 protein subcellular localization in onion cells. A, Cartoon of TDY1 topology showing the N terminus on the luminal side of the membrane and the C terminus on the cytoplasmic face. B, Semiquantitative RT-PCR blots showing expression of Tdy1 and Ubq as a loading control in B73 wild-type tissues. m, Mature leaves; im, immature leaves; e, developing ear; t, developing tassel; r, 1-week-old roots. C to H, Fluorescent proteins transiently expressed in onion epidermal cells. C, ER membrane-targeted GFP-HDEL (Goodin et al., 2007). D, Same cell as in C coexpressing the full-length TDY1-RFP fusion, showing colocalization to the ER. The arrow indicates the perinuclear envelope, which is contiguous with the ER. E, N-terminal amino acids 1 to 71 of TDY1 fused to YFP showing ER localization. F, Same cell as in E coexpressing full-length TDY1-RFP fusion. G, C-terminal amino acids 72 to 272 of TDY1 fused to YFP localizes in the cytoplasm and nucleus. H, Same cell as in G coexpressing soluble RFP in the cytoplasm and nucleus. [See online article for color version of this figure.]
Although proteins similar in structure and showing limited sequence homology to TDY1 are found in dicots, several differences suggest that they may not have a similar function. The most closely related protein in Arabidopsis (Arabidopsis thaliana) shows 23% amino acid identity (42% similarity) to maize TDY1 over the entire protein (Fig. 2D). The Arabidopsis genome contains several genes predicted to encode proteins related to the closest TDY1 homolog, and all contain domains I and II (Supplemental Fig. S1). The functions of these genes are unknown. Although the N terminus is not as conserved in sequence, the At5g66440 protein also contains two stretches of hydrophobic amino acids, and the protein is predicted to show a similar topology to TDY1. However, the contrasting calculated pI of the two proteins suggest that their functions may be different. The pI of TDY1 is basic at 11.4, whereas the Arabidopsis protein pI is acidic at 4.8. In addition, only four of the 10 Pro residues conserved in the middle portion of the TDY1-orthologous proteins from grasses are conserved between maize and Arabidopsis. Furthermore, the Pro residue mutationally defined to be important for function in the tdy1-R allele is not conserved in Arabidopsis. Lastly, we have not observed any visible phenotype in Arabidopsis plants homozygous for a T-DNA or transposon insertion in the protein-coding region of At5g66440 (T.L. Slewinski and D.M. Braun, unpublished data). Hence, it is not clear if TDY1 function is conserved in dicots, although disrupting the functions of multiple related genes may be required to uncover a phenotype.
Transient TDY1 Protein Subcellular Localization
The protein sequence of TDY1 is novel. To gain insight into TDY1 function, we investigated the subcellular localization of the TDY1 protein. Bioinformatic programs (SignalP, Phobius, and TM-HMM) were in agreement that the N terminus of TDY1 contained a predicted signal peptide and a membrane-spanning domain. However, additional analyses utilizing multiple programs (ChloroP, MitoP, TargetP, and PSORT) to predict the subcellular localization of TDY1 yielded contradictory results. To determine the subcellular localization of TDY1, we fused a red fluorescent protein (RFP) to the C terminus of TDY1, which is predicted to be on the cytoplasmic face of the membrane (Fig. 3A). We used particle bombardment to transiently express the TDY1 fusion protein in onion (Allium cepa) epidermal cells. We also heterologously coexpressed TDY1-RFP with various fluorescent protein fusions targeted to known subcellular locations (nucleus, endoplasmic reticulum [ER], Golgi, vacuole, plasma membrane, and cytoplasm). Coexpression of TDY1-RFP with an endomembrane-targeted GFP-HDEL fusion determined that TDY1 localizes to the ER in onion cells (Fig. 3, C and D). Fluorescence was observed in a net-like pattern of strands in the cytoplasm and on the perinuclear envelope, which is contiguous with the ER. We did not observe colocalization of TDY1 with other subcellular markers (data not shown).
Proteins that are localized to the secretory system are synthesized on and trafficked through the ER to reach their cellular destinations. High levels of protein expression have been found to cause ectopic localization of membrane proteins in the ER due to saturation of the target membrane (Gobert et al., 2006). To investigate if this possibility might explain the observed localization of TDY1 to the ER, we examined the localization of a TDY1 fusion to the yellow fluorescent protein (YFP) over time. We found that TDY1 localized exclusively to the ER in onion cells from the earliest time point after transformation, when a low level of protein expression was first detectable, and throughout our observations (Supplemental Fig. S2). TDY1 was never visualized in the plasma membrane or other membrane locations. These data suggest that when heterologously expressed in onion cells, TDY1 resides on the ER membrane and that the protein localization does not result from the fusion protein saturating a different subcellular location and ectopically “backing up” in the endomembrane system. However, in vivo subcellular localization of the endogenous protein in maize is required to confirm these results.
Bioinformatic programs indicated that the subcellular targeting determinants in TDY1 reside in the N terminus of the protein. To test this prediction, we created a protein fusion of the first 71 amino acids of TDY1, which contain the predicted signal peptide and transmembrane domain, translationally fused to YFP. Expressing this construct in onion epidermal cells demonstrated that the protein localized to the ER (data not shown). In addition, this construct was coexpressed with the full-length TDY1-RFP fusion protein. The two fusion proteins showed perfect overlap in their localization (Fig. 3, E and F), indicating that the N terminus of TDY1 was sufficient to confer targeting to the ER in onion cells. To test the C terminal region of TDY1 for subcellular targeting information, we translationally fused the remaining 201 amino acids of TDY1 to YFP and coexpressed the fusion with soluble RFP. Both proteins were similarly distributed in the cytoplasm and able to diffuse into the nucleus (Fig. 3, G and H), indicating that they did not contain any targeting signals. From these data, we infer that the N terminus of TDY1 targets the protein to the ER membrane in onion cells and that the C-terminal domain is present in the cytoplasm.
Tdy1 Reverse Transcription-PCR and RNA in Situ Hybridization Analyses
The primary phenotype of tdy1 mutants is observed in leaf blade tissue (Braun et al., 2006); thus, we predicted that Tdy1 would be expressed only in leaves. To determine which tissues accumulate Tdy1 RNA, we used semiquantitative reverse transcription (RT)-PCR to measure relative transcript abundance. We isolated RNA from mature and immature leaves, developing ears and tassels, and seedling roots. Tdy1 RNA accumulated in all tissues, although at much higher levels in young roots and at the lowest level in mature and immature leaves (Fig. 3B). This was a surprising result, and it suggests that Tdy1 may play a larger role than that defined by mutational analysis.
