Abstract
Plants employ sophisticated mechanisms to recycle intracellular constituents needed for growth, development, and survival under nutrient-limiting conditions. Autophagy is one important route in which cytoplasm and organelles are sequestered in bulk into vesicles and subsequently delivered to the vacuole for breakdown by resident hydrolases. The formation and trafficking of autophagic vesicles are directed in part by associated conjugation cascades that couple the AUTOPHAGY-RELATED8 (ATG8) and ATG12 proteins to their respective targets, phosphatidylethanolamine and the ATG5 protein. To help understand the importance of autophagy to nutrient remobilization in cereals, we describe here the ATG8/12 conjugation cascades in maize (Zea mays) and examine their dynamics during development, leaf senescence, and nitrogen and fixed-carbon starvation. From searches of the maize genomic sequence using Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) counterparts as queries, we identified orthologous loci encoding all components necessary for ATG8/12 conjugation, including a five-member gene family expressing ATG8. Alternative splicing was evident for almost all Atg transcripts, which could have important regulatory consequences. In addition to free ATG8, its membrane-associated, lipidated form was detected in many maize tissues, suggesting that its conjugation cascade is active throughout the plant at most, if not all, developmental stages. Levels of Atg transcripts and/or the ATG8-phosphatidylethanolamine adduct increase during leaf senescence and nitrogen and fixed-carbon limitations, indicating that autophagy plays a key role in nutrient remobilization. The description of the maize ATG system now provides a battery of molecular and biochemical tools to study autophagy in this crop under field conditions.
The recycling of intracellular constituents is critical to all organisms for optimal growth and development and survival under nutrient-limiting conditions. This recycling is especially important throughout a plant's life cycle, especially as plants scavenge available nutrients from storage tissues and older senescing leaves for the synthesis of reproductive organs and new leaves and during whole plant senescence as fixed carbon (C) and nitrogen (N) are mobilized to reproductive or storage organs (Vierstra, 1996; Hopkins et al., 2007; Lim et al., 2007). The process is initiated in source tissues by dramatic changes in gene expression, in which genes involved in basic metabolism, including photosynthesis and protein biosynthesis, are down-regulated while those involved in programmed cell death and the stress response and/or encoding various hydrolytic enzymes are up-regulated (Hopkins et al., 2007; Lim et al., 2007). Nutrient mobilization in plants, as in yeast and animals, involves a battery of catabolic processes, including a sophisticated recycling system called autophagy or “self-eating.” Here, cytoplasm and organelles are sequestered in bulk into special autophagic vesicles, which then deliver their contents to the vacuole (or lysosome) for breakdown by resident hydrolases (Thompson and Vierstra, 2005; Bassham, 2007). The release products can be either consumed by the host cell or transported to other tissues and organs for reuse.
In yeast (Saccharomyces cerevisiae) and animal cells, autophagy (also referred to as macroautophagy) begins with the formation of a sequestering membrane called the phagophore (or isolation membrane), which engulfs portions of the cytoplasm to form the double membrane-bound autophagosome vesicle (Ohsumi, 2001; Klionsky, 2007; Mizushima, 2007). The outer membrane of the autophagosome fuses with the membrane of the lytic compartment (i.e. vacuole in yeast and lysosome in animals), and the inner vesicle is released into the lytic compartment as an autophagic body. Both the limiting membrane and contents of the autophagic body are then degraded by a cache of hydrolases. While a number of studies have identified autophagic bodies and autophagosome-like compartments in plants, including those containing various organelles (Moriyasu et al., 2003; Yoshimoto et al., 2004; Inoue et al., 2006), the dynamics of the phagophore remain elusive.
Despite its relevance to the C/N economy of plants, we are only now beginning to appreciate autophagy at the molecular level. Key to this understanding was the discovery of an elaborate recycling system in yeast involving a suite of autophagy-related (ATG) proteins (Ohsumi, 2001; Thompson and Vierstra, 2005; Klionsky, 2007; Mizushima, 2007). Important factors include the ATG1 kinase complex, which responds to various nutrient sensors such as the target-of-rapamycin and Snf1 kinases, and the downstream ATG6/Beclin1 phosphatidylinositol 3-kinase complex. These kinases then modulate two ubiquitin (Ub)-like conjugation cascades, which work sequentially to help assemble the engulfing phagophore, fusion of the autophagosome with the vacuolar membrane, and the subsequent deposition of autophagic bodies into the vacuolar lumen.