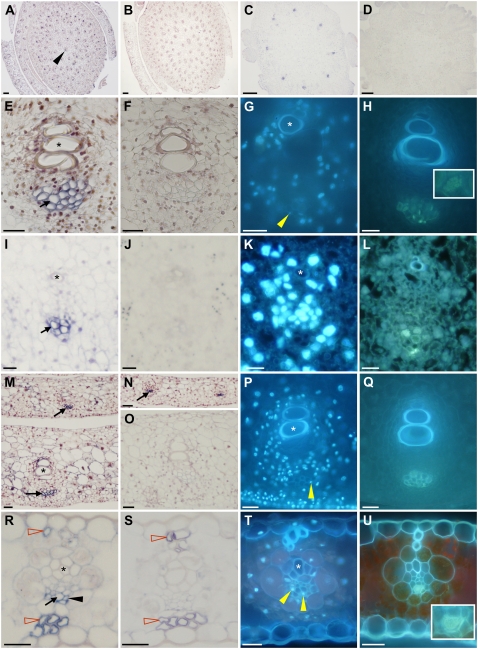
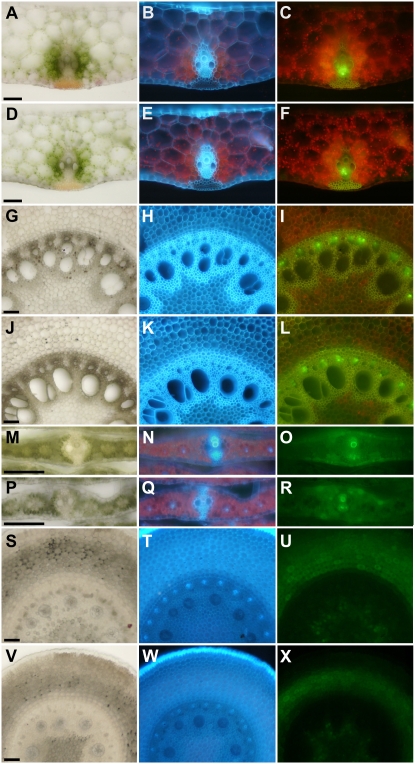
From a clonal mosaic analysis, we determined that Tdy1 functioned in the middle tissue layer of leaves, composed of the interveinal M, BS, and vascular cells (Baker and Braun, 2007). Based on these and other studies, we hypothesized that Tdy1 may act within the veins to promote phloem loading of Suc. To ascertain if Tdy1 was expressed in all cell types or showed cell-specific expression, we performed RNA in situ hybridization. In agreement with the conclusions from the mosaic analysis, Tdy1 RNA was localized exclusively to the veins of immature leaves, stems, and ears (Fig. 4, A and C). Tdy1 RNA accumulated only in protophloem cells in these tissues (Fig. 4, E, I, and M). Interestingly, Tdy1 RNA was present in protophloem cells upon differentiation and prior to protoxylem differentiation (Fig. 4N). The great majority of protophloem cells in maize are SE that lack associated CC (Sharman, 1942; Esau, 1943; Evert et al., 1996). To determine the identity of the Tdy1-expressing cells, we stained comparable sections with 4′,6-diamidino-2-phenylindole (DAPI), which binds to DNA and therefore stains CC (Lalonde et al., 2003b), or separately with aniline blue, which binds callose in the sieve plates of SE (Lalonde et al., 2003b). Because the veins of immature stems, ears, and leaves were found to contain few DAPI-positive cells (Fig. 4, G, K, and P), we infer that most of the Tdy1-expressing cells are protophloem SE. Aniline blue staining confirmed that these cells contained sieve plates (Fig. 4, H, L, and Q). This RNA localization pattern suggests that Tdy1 function may be present as soon as protophloem SE become competent for transport. To examine expression in mature tissue, we analyzed fully expanded source leaves (Fig. 4R). Tdy1 RNA was present only in phloem cells, although both the antisense and sense strand probes showed nonspecific hybridization to the thick-walled hypodermal sclerenchyma cells (Fig. 4, R and S). DAPI and aniline blue staining revealed that Tdy1 was expressed in both metaphloem CC and SE (Fig. 4, T and U). No expression was detected in tissues from plants homozygous for the tdy1-D6 deletion allele (Fig. 4D; Supplemental Fig. S3) or in wild-type tissues using the sense strand control (Fig. 4, B, F, J, O, and S), demonstrating that the antisense probe was specific. These data indicate that Tdy1 RNA specifically accumulates in phloem cells in all tissues and that TDY1 may play a role in vascular function upon phloem cell differentiation.
Figure 4.
Tdy1 RNA in situ hybridizations. All tissues are from B73 wild-type plants except D, which is from a tdy1-D6 deletion control. A, C, D, E, I, M, N, and R, Tissue sections probed with antisense Tdy1 RNA. Blue precipitate indicates signal (black arrows). B, F, J, O, and S, Tissue sections probed with Tdy1 sense strand control. A and B, Young stems and surrounding immature leaves. The black arrowhead indicates a vein. C and D, Developing ears. E to H, Veins in developing stem tissue. I to L, Veins in developing ears. M to Q, Veins in immature leaves. R to U, Veins in mature leaves. G, K, P, and T, DAPI-stained veins. Yellow arrowheads indicate DAPI-positive CC nuclei. H, L, Q, and U, Aniline blue-stained veins. Insets in H and U show aniline blue-stained sieve plates. Asterisks indicate xylem elements. N, Tdy1 expression in protophloem cells prior to protoxylem differentiation. R, Tdy1-expressing CC (black arrowhead) and SE (black arrow) as judged by their appearance, position, and relative size within the phloem. Red-outlined arrowheads indicate nonspecific signal in thick-walled hypodermal sclerenchyma cells. Bars = 100 μm in A to D and 25 μm in E to U.
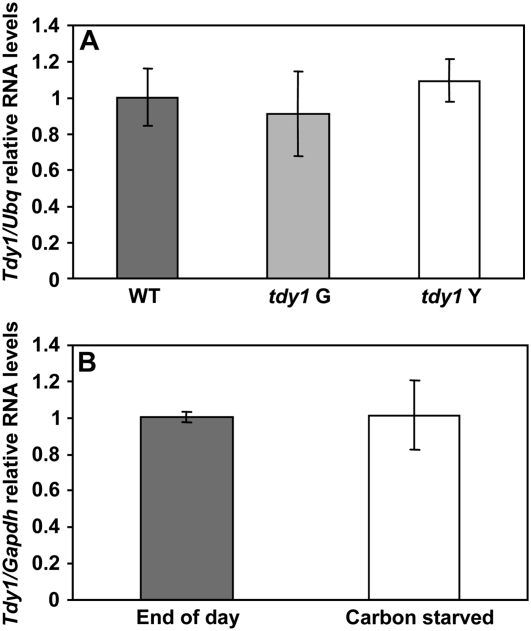
We previously proposed that one potential function of TDY1 may be in a Suc-sensing signal transduction pathway (Ma et al., 2008). To test whether the Tdy1 gene was transcriptionally induced in response to excess carbohydrates, we utilized the tdy1-R allele, which has a single base pair change in the coding region (Fig. 1F; Supplemental Table S1). This allele would be expected to normally produce RNA but not functional protein. As previously mentioned, tdy1 mutants produce variegated leaves with normal-appearing green and chlorotic regions (Fig. 1B). The tdy1-R chlorotic leaf regions hyperaccumulate Suc, Glc, Fru, and starch and therefore experience excess sugar stress, while the tdy1-R green regions contain carbohydrate levels similar to those of wild-type plants (Braun et al., 2006). RNA was isolated from wild-type, tdy1-R green, and tdy1-R chlorotic leaf regions. Quantitative RT-PCR (qRT-PCR) experiments showed that Tdy1 RNA was expressed at similar levels in all three tissues relative to Ubiquitin (Ubq), indicating that Tdy1 was not transcriptionally induced by sugar stress (Fig. 5A). As a positive control, a gene that shows highly induced expression under excess sugar conditions (glucose 6-phosphate/phosphate translocator [G6PT]; Lloyd and Zakhleniuk, 2004; Gonzali et al., 2006) was strongly up-regulated in tdy1 chlorotic tissue but not in tdy1 green or wild-type tissue (Supplemental Fig. S4). To examine if Tdy1 RNA might instead be induced by carbohydrate depletion, we transferred greenhouse-grown wild-type plants into a dark room for 3 d. This amount of time in darkness is sufficient to eliminate carbon reserves in leaves (Slewinski et al., 2008). For this experiment, Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used as the reference gene, since we determined that Ubq was not a suitable control under these conditions, potentially due to increased proteolysis upon carbon starvation (see “Materials and Methods”). Comparing Tdy1 expression in wild-type leaves collected at the end of the day versus plants maintained in the dark revealed no change in Tdy1 RNA (Fig. 5B), suggesting that Tdy1 RNA is not induced by carbon starvation. Collectively, these data suggest that Tdy1 expression is not regulated by the carbohydrate status of the tissue. Therefore, if Tdy1 functions in a sugar-sensing pathway, it likely does so posttranscriptionally.