The conjugation cascades center on two Ub-fold proteins, ATG8 and ATG12, which become covalently attached to their respective targets, the lipid phosphatidylethanolamine (PE) and the ATG5 protein, by a process mechanistically similar to ubiquitination (Ohsumi, 2001). Through a common ATP-dependent E1 activating enzyme, ATG7, both ATG8 and ATG12 become adenylated at their C-terminal glycines and then are bound by a thiolester bond to the active-site Cys in ATG7. Activated ATG8 and ATG12 are then donated by transesterification to the active-site Cys in their respective E2 conjugating enzymes, ATG3 and ATG10. Finally, ATG8 and ATG12 are linked via their C termini to PE and ATG5 through peptide bonds. For ATG8, the amino group of PE is the attachment site, whereas for ATG12, the ɛ-amino group of a conserved Lys in ATG5 is the attachment site. The ATG4 processing protease is also essential for ATG8 activity; it exposes the C-terminal Gly in the longer ATG8 precursor and recycles mature ATG8 by releasing it from the ATG8-PE adduct, presumably before fusion of the autophagosome with the vacuolar membrane.
The biochemical roles of ATG8-PE and ATG12-ATG5 conjugates have recently been revealed. Based on cell-free studies, the ATG8-PE adduct appears to promote the expansion of autophagosomal membrane and hemifusion of this membrane with the vacuolar membrane (Nakatogawa et al., 2007). ATG8-PE assembly is stimulated in vitro by a complex containing the ATG12-ATG5 adduct and ATG16, suggesting that the ATG12-ATG5 adduct has an E3-like catalytic role in ATG8 conjugation (Hanada et al., 2007; Fujita et al., 2008). Whereas the ATG12-ATG5 conjugate is released from the autophagosome before deposition in the vacuole, at least some ATG8-PE enters the vacuole attached to the autophagic body and is catabolized.
Over the past few years, obvious orthologs of various yeast ATG proteins have been detected in both higher and lower plants, indicating that a similar autophagic system likely exists throughout the plant kingdom. The Arabidopsis (Arabidopsis thaliana) genome, for example, encodes proteins related to subunits of the yeast ATG1 kinase and ATG6/Beclin1 phosphatidylinositol 3-kinase complexes and all components of the ATG8/12 conjugation cascades, including small gene families encoding the two tags (Doelling et al., 2002; Hanaoka et al., 2002; Surpin et al., 2003; Yoshimoto et al., 2004; Liu et al., 2005; Inoue et al., 2006; Fujiki et al., 2007; Xiong et al., 2007; Phillips et al., 2008; A. Suttangkakul and R.D. Vierstra, unpublished data). Using a recombinant system, Fujioka et al. (2008) recently succeeded in assembling a functional Arabidopsis ATG8 conjugation cascade in vitro by adding ATP to reactions containing ATG7, ATG3, and liposomes prepared with PE. Reverse genetic analyses of several Arabidopsis components have linked the ATG system, and by inference autophagy, to senescence, survival under N- and fixed-C-limiting growth conditions, the stress response, and programmed cell death triggered by pathogen resistance (Liu et al., 2005; Xiong et al., 2007; Phillips et al., 2008). For instance, plants missing ATG4a/b, ATG5, ATG7, or ATG10 senesce faster and are hypersensitive to N starvation and extended darkness that limits the availability of fixed C (Doelling et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008). Using GFP-ATG8 fusions as markers, it was also shown that these atg mutants block the vacuolar accumulation of autophagic bodies during N and C stress (Yoshimoto et al., 2004; Sláviková et al., 2005; Thompson et al., 2005; Yano et al., 2007; Phillips et al., 2008).
While our understanding of the Arabidopsis ATG system is progressing, little is currently known about this system or autophagy in other plant species. Given the potential significance of autophagic recycling to many agriculturally relevant processes, including seed germination, N and C mobilization from leaves into seeds, fruits, and roots, survival under suboptimal growth conditions, pathogen protection, and senescence (Liu et al., 2005; Thompson and Vierstra, 2005; Bassham, 2007), information is especially needed for crop plants. Such knowledge may reveal important control points in autophagy that could be manipulated in both food and biofuel crops to enhance nutrient use efficiency or to better allocate C and N to specific organs for improved yield. As the first step toward understanding the importance of autophagy in crops, we describe here a collection of components that participate in the ATG8/12 conjugation cascades in both rice (Oryza sativa) and maize (Zea mays) and characterize the expression of the maize Atg genes and the lipidation of ATG8 during development and in response to nutrient limitations. The accumulation of the ATG8-PE adduct in particular correlates with senescence and N availability in maize, suggesting that it could be a sensitive indicator of a crop's nutritional status.