Figure 5.
qRT-PCR analyses of Tdy1 RNA accumulation. A, Tdy1 RNA levels relative to Ubq. Wild type (WT) is shown by the dark gray bar, tdy1-R green (G) regions are shown by the light gray bar, and tdy1-R chlorotic (Y), carbon-hyperaccumulating regions are shown by the white bar. B, Tdy1 RNA levels relative to Gapdh. RNA was isolated from leaves of wild-type plants grown in the greenhouse at the end of the day (dark gray bar) or from carbohydrate-depleted plants kept in a dark room for 3 d (white bar). For both A and B, bars represent means and error bars represent the se. Samples were not statistically different at P ≤ 0.05 using Student's t test.
tdy1 Mutants Have Reduced Root Mass and Sugar Content When Grown in High Light
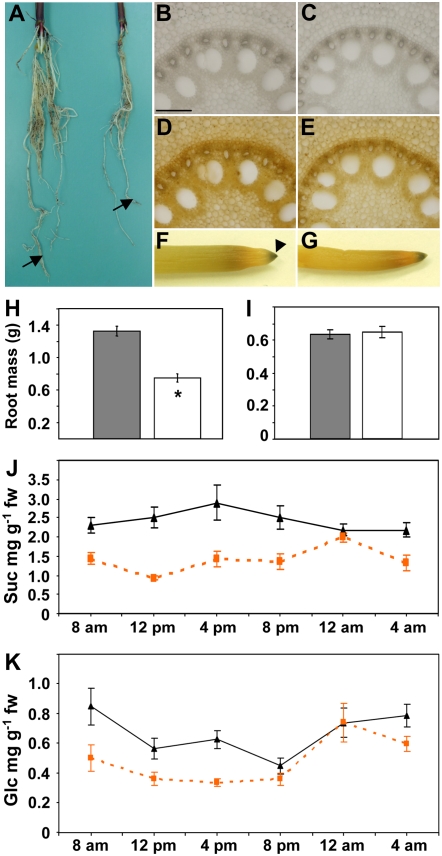
Tdy1 RNA accumulated to the highest level in roots, suggesting that Tdy1 may play an important role in this organ. To determine the consequences of the loss of Tdy1 function in roots, we compared wild-type and tdy1 root morphology, mass, and starch accumulation. We grew plants for 10 d in a high-light growth chamber in which the tdy1 chlorotic leaf phenotype was strongly expressed. We found that the root system of tdy1 plants was significantly smaller than that of wild-type siblings (Fig. 6, A and H). All classes of roots were present in tdy1 mutants, indicating that the reduced root mass was not caused by a failure to elaborate a particular type of root. To determine if the reduced root mass was due to altered carbohydrate partitioning, we stained root cross-sections and whole roots from wild-type and tdy1 mutant plants with iodine. No differences in tissue histology or starch deposition were observed (Fig. 6, B–G). To examine if there might be differences in the levels of sugars or starch in the roots of wild-type and tdy1 mutant plants, we quantified their abundance over a diurnal cycle. tdy1 mutant roots contained significantly less Suc at all time points except midnight compared with wild type (Fig. 6J). Glc accumulation was also lower in tdy1 roots relative to wild type, except at 8 pm and 12 am (Fig. 6K). Fru and starch levels were low in both tdy1 and wild-type roots and not significantly different at any time point (Supplemental Fig. S5). To determine if tdy1 plants might have any root phenotype when carbohydrates did not hyperaccumulate in leaves, we grew wild-type and mutant plants under low-light conditions that did not induce any visible tdy1 phenotype. Comparison of root biomass between wild-type and tdy1 plants showed no difference (Fig. 6I), suggesting that carbohydrates were normally translocated to the roots in mutant plants and that there was no apparent phenotype when grown under low light. The reduced tdy1 root mass observed during growth in high light is consistent with the previously reported reductions in plant height and inflorescence size and the delayed time to flowering seen in tdy1 mutants in comparison with wild-type siblings (Braun et al., 2006). We propose that these phenotypes are due to retention of excess carbohydrates in tdy1 leaf tissues.
Figure 6.
Root anatomy, starch accumulation, biomass, and sugar levels over a diurnal cycle of wild-type and tdy1 plants. All data were collected from high-light-grown plants, except I. A, Photograph of wild-type (left) and tdy1-R mutant (right) roots of 10-d-old seedlings. Arrows indicate primary roots. B, D, and F, Wild type. C, E, and G, tdy1-R mutant. B and C, Root cross-sections viewed in bright field. D and E, Same sections after starch staining and viewed in bright field. F and G, Photographs of starch-stained roots. The arrowhead indicates starch accumulation in the root cap. Bar in B = 200 μm. H and I, Root biomass of high- and low-light-grown plants, respectively. Wild type is shown by the dark gray bars, and tdy1 mutant is shown by the white bars. Bars represent mean (n = 10) and error bars represent the se. The asterisk indicates that the value is significantly different from the wild-type value at P ≤ 0.05 using Student's t test. J and K, Sugar accumulation in roots over a diurnal cycle. Wild type is shown by black triangles and tdy1 is shown by orange squares (n = 9). J, Suc. K, Glc. fw, Fresh weight. At all time points, tdy1 values were significantly different from wild-type values except at 12 am for both Suc and Glc and at 8 pm for Glc, as determined by Student's t test at P ≤ 0.05. [See online article for color version of this figure.]
Carboxyfluorescein Transport Studies
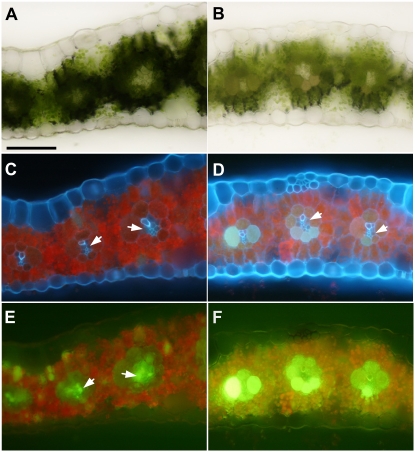
tdy1 mutants accumulate excess carbohydrates in their leaves (Braun et al., 2006). To determine whether any physical blockage of plasmodesmata could explain the carbon hyperaccumulation, we previously examined the symplastic transport route from the photosynthetic cells to the vein for callose deposition and plasmodesmatal ultrastructural changes that would prevent Suc movement. No alterations or structural perturbations were found, suggesting that the blockage to Suc transport occurs at the apoplastic step (Ma et al., 2008). If the apoplastic step is impeded, reduced solute mobility into the vein would be predicted. To test whether mobility into the phloem is reduced, we applied the fluorescent dye 5,6-carboxyfluorescein (CF) directly to abraded leaves to monitor dye transport from the photosynthetic cells into the vein. In wild-type leaves, the dye was taken up by M cells, transported along the symplastic pathway into BS cells, and preferentially accumulated in the VP cells (Fig. 7, A, C, and E). CF staining in green regions of tdy1 mutant leaves appeared identical to that in wild-type leaves (data not shown). However, we found that the CF mobility into the vein was reduced in tdy1 chlorotic regions. CF was taken up by the M cells and moved into the BS cells, where it accumulated, but less dye entered the VP cells (Fig. 7, B, D, and F). To examine phloem transport of the dye, we measured the rate of CF movement in wild-type and tdy1 leaves. Although the value for tdy1 mutants was slightly slower than in wild type (tdy1 = 107.3 ± 9.7 mm h−1, wild type = 119.0 ± 7.6 mm h−1), no significant difference was detected. These data suggest that the principal defect in CF movement in tdy1 chlorotic leaf regions is impeded solute flow from the photosynthetic cells toward the vein.
Figure 7.
CF movement along the symplastic pathway is retarded in tdy1 leaves. A, C, and E, Wild type. B, D, and F, tdy1-R chlorotic region. A and B, Transverse sections of mature leaf blades under bright field. C and D, Same sections under UV illumination. E and F, Same sections under blue light to image CF. Arrows indicate VP cells. Bar = 100 μm. [See online article for color version of this figure.]
Tdy1 RNA accumulates in the phloem in all tissues. To investigate whether Tdy1 may function in the transport and release phloem, we fed wild-type and mutant leaves the phloem-mobile tracer CF diacetate (CFDA). CFDA is a nonfluorescent form of the dye that can cross cell membranes. Upon entering a cell, nonspecific esterases remove the diacetate groups, releasing CF, which is fluorescent and charged and therefore remains in the cell (Wright and Oparka, 1996). We used CFDA to specifically image CF after entering into phloem cells. CF was transported through the veins to distal parts of the plant. In the transport phloem of the leaf sheath of the CF-fed leaf, the dye was similarly present in the phloem cells (Fig. 8, A–F). In the transport phloem of roots, the dye was equally present in wild-type and mutant plants (Fig. 8, G–L). In addition, we did not detect any difference in phloem unloading in the release phloem of immature leaves (Fig. 8, M–R) or in the unloading domain of the roots (Fig. 8, S–X). Taken together, these data suggest that tdy1 mutants have phloem transport capabilities similar to wild type and that reduced phloem transport capacity to distal sink tissues does not account for the hyperaccumulation of carbohydrates in leaves. These data also suggest that Tdy1 does not play a major role in the transport or release phloem.
Figure 8.
CF movement is normal in the transport and release phloem of tdy1 mutants. A to C, G to I, M to O, and S to U, Wild type. D to F, J to L, P to R, and V to X, tdy1-R mutant. A, D, G, J, M, P, S, and V, Transverse sections under bright field. B, E, H, K, N, Q, T, and W, Same sections under UV illumination showing cellular anatomy. C, F, I, L, O, R, U, and X, Same sections under blue light to image CF fluorescence. A to F, Leaf sheath of CF-fed leaf. G to L, Mature root above the zone of differentiation. M to R, Immature sink leaf enclosed in the whorl. S to X, Root cross-sections in the unloading domain determined by CF diffusion into cortex cells. Bars = 100 μm. [See online article for color version of this figure.]
DISCUSSION
The molecular mechanisms governing carbon partitioning in plants are largely unknown. In this paper, we cloned and characterized the maize Tdy1 gene, which functions to promote carbohydrate export from leaves. We found that Tdy1 encodes a novel protein. Orthologous proteins were identified in grasses but not in more distantly related plants. However, we identified two domains of TDY1 that are evolutionarily conserved across angiosperms and therefore may be important for the function of these proteins. Additionally, using heterologous transient expression of a fluorescent reporter protein fusion, we determined that TDY1 localizes to the ER. Furthermore, we showed that Tdy1 RNA is exclusively expressed in phloem cells. Although Tdy1 RNA is highly abundant in sink tissues, dye labeling studies suggest that Tdy1 may function in phloem loading but does not seem to play a substantial role in the transport or release phloem.
Insights into Potential TDY1 Functions Based on Protein Sequence and Transient Subcellular Localization
The deduced amino acid sequence of TDY1 does not share similarity to any proteins of known function; however, two conserved domains were identified. Intriguingly, the first conserved protein domain is located within the predicted transmembrane domain and consists of hydrophobic and noncharged amino acids. Several possibilities may explain the function of this domain. It may be that this domain represents a binding site for a lipid or lipid-soluble effector molecule(s). Alternatively, this domain could be important for protein-protein interaction (e.g. association with other membrane proteins). A third possibility for the function of this domain is that it may be a target sequence for regulated intramembrane proteolysis (Brown et al., 2000; Wolfe and Kopan, 2004). Recently, it was shown in animals, yeast, and plants that ER-localized membrane proteins can be released from the membrane by regulated proteolysis. For example, the unfolded protein stress response results in the proteolytic release of an ER membrane-tethered basic domain Leu zipper (bZIP) transcription factor, AtbZIP28 (Liu et al., 2007). This processing involves two step-wise proteolytic cleavages by the site-1 protease followed by the site-2 protease (S2P), which cleaves within the transmembrane domain. Mammalian S2P recognizes a NP dipeptide within the transmembrane domain (Ye et al., 2000a), although substitutions with some other residues are tolerated (Ye et al., 2000b). In addition, S2P requires a SR dipeptide just outside of the transmembrane domain containing the target site (Ye et al., 2000b). Interestingly, TDY1-orthologous proteins from grasses contain both a NA dipeptide within the conserved domain I and a SR dipeptide immediately following the transmembrane domain. The N residue in domain I is invariant in TDY1-related proteins, although the A is substituted by similar noncharged residues. Hence, conserved domain I might represent a protease target sequence; however, the SR dipeptide is not conserved in the closest Arabidopsis protein. This could indicate that the Arabidopsis protein is not a substrate for regulated proteolysis. If true, it may support the hypothesis that the Arabidopsis protein does not share the same function as the maize TDY1 protein. Whether the C terminus of TDY1 is released from the ER membrane in vivo remains to be determined. The C terminal cytoplasmic portion of TDY1 is unlikely to function as a putative transcriptional regulator, since expressing this part of the protein as a soluble fusion in onion cells did not lead to preferential localization in the nucleus as in the case of AtbZIP28 (Liu et al., 2007). Moreover, the C-terminal domain of TDY1 does not show any homology to known transcription factors.