RESULTS
Identification of Rice and Maize Atg Genes
Previous genome searches of Arabidopsis identified a suite of proteins structurally and functionally related to many ATG components present in yeast (Thompson and Vierstra, 2005; Bassham, 2007). To facilitate the search for maize orthologs, we first identified a collection of Atg genes from the more complete and better annotated japonica rice cDNA and genomic databases, using the yeast and Arabidopsis counterparts as queries (Table I This rice collection contained easily recognizable relatives (typical E values of less than e−50 by TBLASTN) of most Arabidopsis ATG genes, with a number of components similarly encoded by small gene families (e.g. five rice loci encoding ATG8 versus nine loci in Arabidopsis). The Arabidopsis and rice collections were then used as queries in TBLASTN searches of the various DNA sequence databases for the maize B73 inbred, including the cDNA, EST, EST assembly (e.g. ZmGI), genome survey sequence, and genomic assembly (e.g. AZM) databases. In most cases, alignments of the cDNA and genomic sequences were used to establish the Zm Atg gene models. However, in the absence of mRNA sequences, further BLASTX alignments with Arabidopsis, rice, and yeast ATG proteins were used to predict the full-length protein sequences in the Zm Atg gene models. As high-throughput B73 genome sequences became available later, gaps in the Zm Atg gene models were eliminated by sequence analysis of corresponding bacterial artificial chromosome (BAC) clones (Table II).
Table I.
Collection of rice Atg genes
N.S., Not significantly similar in a pairwise BLAST analysis.
| Gene | TIGR Locus | GenBank Accession No. for Corresponding cDNA | No. of Amino Acid Residues | Identity/Similarity to Yeast; Arabidopsisa |
|---|---|---|---|---|
| Atg3a | Os01g10290 | AK067737 | 314 | 32/51%; 73/86% |
| Atg3b | Os10g41110 | AK241567 | 314?b | 46/56%; 77/90% |
| Atg4a | Os03g27350 | AK242832 | 474 | 27/39%; 51/68% |
| Atg4b | Os04g58560 | AK069012 | 478 | 26/43%; 50/67% |
| Atg5 | Os02g02570 | AK063557 | 380 | 21/41%; 52/67% |
| Atg7 | Os01g42850 | AK067422 | 1,042 | 29/48%; 47/63% |
| Atg8a | Os07g32800 | AK059939 | 119 | 73/92%; 84/94% |
| Atg8b | Os04g53240 | AK121268 | 119 | 74/91%; 84/92% |
| Atg8c | Os08g09240 | AK121169, AK062573 | 120 | 74/91%; 87/95% |
| Atg8d | Os11g01010 | CI298209, CF992091, CI034410, EE591899 | 118 | 51/74%; 53/71% |
| Atg8e | Os02g32700 | No supporting EST | 107?b | 52/78%; 51/75% |
| Atg10a | Os04g41990 | AK241831 | 198?b | N.S.; 42/56% |
| Atg10b | Os12g32210 | AK099684 | 110 | N.S.; 33/55% |
| Atg12 | Os06g10340 | AK243073 | 93 | 35/67%; 76/88% |
Sequence compared with the single gene product in yeast and the a isoform from Arabidopsis.
In the absence of supporting full-length cDNA or EST data, the amino acid sequence length was predicted by comparing the corresponding genomic sequence with the amino acid sequence of its paralogous protein.
Table II.
Collection of maize Atg genes
N.S., Not significantly similar in a pairwise BLAST analysis.
| Gene | BAC Clones | ZmGI No. | No. of Amino Acid Residues | Identity/Similarity to Yeast; Arabidopsis; Ricea |
|---|---|---|---|---|
| Atg3 | AC204922, AC197919 | TC316720 | 311 | 33/49%; 71/84%; 89/94% |
| Atg4a | AC190579 | TC324693 | 492 | 28/43%; 50/67%; 75/81% |
| Atg4b | AC213882 | TC336458 | 492 | 29/46%; 50/66%; 73/81% |
| Atg5 | AC197692 | TC337283 | 374 | 23/44%; 50/68%; 76/85% |
| Atg7 | AC193430 | TC366712 | 1,021 | 30/49%; 48/63%; 74/83% |
| Atg8a | AC204058 | TC329822 | 119 | 73/90%; 85/94%; 94/97% |
| Atg8b | AC202429 | TC326171 | 120 | 74/91%; 86/94%; 86/96% |
| Atg8c | AC195583 | TC352053 | 120 | 74/91%; 86/94%; 86/96% |
| Atg8d | AC198749 | TC337655 | 119 | 75/92%; 86/94%; 88/96% |
| Atg8e | TC330328 | 119 | 75/92%; 85/94%; 88/96% | |
| Atg10 | AC205481 | TC320991 | 215 | N.S.; 44/60%; 63/74% |
| Atg12 | AC188134 | TC368886 | 91 | 36/60%; 82/89%; 77/89% |
Sequence compared with the single gene product in yeast and the a isoform from Arabidopsis and rice.