The middle portion of TDY1-like proteins is the most variable and does not contain a conserved domain. Within grasses, a series of conserved Pro residues in this region may be important for secondary structure. However, the majority of these Pro residues are not conserved in the closest Arabidopsis homolog. If this protein in Arabidopsis has a similar function to maize TDY1, the middle region of the protein might serve as a spacer or possibly a hinge region.
The C terminus of TDY1 contains the second conserved domain. This domain is composed of many charged residues (13 of 27 in maize TDY1) and is likely to be located on the surface of the protein. Within domain II is the conserved sequence EAFIXK, where X is any residue. Using PSI-BLAST analysis, we found that this motif is present within a plant-specific domain of unknown function (DUF761; pfam05553). None of the proteins containing this domain have a characterized function. Two tdy1 mutant alleles encode proteins predicted to lack this domain due to a small deletion (tdy1-shadow) or an upstream frameshift mutation (tdy1-PM). These alleles confer a tdy1 mutant phenotype indistinguishable from the tdy1-D6 complete deletion allele, implying that this part of the protein is critical for its function.
Using a transient protein expression assay, we determined that TDY1 localized to the ER in onion cells. As the ER is the organelle responsible for the synthesis of proteins in the secretory system, it is possible that TDY1 functions in protein trafficking (Lee et al., 2003). The ER was found to be continuous between the CC and SE in tobacco (Nicotiana tabacum; Martens et al., 2006), and protein movement between the CC and SE was shown (Thompson and Wolniak, 2008). These data suggest that the TDY1 protein could function in both of these cell types.
An interesting parallel to TDY1 localization and potential function may be found in comparison with the maize Floury1 (Fl1) gene. FL1 is a novel plant-specific, ER-localized membrane protein that contains a domain of unknown function in its C terminus that is different from the one present in TDY1 (Holding et al., 2007). fl1 mutants are so named because they have an opaque endosperm. In addition, fl1 mutants display uniform localization of the 22-kD α-zein seed storage proteins throughout the protein bodies, in contrast to the specific localization seen in wild type. It was suggested that FL1 may act to facilitate the interaction of 22-kD α-zeins with chaperone proteins involved in their correct targeting within protein bodies. Although not related at the sequence level, it is tempting to speculate that since TDY1 has a similar protein architecture and putative localization, it might play a similar role as FL1. It is possible that TDY1 acts as a chaperone to assist in the correct protein targeting of phloem membrane/secretory proteins. Future work will characterize the functions of TDY1-interacting proteins and the localization of phloem membrane/secretory proteins in wild-type and tdy1 plants to test this hypothesis.
RNA Localization and CF Transport Studies Suggest TDY1 Functions in Phloem Loading
Several inferences can be drawn from the Tdy1 RNA localization pattern. (1) Tdy1 is expressed in protophloem cells upon differentiation, suggesting that Tdy1 function may be required as soon as these cells become competent for phloem transport. To our knowledge, Tdy1 is one of the earliest known maize genes specifically expressed in protophloem. Tdy1 expression may be useful to mark this tissue, and the Tdy1 promoter could have applications in driving gene expression from the onset of phloem function. (2) We found that Tdy1 is expressed strongly in the phloem cells of sink tissues and is also expressed in mature source leaves. This suggests that Tdy1 could function in sink and source tissues and possibly play a role in both phloem loading and unloading. However, the tdy1 carbohydrate-hyperaccumulation phenotype is only evident in leaves, and no defect in CF unloading in tdy1 mutants was observed. One explanation for these results could be that genetic redundancy masks any phenotype in sink tissues (see below). Another intriguing possibility is that the Tdy1 RNA is mobile in the phloem, which could account for the high level of expression detected in roots, ears, and other sink tissues. Analysis of where the Tdy1 promoter is transcriptionally active will be required to explore this speculation. (3) Tdy1 expression in phloem cells indicates that the decrease in chlorophyll observed in tdy1 chlorotic leaf regions is an indirect effect likely caused by excess carbohydrates repressing photosynthetic gene expression (Sheen, 1990; Goldschmidt and Huber, 1992; Krapp and Stitt, 1995; Koch, 1996). This is in agreement with an earlier study that showed that starch accumulation preceded leaf chlorosis in tdy1 emerging leaves (Braun et al., 2006).
Dye transport assays suggest that Tdy1 functions in the collection phloem but does not have an essential role in the transport or release phloem. In wild-type leaves, the high level of CF detected in the VP cells reflects solute flow from the M cells toward the vein following symplastic Suc movement. As no transporter to export the xenobiotic dye to the apoplast is likely to exist, CF accumulates in the VP cells. In contrast, in tdy1 mutant leaves, we observed that CF movement from tdy1 chlorotic tissue into the veins was retarded. CF accumulated to higher levels in both the tdy1 BS and M cells but had diminished levels in the VP cells compared with wild type. These data suggest that tdy1 chlorotic leaf regions have reduced symplastic solute flow leading to decreased phloem loading, which would explain the reduced amount of Suc, Glc, and biomass observed in tdy1 mutant roots relative to wild type. One possibility to account for these data is that some type of plasmodesmatal restriction at the BS-VP cell interface constrains symplastic flow. However, from former investigations, no alterations or blockages of the plasmodesmata were detected in tdy1 leaves (Ma et al., 2008). Thus, these data suggest that any limitation in symplastic solute movement may be caused by a partial closure of the plasmodesmata rather than by a physical obstruction. In addition, Tdy1 RNA is expressed in the CC and SE, suggesting that a BS-VP plasmodesmatal constriction would likely be an indirect and downstream effect. Further studies are necessary to establish the molecular function of TDY1 and its role in promoting Suc loading into the collection phloem.
Based on the carbohydrate hyperaccumulation and other previous phenotypic characterizations, we hypothesized that Tdy1 may function to promote carbon export from leaves, possibly by inducing SUT activity (Braun et al., 2006). However, a surprising finding was the high level of Tdy1 RNA accumulation detected in roots. Phloem unloading in maize roots occurs symplastically (Giaquinta et al., 1983) and therefore does not require SUT function. These data raise the possibility that, at least in roots, TDY1 function is independent of SUTs or that SUT regulation may be but one component of Tdy1 function. The expression data also suggest the alternative possibility that the tdy1 mutation may have uncovered only a limited aspect of Tdy1 function (i.e. no phenotype was observed in sink tissues with high levels of Tdy1 RNA due to genetic redundancy). We previously characterized an independent locus, Tdy2, with a nearly identical mutant phenotype and proposed that it may have a related function to Tdy1 (Baker and Braun, 2008). In addition, genetic modifiers of tdy1, distinct from tdy2, are currently being isolated that enhance the severity of the tdy1 phenotype and may represent genes with partially overlapping functions. Finally, sequences related to Tdy1 are present in the maize genome. Future research to characterize the functions of these genes and their overlap with Tdy1 may uncover additional roles for Tdy1 in sink tissues.
In summary, Tdy1 encodes a novel protein expressed in the phloem. Orthologous proteins are present only in grasses, but two domains conserved in angiosperms were identified. Based on the wild-type Tdy1 RNA localization pattern, the putative TDY1 protein subcellular localization, the defects in CF movement into the veins, and the carbohydrate hyperaccumulation in tdy1 chlorotic leaves, we hypothesize that Tdy1 acts in an ER-localized process that promotes phloem loading of Suc. If so, it is somewhat surprising that TDY1 sequence would not be more conserved in dicots and perhaps more distantly related vascular plants. In this respect, it is interesting that monocots and dicots contain evolutionarily distinct clades of SUTs (Sauer, 2007; Braun and Slewinski, 2009). TDY1 may function in a grass-specific aspect of phloem biology, possibly by interacting with, and having coevolved with, grass-specific SUTs or other phloem proteins.
MATERIALS AND METHODS
Growth Conditions
Maize (Zea mays) plants were grown in the summer at the Rock Springs Agronomy Farm at Pennsylvania State University. For CF transport and high-light studies, plants were grown in a growth chamber illuminated with incandescent and fluorescent light (650 μmol m−2 s−1) under an 18-h-light (28°C)/6-h-dark (25°C) regime. Plants used for RNA isolation and carbohydrate quantification experiments were grown in a greenhouse supplemented with sodium vapor and metal halide lamps at 1,400 μmol m−2 s−1 under a 16-h-day (30°C)/8-h-night (20°C) cycle. Dawn was at 6 am and dusk was at 10 pm. For carbon-starvation treatment, greenhouse-grown plants were shifted to a dark room maintained at 25°C for 3 d. For low-light growth, plants were grown under an 8-h-day (25°C)/16-h-night (20°C) cycle with 100 μmol m−2 s−1 provided by fluorescent bulbs.
Genetic Stocks, Transposon Tagging, and Mutant Alleles
The tdy1-R allele has been described previously (Braun et al., 2006). Highly Mu-active stocks were crossed by tdy1-R homozygous plants. Four new mutant alleles were recovered from approximately 50,000 F1 plants. The new alleles were repeatedly outcrossed to the B73 inbred line and to the bz1-mum9 and a1-mum2 genetic testers to segregate away unlinked transposons and monitor for loss of Mu activity. Plants were genotyped with the tightly linked molecular marker umc1653 to distinguish between the tdy1-R, new Mu-insertion, and wild-type alleles. After five generations of outcrossing, plants heterozygous for a new Mu allele were crossed by homozygous tdy1-R plants to generate families used for cosegregation analyses.
DNA was extracted from tdy1 mutant and wild-type plants, digested with BamHI, separated on a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with a series of Mu-specific probes. An approximately 5.5-kb Mu1 fragment that perfectly cosegregated with the tdy1-D190 mutant allele was identified. DNA from this size region was isolated from a preparative gel and ligated into BamHI-cut λZAP Express vector (Stratagene). The subgenomic DNA library was screened with the Mu1 probe, and the approximately 5.5-kb Mu1-containing plasmid was recovered and sequenced. Other alleles were found to contain Mu insertions by PCR amplification with a gene-specific primer and a Mu primer facing out from the terminal inverted repeat (see Supplemental Table S3 for all primers). Alleles not containing a Mu insertion were amplified by PCR and sequenced using gene-specific primers. Tdy1 PCR typically contained 10% dimethyl sulfoxide and 3% glycerol due to the high GC content, and all sequencing was performed by the Pennsylvania State University Nucleic Acids Facility.
TDY1 Sequence Alignments
Tdy1 DNA and protein sequences were used to query public EST, genomic DNA, and protein databases by BLAST analyses (Altschul et al., 1990). Protein alignment was performed using Align X (Vector NTI; Invitrogen).
Protein Subcellular Localization
Tdy1 contains no introns. The protein-coding region was PCR amplified from B73 genomic DNA using a proofreading DNA polymerase (Pfu; Stratagene), cloned into the pENTR/SD/D-TOPO vector (Invitrogen), and sequenced. To generate YFP translational fusions to TDY1, the entry clone was recombined with the pEarleyGate 101 destination vector (Earley et al., 2006). For RFP fusions, a modified pEarleyGate vector containing mCherry was used (Skirpan et al., 2008). Construction of the N-terminal 71-amino acid or C-terminal 201-amino acid fusions was similarly performed. Constructs were transformed into onion (Allium cepa) bulb epidermal cells by biolistic particle bombardment using a Bio-Rad PDS-1000/He system according to the manufacturer's protocol. Fluorescent proteins were observed using a Nikon Eclipse 80i fluorescence microscope with a 100-W mercury lamp, and images were captured using a DXM1200F digital camera. YFP/GFP were imaged using a 465- to 495-nm excitation filter and a 515- to 555-nm band-pass emission filter. RFP was imaged using a 530- to 560-nm excitation filter and a 590- to 650-nm band-pass emission filter. Control experiments determined that no YFP/GFP fluorescence was observed using the RFP filter set, nor was RFP detected using the YFP/GFP filter set (data not shown).
RT-PCR and RNA in Situ Hybridization Analyses
RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) from fully mature adult leaves, young, etiolated, immature leaves not emerged from the whorl, approximately 2.5-cm-long developing ears and tassels, and 1-week-old seedling roots from seeds germinated in the dark on moist paper towels. RNA was treated with RQ1 DNAse (Promega), the integrity and purity were assessed by agarose gel electrophoresis, and the concentration was determined with a NanoDrop spectrophotometer (Thermo Scientific). cDNA was synthesized following the manufacturer's instructions from 1 μg of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For semiquantitative RT-PCR assays, 1 μL of cDNA was used as template in a PCR with 20 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. The products were separated on a 1.5% agarose gel, transferred to a nylon membrane, hybridized with a corresponding 32P-labeled DNA probe, and washed at high stringency, and the blots were exposed to x-ray film. For qRT-PCR assays, the cDNA was diluted 5-fold and used as templates in an ABI 7500 Fast Real-Time PCR System with SYBR Green detection. In each qRT-PCR experiment, similar threshold cycle values of the reference genes were observed across all samples, indicating their suitability as appropriate standards. To test sugar induction of Tdy1 expression, Ubq was used as the reference gene. However, Gapdh was used as the reference gene to test Tdy1 regulation by carbohydrate depletion, as we found that the threshold cycle values for Ubq varied between treatments. Melting curve and gel electrophoresis analyses determined that a single product of the expected size was amplified for all PCR. Three independent RNA isolations (biological replicates) were analyzed with three technical replicates each for qRT-PCR experiments. As a negative control, separate reactions containing RNA isolated from tdy1-D6 leaves were performed in all qRT-PCR experiments to ensure specificity and that no contamination occurred. The relative quantification was calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). As a positive control for sugar-induced gene expression, standard RT-PCR was performed on G6PT using the same cDNA pools with the following conditions: 95°C for 20 s, 60°C for 20 s, 72°C for 20 s, 32 cycles. To independently verify the normalization of the cDNA pools, β-tubulin primers were used with the same amplification conditions except for 31 cycles.