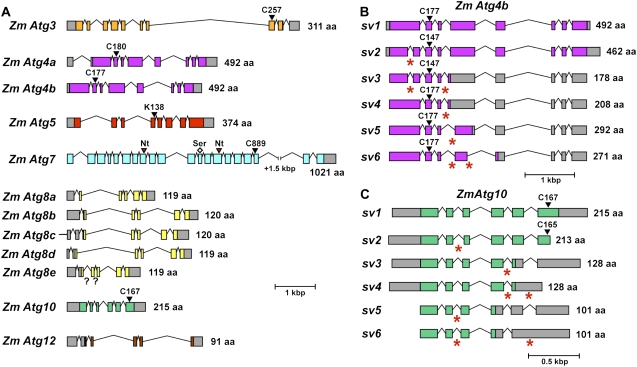
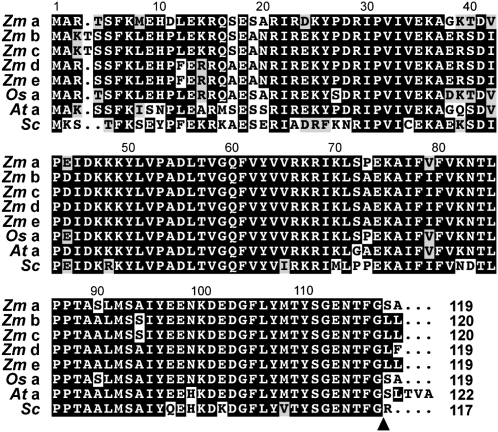
As shown in Figure 1A and Table II, this survey identified five maize loci encoding the ATG8 protein tag, two encoding the ATG4 processing protease, and one gene each encoding the ATG12 protein tag, the common ATG7 E1 activating enzyme, the ATG3 and ATG10 E2 conjugating enzymes specific for ATG8 and ATG12, respectively, and the ATG5 target of ATG12. For almost all loci, strong amino acid sequence conservation was evident when compared with their rice, Arabidopsis, and yeast orthologs (Table II). For example, the five maize Atg8 genes (Zm Atg8a to Zm Atg8e), which are 100% to 95%/100% to 84% similar/identical to each other, are 94%/86% to 85% and 92% to 90%/75% to 73% similar/identical to Arabidopsis ATG8a and yeast ATG8, respectively (Fig. 2; Table II). The Zm Atg8b and Zm Atg8c loci are on different chromosomes but encode identical proteins, suggesting that this pair arose by a recent gene duplication. Furthermore, key amino acids necessary for the function of each component were detected, indicating that the maize conjugation cascades are mechanistically identical to those present in yeast and Arabidopsis. These residues include the active-site Cys residues in ATG3, ATG7, and ATG10 that form the thiolester adducts with ATG8 and/or ATG12, the Lys acceptor site in ATG5, the positionally conserved Glys in ATG8 and ATG12 that directly participate in covalent binding, and the Cys essential for the proteolytic activity of ATG4 (Fig. 1A).
Figure 1.
Description of the maize Atg genes that participate in the ATG8/12 conjugation cascades. A, Diagram of the maize Atg genes. Colored and gray boxes represent coding and untranslated regions, respectively. Solid lines represent introns, and question marks indicate sequence gaps in introns. The predicted amino acid (aa) length for each of the corresponding proteins is shown at right. The conserved active-site Cys residues (C) for ATG3, ATG4a, ATG4b, ATG7, and ATG10 and the conserved Lys (K) acceptor site in ATG5 for ATG12 attachment are indicated by the black arrowheads. The predicted nucleotide-binding pockets (Nt) in ATG7 are represented by red arrowheads. The diamond indicates the Ser residue within the degenerated active site in the first duplicated C-terminal domain of ATG7 (see Fig. 3). B and C, Splice variants (SVs) of maize Atg4b (B) and Atg10 (C). Red asterisks represent alternative 5′ (for SV6 in Atg4b and SV3 and SV4 in Atg10) or 3′ (for SV3, SV4, SV5, and SV6 in Atg4b and SV2, SV5, and SV6 in Atg10) splice sites or sites of intron retention (for SV2 and SV3 in Atg4b and SV4 and SV6 in Atg10). See Supplemental Table S1 for EST and cDNA sequences supporting each SV.
Figure 2.
Amino acid sequence comparisons of the five maize (Zm) ATG8 isoforms with rice (Os) ATG8a, Arabidopsis (At) ATG8a, and yeast (Sc) ATG8. Identical and similar amino acids are in white letters and gray boxes, respectively. The processing site by the ATG4 protease that exposes the C-terminal Gly of ATG8 for lipidation is identified by the arrowhead. The number of amino acids for each protein is indicated at the end of the sequence.
Our maize gene models were further validated by sequence analysis of a collection of transcripts for each Zm Atg gene (Table II). With the exception of Zm Atg5, for which we were unable to generate a cDNA by reverse transcription (RT)-PCR, presumably due to the high GC content of its 5′ end, cDNAs encompassing the full coding region for each locus were obtained. These cDNA sequences not only confirmed that each Zm Atg gene is transcriptionally active but also identified a number of splice variants for many loci (Fig. 1, B and C; Table III; Supplemental Table S1). The number of Zm Atg genes with splice variants (11 of 12 genes) was higher than for Arabidopsis (seven of 17) or rice (11 of 14; Table III). The average number of splice variants per gene was also higher in maize, suggesting that alternative pre-mRNA splicing is more extensive in this species.
Table III.
Splice variants (SVs) of selected Arabidopsis (At), rice (Os), and maize (Zm) Atg genes
| Gene | At | SVs without the Essential Codona | Os | SVs without the Essential Codona | Zm | SVs without the Essential Codona |
|---|---|---|---|---|---|---|
| Atg3(a)b | 1 | – | 3 | SV2, SV3 | 2 | SV2 |
| Atg3bb | – | – | 2 | SV1, SV2 | – | – |
| Atg4a | 2 | None | 2 | SV2 | 2 | None |
| Atg4b | 3 | None | 1 | – | 6 | None |
| Atg5 | 1 | – | 2 | None | 2 | None |
| Atg7 | 1 | – | 5 | SV2, SV3, SV4, SV5 | 1 | – |
| Atg8a | 2 | None | 2 | SV2 | 2 | SV2 |
| Atg8b | 2 | None | 4 | SV4 | 4 | SV3, SV4 |
| Atg8c | 1 | – | 3 | SV2, SV3 | 3 | SV2, SV3 |
| Atg8d | 1 | – | 3 | SV2, SV3 | 4 | SV2?, SV3, SV4 |
| Atg8e | 2 | None | 1 | – | 3 | None |
| Atg8f | 2 | None | – | – | – | – |
| Atg8g | 1 | – | – | – | – | – |
| Atg8h | 1 | – | – | – | – | – |
| Atg8i | 1 | – | – | – | – | – |
| Atg10(a)b | 2 | None | 1 | – | 6 | SV3, SV4, SV5, SV6 |
| Atg10bb | – | – | 2 | SV1 | – | – |
| Atg12a | 1 | – | 2 | SV2 | 5 | SV2, SV3, SV4, SV5 |
| Atg12b | 1 | – | – | – | – | – |
Criteria for SVs without the essential codon solely depend on amino acid sequences deduced from SVs and vary for different Atg genes. For Atg3, Atg7, and Atg10, the essential codon means one that encodes a conserved active-site Cys in the corresponding protein. For Atg8 and Atg12, a codon for the C-terminal Gly residue is important, while the essential codon for Atg4 genes encodes an N-terminal active-site Cys. For Atg5, the codon for a Lys acceptor is essential. See Figure 1 for the positions of these codons within the predicted full-length gene.
These genes are single copy in Arabidopsis and maize but duplicated in rice.
Some of the splice variants substantially changed the corresponding reading frames, indicating that the resulting mRNAs would direct the synthesis of altered proteins if translated. For example, several splice variants of Zm Atg4b could affect the activity of the encoded protease by removing a stretch of amino acids near its active-site Cys (Fig. 1B). Additionally, several splice variants of Zm Atg10 eliminate the exonic region encoding the active-site Cys that binds ATG12 during its conjugation cascade (Fig. 1C), thus inactivating the variant ATG10 protein. Whether any of these variant proteins actually accumulate and adversely affect the ATG8 or ATG12 conjugate cascades is not yet known.