For RNA in situ hybridization studies, an approximately 0.3-kb fragment containing the 3′ end of the Tdy1 coding and untranslated regions was subcloned into the pGEM-T Easy vector (Promega). The plasmid was linearized with NdeI and transcribed with T7 RNA polymerase to produce a digoxigenin-labeled antisense probe. To produce the sense strand probe, the plasmid was linearized with NcoI and transcribed with SP6 RNA polymerase. RNA in situ hybridization was carried out as described (Jackson et al., 1994; Langdale, 1994). To identify phloem cell types, tissue sections were stained with DAPI (1 μg mL−1 in water) or aniline blue (0.05% [w/v] in potassium phosphate buffer, pH 9.0) and viewed under UV light. DAPI was visualized with a 340- to 380-nm excitation filter and a 435- to 485-nm band-pass emission filter. Aniline blue was imaged with a 360- to 370-nm excitation filter and a 420-nm long-pass emission filter.
CF Transport Assays
To monitor CF symplastic movement, the adaxial surface of mature leaves was gently abraded with sandpaper, and 75 μL of 50 μg mL−1 CF prepared according to Grignon et al. (1989) was applied for 1 h as described (Haupt et al., 2001; Botha, 2005). To measure CF transport rates in leaves, the fourth or fifth mature leaf from 3- to 4-week-old plants was abraded 200 mm distal to the ligule, 50 μL of CF was applied, and the application site was covered with Parafilm. Preliminary experiments examined CF movement at multiple time points after application. One hour was selected as the optimal assay time as the dye had not exited the leaf and was predominantly present in phloem cells 2 to 3 cm proximal to the application site at that time. After 1 h, the leaf was dissected and analyzed by free-hand cross-sections to assess dye movement in the phloem. For both wild-type and tdy1 plants, three leaves were assayed and the experiment was repeated three times. Data for one replicate are presented. For studies of CF movement in the transport and release phloem, 50 μg mL−1 CFDA (Invitrogen) was fed to the cut end of mature leaf 4 as described (Aoki et al., 2004). Leaf sheaths and roots were examined at 3 h after feeding, and immature sink leaves were inspected after 6 h. To visualize CF, free-hand sections were examined using either a 450- to 490-nm excitation filter and a 515-nm long-pass emission filter or a 465- to 495-nm excitation filter and a 515- to 555-nm band-pass emission filter to reduce tissue autofluorescence.
Starch Staining, Carbohydrate Quantification, and Root Mass Determination
Starch was detected in cleared leaves and roots by staining with iodine potassium iodide (Braun et al., 2006). Carbohydrates were quantified according to Dinges et al. (2001) from 100 mg of root tissue harvested from the middle of the root system of 10-d-old seedlings (n = 3). Tissue was collected every 4 h over a diurnal cycle, and the experiment was replicated three times. Data presented are for all measurements. Root mass was determined by weighing entire root systems isolated at the seed attachment point of 10-d-old seedlings (n = 10). For both high- and low-light-grown plants, the experiment was replicated twice with similar results, and one repeat is shown.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number FJ376984.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of Arabidopsis TDY1-like proteins.
Supplemental Figure S2. Time course of TDY1-YFP localization.
Supplemental Figure S3. Tdy1 RT-PCR of wild-type and tdy1-D6 tissues.
Supplemental Figure S4. G6PT induction in tdy1 yellow tissue.
Supplemental Figure S5. Fru and starch levels in wild-type and tdy1 roots.
Supplemental Table S1. tdy1 allele characterization.
Supplemental Table S2. Genes deleted in the tdy1-D6 allele.
Supplemental Table S3. List of primers used.
Supplementary Material
Acknowledgments
We thank Tony Omeis and Scott Harkcom for excellent plant care. We thank Mike Freeling for support during the early stages of this work and for providing the Mu-active stocks. We appreciate the generosity of several Maize Genetics Cooperators in donating tdy1 alleles: Paula McSteen (tdy1-PM), Chris Carson (tdy1-shadow), and Masaharu Suzuki and Don McCarty (tdy1-UMu, Uniform Mu). We thank Stephen Howell for the suggestion of regulated intramembrane proteolysis. We thank Michael Goodin for the ER-GFP construct, Paula McSteen for guidance with the in situ hybridizations, and Sarah Swanson, Simon Gilroy, and Richard Cyr for assistance with fluorescent protein imaging experiments. We are especially grateful to Dan Bush, two anonymous reviewers, and members of the Braun and McSteen laboratories for discussions of the data and comments on the manuscript.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant nos. 2008–35304–04597 and 2004–35304–14948 to D.M.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David M. Braun (dbraun@psu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Aoki N, Scofield GN, Wang XD, Patrick JW, Offler CE, Furbank RT (2004) Expression and localisation analysis of the wheat sucrose transporter TaSUT1 in vegetative tissues. Planta 219 176–184 [DOI] [PubMed] [Google Scholar]
- Baker RF, Braun DM (2007) tie-dyed1 functions non-cell autonomously to control carbohydrate accumulation in maize leaves. Plant Physiol 144 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RF, Braun DM (2008) tie-dyed2 functions with tie-dyed1 to promote carbohydrate export from maize leaves. Plant Physiol 146 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J, Springer PS, Cresse AD, Hendrickx M (1993) Specificity and regulation of the Mutator transposable element system in maize. Crit Rev Plant Sci 12 57–95 [Google Scholar]
- Botha CEJ (2005) Interaction of phloem and xylem during phloem loading: functional symplasmic roles for thin- and thick-walled sieve tubes in monocotyledons. In NM Holbrook, MA Zwieniecki, eds, Vascular Transport in Plants. Elsevier Academic Press, Amsterdam, pp 115–130
- Braun DM, Ma Y, Inada N, Muszynski MG, Baker RF (2006) tie-dyed1 regulates carbohydrate accumulation in maize leaves. Plant Physiol 142 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Slewinski TL (2009) Genetic control of carbon partitioning in grasses: roles of Sucrose Transporters and Tie-dyed loci in phloem loading. Plant Physiol 149 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Ye J, Rawson RB, Goldstein J (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100 391–398 [DOI] [PubMed] [Google Scholar]
- Bürkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB (1998) The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol 118 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush DR (1993) Proton-coupled sugar and amino acid transporters in plants. Annu Rev Plant Physiol Plant Mol Biol 44 513–542 [Google Scholar]
- Chandler VL, Hardeman KJ (1992) The Mu elements of Zea mays. Adv Genet 30 77–122 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Geiger D, Fromm J, Koroleva O, Ache P, Langenfeld-Heyser R, Sauer N, May S, Hedrich R (2002) Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216 334–344 [DOI] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, Myers AM, James MG (2001) Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol 125 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Esau K (1943) Ontogeny of the vascular bundles in Zea mays. Hilgardia 15 327–368 [Google Scholar]
- Esau K (1977) Anatomy of Seed Plants, Ed 2. John Wiley & Sons, New York
- Evert RF, Eschrich W, Heyser W (1978) Leaf structure in relation to solute transport and phloem loading in Zea mays L. Planta 138 279–294 [DOI] [PubMed] [Google Scholar]
- Evert RF, Russin WA, Bosabalidis AM (1996) Anatomical and ultrastructural changes associated with sink-to-source transition in developing maize leaves. Int J Plant Sci 157 247–261 [Google Scholar]
- Fritz E, Evert RF, Heyser W (1983) Microautoradiographic studies of phloem loading and transport in the leaf of Zea mays L. Planta 159 193–206 [DOI] [PubMed] [Google Scholar]
- Fritz E, Evert RF, Nasse H (1989) Loading and transport of assimilates in different maize leaf bundles: digital image analysis of 14C microautoradiographs. Planta 178 1–9 [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Plant comparative genetics after 10 years. Science 282 656–659 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K (2007) Plant proton pumps. FEBS Lett 581 2204–2214 [DOI] [PubMed] [Google Scholar]
- Giaquinta RT, Lin W, Sadler NL, Franceschi VR (1983) Pathway of phloem unloading of sucrose in corn roots. Plant Physiol 72 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Park G, Amtmann A, Sanders D, Maathuis FJM (2006) Arabidopsis thaliana Cyclic Nucleotide Gated Channel 3 forms a non-selective ion transporter involved in germination and cation transport. J Exp Bot 57 791–800 [DOI] [PubMed] [Google Scholar]
- Goldschmidt EE, Huber SC (1992) Regulation of photosynthesis by end-product accumulation in leaves of plants storing starch, sucrose, and hexose sugars. Plant Physiol 99 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJR, Braun D, Meeley R, Cande WZ (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119 3306–3315 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P (2006) Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J Plant Res 119 115–123 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Chakrabarty R, Yelton S, Martin K, Clark A, Brooks R (2007) Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant-adapted rhabdovirus. J Gen Virol 88 1810–1820 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignon N, Touraine B, Durand M (1989) 6(5)Carboxyfluorescein as a tracer of phloem sap translocation. Am J Bot 76 871–877 [Google Scholar]
- Haupt S, Duncan GH, Holzberg S, Oparka KJ (2001) Evidence for symplastic phloem unloading in sink leaves of barley. Plant Physiol 125 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstra G, Nelson C (1969) The translocation of photosynthetically assimilated 14C in corn. Can J Bot 47 1435–1442 [Google Scholar]
- Holding DR, Otegui MS, Li B, Meeley RB, Dam T, Hunter BG, Jung R, Larkins BA (2007) The maize Floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413 [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47 509–540 [DOI] [PubMed] [Google Scholar]
- Krapp A, Stitt M (1995) An evaluation of direct and indirect mechanisms for the “sink-regulation” of photosynthesis in spinach: changes in gas exchange, carbohydrates, metabolites, enzyme activities and steady-state transcript levels after cold-girdling source leaves. Planta 195 313–323 [Google Scholar]
- Kühn C, Quick WP, Schulz A, Riesmeier JW, Sonnewald U, Frommer WB (1996) Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant Cell Environ 19 1115–1123 [Google Scholar]
- Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW (2003. a) Phloem loading and unloading of sugars and amino acids. Plant Cell Environ 26 37–56 [Google Scholar]
- Lalonde S, Weise A, Walsh R, Ward J, Frommer W (2003. b) Fusion to GFP blocks intercellular trafficking of the sucrose transporter SUT1 leading to accumulation in companion cells. BMC Plant Biol 3 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Wipf D, Frommer WB (2004) Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol 55 341–372 [DOI] [PubMed] [Google Scholar]
- Langdale J (1994) In situ hybridization. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 165–180
- Lee JY, Yoo BC, Rojas MR, Gomez-Ospina N, Staehelin LA, Lucas WJ (2003) Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299 392–396 [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55 1221–1230 [DOI] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57 203–232 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Furbank RT (1999) Tansley Review No. 105. Sucrose biosynthesis in C4 plants. New Phytol 143 221–237 [Google Scholar]
- Ma S, Quist TM, Ulanov A, Joly R, Bohnert HJ (2004) Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. Plant J 40 845–859 [DOI] [PubMed] [Google Scholar]
- Ma Y, Baker RF, Magallanes-Lundback M, DellaPenna D, Braun DM (2008) Tie-dyed1 and Sucrose export defective1 act independently to promote carbohydrate export from maize leaves. Planta 227 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A (2006) Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiol 142 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher LM, Miao L, Sinha N, Lucas WJ (2001) Sucrose export defective1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell 13 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom-Hodgkins W, Vaughn M, Bush D (2003) Protein phosphorylation plays a key role in sucrose-mediated transcriptional regulation of a phloem-specific proton-sucrose symporter. Planta 217 483–489 [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB (1994) Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J 13 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SH, Evert RF (1985) Leaf vasculature in Zea mays L. Planta 164 448–458 [DOI] [PubMed] [Google Scholar]
- Russin WA, Evert RF, Vanderveer PJ, Sharkey TD, Briggs SP (1996) Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. Plant Cell 8 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N (2007) Molecular physiology of higher plant sucrose transporters. FEBS Lett 581 2309–2317 [DOI] [PubMed] [Google Scholar]
- Sharman BC (1942) Developmental anatomy of the shoot of Zea mays L. Ann Bot (Lond) 6 245–284 [Google Scholar]
- Sheen J (1990) Metabolic repression of transcription in higher plants. Plant Cell 2 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan A, Wu X, McSteen P (2008) Genetic and physical interaction suggest that BARREN STALK1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J 55 787–797 [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Ma Y, Baker RF, Huang M, Meeley R, Braun DM (2008) Determining the role of Tie-dyed1 in starch metabolism: epistasis analysis with a maize ADP-glucose pyrophosphorylase mutant lacking leaf starch. J Hered 99 661–666 [DOI] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG (2008) Functional characterization of the Arabidopsis thaliana AtSUC2 Suc/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 147 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N (2005) Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J 41 319–331 [DOI] [PubMed] [Google Scholar]
- Thompson MV, Wolniak SM (2008) A plasma membrane-anchored fluorescent protein fusion illuminates sieve element plasma membranes in Arabidopsis and tobacco. Plant Physiol 146 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJ (2003) Phloem, a miracle of ingenuity. Plant Cell Environ 26 125–149 [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS, Kopan R (2004) Intramembrane proteolysis: theme and variations. Science 305 1119–1123 [DOI] [PubMed] [Google Scholar]
- Wright K, Oparka KJ (1996) The fluorescent probe HPTS as a phloem-mobile, symplastic tracer: an evaluation using confocal laser scanning microscopy. J Exp Bot 47 439–445 [Google Scholar]
- Ye J, Dave U, Grishin N, Goldstein J, Brown M (2000. a) Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc Natl Acad Sci USA 97 5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL (2000. b) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6 1355–1364 [DOI] [PubMed] [Google Scholar]
- Yu F, Fu A, Aluru M, Park S, Xu Y, Liu H, Liu X, Foudree A, Nambogga M, Rodermel S (2007) Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ 30 350–365 [DOI] [PubMed] [Google Scholar]
- Zhao R, Dielen V, Kinet JM, Boutry M (2000) Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.