Maize and Rice ATG7 Has a Duplicated C-Terminal Region
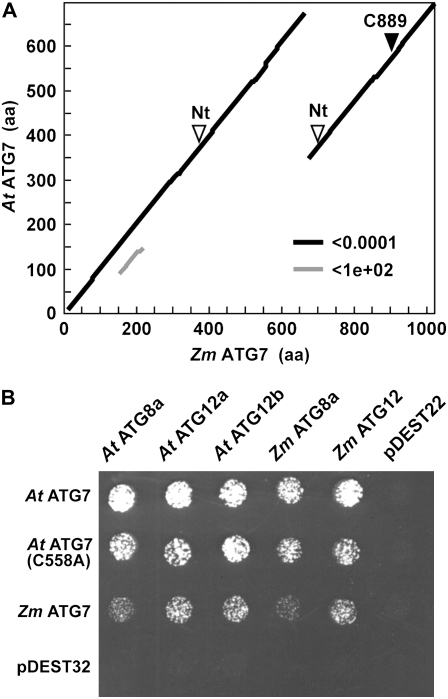
Initially, there was no clear full-length cDNA model for Zm Atg7, even though our BLAST analyses detected ESTs corresponding to the 3′ end of the transcript. Fortunately, we were able to generate a putative full-length Atg7 cDNA by RT-PCR using a forward primer designed from a partial 5′ genomic sequence and a reverse primer based on the partial 3′ EST. This longer Zm Atg7 cDNA matched (1) our partial maize gene models, (2) a full-length gene model generated from a more recently released BAC sequence of maize, and (3) a predicted full-length rice Atg7 transcript. Surprisingly, when aligned with the Arabidopsis ATG7 coding region, we found that the rice and maize transcripts encoded substantially longer polypeptides (1,021 and 1,042 residues for maize and rice ATG7, respectively, versus 697 residues for Arabidopsis ATG7).
Dot plot amino acid sequence comparisons of the maize and rice polypeptides with Arabidopsis ATG7 revealed that the increased length was created solely by a sizable duplication of the C-terminal region of the maize and rice proteins (Fig. 3A; data not shown). This arrangement has not yet been described in other plants, fungi, or animals, implying that it was generated by a cereal-specific tandem duplication. The region involved in the duplication is essential for the E1 activating activity of ATG7, as it contains both the nucleotide-binding pocket and the conserved Cys that binds ATG8/12 via thiolester bond (Doelling et al., 2002; Fujioka et al., 2008). In maize and rice, the more C-terminal duplicated region retained both the nucleotide-binding pocket and the essential Cys, whereas the more internal duplicated region had the essential Cys replaced by a Ser, implying that the internal segment cannot be catalytically active by itself (Fig. 3A).
Figure 3.
The maize ATG7 protein contains a C-terminal duplication. A, A PLALIGN dot plot of local sequence similarity for maize (Zm) ATG7 versus Arabidopsis (At) ATG7 reveals the C-terminal duplicated region. The duplicated nucleotide-binding pocket (Nt) and the active-site Cys (C889) are indicated by the arrowheads. aa, Amino acids. B, Zm ATG7 retains its ability to interact with ATG8 and ATG12 by Y2H analysis despite the C-terminal duplication. Binding of full-length Zm and At ATG7 proteins with maize and Arabidopsis versions of ATG8 and ATG12 was demonstrated by growth on selection medium lacking His, Leu, and Trp and containing 3-amino-1′,2′,4′-triazole. At ATG7(C558A) represents an enzymatically inactive mutant in which the active-site Cys (C558) that interacts with ATG8 and ATG12 via a thiolester linkage was substituted for an Ala. pDEST22 and pDEST32 represent empty vector controls for either the activation domain or the binding domain construction.
To test whether the extended maize and rice ATG7 proteins retained their ability to interact with ATG8/12, we examined by yeast-two hybrid (Y2H) analysis the interaction of Zm ATG7 with Arabidopsis and maize ATG8 and ATG12. As shown in Figure 3B, Zm ATG7, like its Arabidopsis ortholog, could bind both tags, including the At ATG8a and Zm ATG8a isoforms, Zm ATG12, and both isoforms of At ATG12. The interactions of the tags with At ATG7 were not abolished by the C558A variant, in which the active-site Cys was substituted for an Ala (Fig. 3B), indicating that the binding was likely to be noncovalent and did not require formation of the thiolester adduct. Taken together, we conclude that the maize (and likely rice) ATG7 protein retained its ability to interact with its ATG8 and ATG12 substrates despite the C-terminal duplication.
Maize ATG8 Is Lipidated in Planta
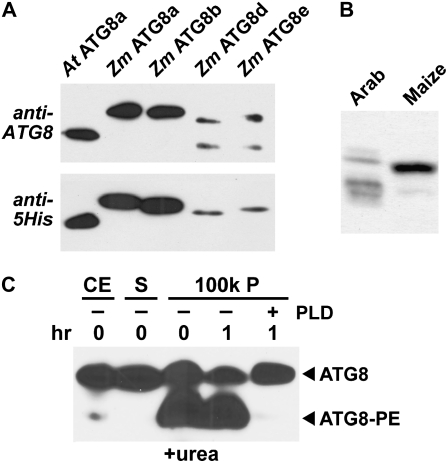
The modification of ATG8 with PE is an important molecular and cytological marker for autophagic compartments (Ohsumi, 2001, Mizushima, 2007). Whereas the free form remains soluble, the lipidated form becomes bound to the limiting membranes of autophagosomes and autophagic bodies via the PE anchor. In an attempt to detect the ATG8-PE adduct in maize, we first confirmed that the antibodies against Arabidopsis ATG8a could recognize its maize orthologs. As shown in Figure 4A, four of the five recombinant Zm ATG8 proteins tagged with 6His were easily detected by immunoblot analysis following SDS-PAGE. (Zm ATG8c was not tested, since its amino acid sequence is identical to that of Zm ATG8b [Fig. 2].) When compared with their recognition by anti-5His antibodies, it appeared that the sensitivities of the antibodies toward Zm ATG8a to Zm ATG8e were not significantly different from that toward the At ATG8a antigen, consistent with the strong amino acid sequence identities among the group (Fig. 2). Whereas these antibodies detected several members of the Arabidopsis ATG8 family in crude seedling extracts following SDS-PAGE (Fig. 4B; Thompson et al., 2005; Phillips et al., 2008), a single species was prominent in crude extracts from maize seedlings (Fig. 4B), which is consistent with the similar size and charge of the maize ATG8 proteins.
Figure 4.
Detection of free ATG8 and the ATG8-PE adduct in maize by immunoblot analysis with anti-At ATG8a antibodies. A, Cross-reactivity of anti-At ATG8a antibodies with Zm ATG8a, ATG8b, ATG8d, and ATG8e. Each lane represents an aliquot of a crude E. coli extract expressing the various 6His-tagged recombinant proteins that was separated by SDS-PAGE without urea. (Zm ATG8c, which is identical in amino acid sequence to Zm ATG8b, was not tested.) The ATG8 proteins were then detected by immunoblot analysis with either anti-At ATG8a antibodies (top) or anti-5His antibodies (bottom). B, Detection of Zm ATG8 in crude extracts with anti-At ATG8 antibodies. Crude extracts prepared from Arabidopsis and maize seedlings were separated by SDS-PAGE without urea. C, Detection of the ATG8-PE adduct in maize. Crude extracts (CE) prepared from maize roots were centrifuged at 100,000g to collect the soluble (S) and membrane (100k P) fractions. The 100k P was solubilized with Triton X-100 and incubated for 1 h with or without PLD. The samples were then subjected to SDS-PAGE in the presence of 6 m urea. The free and lipidated forms of ATG8 are indicated by the arrowheads.
To study the lipidation of maize ATG8, we exploited the assay developed by Yoshimoto et al. (2004) that enriches for the ATG8-PE adduct by the centrifugal isolation of a crude membrane fraction, solubilizes the membranes with Triton X-100, and then separates lipidated ATG8 from the free form by SDS-PAGE in the presence of 6 m urea. Lipidation of ATG8 can then be confirmed by treatment of the solubilized membranes with phospholipase D (PLD), which converts the faster migrating ATG8-PE adduct into the slower migrating free form (Tanida et al., 2004; Fujioka et al., 2008). When the maize samples were subjected to SDS-PAGE in the presence of urea, we detected a faster migrating species in addition to the slower migrating free ATG8 protein (Fig. 4C; data not shown). This faster migrating species could even be detected in crude extracts prior to membrane isolation and became highly enriched in the solubilized membranes, as expected for a membrane-anchored protein. The abundance of this faster migrating form was substantially reduced upon incubation of the solubilized membranes with PLD, indicating that this species is a lipidated form of Zm ATG8 (Fig. 4C). While not proven here, this species likely represents the PE adduct (Fujioka et al., 2008).
Spatial and Temporal Patterns of Zm Atg Gene Expression and ATG8 Lipidation
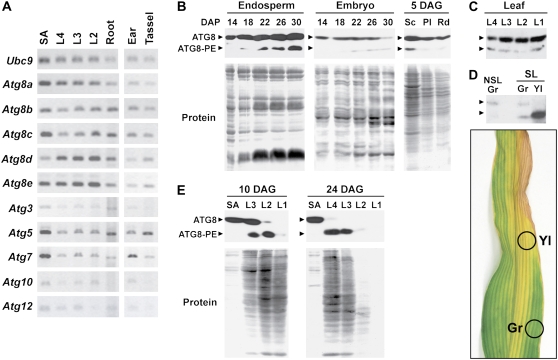
Previous studies with Arabidopsis demonstrated that the genes encoding the ATG8/12 conjugation cascades are ubiquitously expressed, with senescence and nutrient limitations increasing transcript abundance (Contento et al., 2004; Yoshimoto et al., 2004; Buchanan-Wollaston et al., 2005; Sláviková et al., 2005; Thompson et al., 2005; Rose et al., 2006; van der Graaff et al., 2006; Osuna et al., 2007; Peng et al., 2007; Phillips et al., 2008). Similar widespread expression was also apparent for the orthologous maize loci. Using semiquantitative RT-PCR, we detected mRNAs for all Zm Atg genes assayed in a variety of tissues, including the shoot apex (Fig. 5A). Slight variations in the expression levels of the five Zm Atg8 loci were observed, which could reflect tissue-specific functions of the corresponding isoforms (e.g. Zm Atg8c was best expressed in the shoot apex, while Zm Atg8d was better expressed in older leaves; Fig. 5A).
Figure 5.
Developmental patterns of maize Atg gene expression and ATG8 lipidation. A, Zm Atg transcripts are expressed ubiquitously in various maize tissues. The shoot apex (SA), L4, L3, and L2 seedling leaves (leaf numbers correspond to the order of appearance), and roots were collected from seedlings grown hydroponically for 20 d with MS medium. The nonpollinated ears and tassels were collected from a mature plant grown in soil. Total RNA was subjected to semiquantitative RT-PCR using primers specific for various maize Atg genes or the Ubc9 control gene. B to E, Analysis of free ATG8 and the ATG8-PE adduct. Crude extracts were prepared from various tissues and subjected to SDS-PAGE with 6 m urea followed by immunoblot analysis with anti-At ATG8a antibodies. B, Endosperm and embryos at various DAP from a soil-grown plant and tissues dissected from seedlings germinated for 5 d on MS medium. Sc, Scutellum; Pl, plumule; Rd, radicle. C, Leaves from seedlings grown in soil for 20 DAG. D, Naturally senescing leaves. Samples were collected from a nonsenescing leaf (NSL) and yellow (Yl) and green (Gr) sectors from a subtending senescing leaf (SL; see photograph for the sampled areas) from a flowering plant grown in soil. E, Shoot apex and leaves of various ages (L1 older to L4 younger) from seedlings grown hydroponically on MS medium for the indicated DAG. C and D contain 20 μg of total protein per lane. The lanes in B and E were loaded on an equal fresh weight basis except for Pl, Rd, and SA. Protein profiles in the lower panels of B and E were detected by Ponceau S staining.
To gain further insights into the regulation of ATG8 conjugation during development, we examined the levels of the free ATG8 protein and its lipid adduct in selected maize tissues at various stages of development. In most cases, enrichment of the ATG8-PE adduct by membrane isolation was not needed to enable detection of the faster migrating lipidated form, indicating that some maize tissues accumulate high levels of this species. For example, both free ATG8 and the ATG8-PE adduct were easily detected during seed/seedling development in crude extracts prepared from both the endosperm and embryo at increasing times of maturity and in several parts of the seedling at 5 d after germination (DAG; Fig. 5B). In young embryos, almost all ATG8 was present in the free form, but as they developed, the ATG8-PE adduct appeared around 22 d after pollination (DAP). In mature embryos at 30 DAP, the level of free ATG8 was substantially reduced, while the level of ATG8-PE remained the same. The endosperm also accumulated higher levels of the ATG8-PE adduct as it matured, suggesting that autophagy may participate in programmed cell death during endosperm development. A high level of adduct was evident in the scutellum, but not in the plumule and radicle, during germination, which could reflect the autophagic mobilization of stored reserves in the scutellum to support new growth (Fig. 5B). Leaves harvested from soil-grown, well-fertilized seedlings also contained both the free and lipidated forms of ATG8, with a similar ratio evident in green leaves of various ages, as categorized by their positions on the plant (Fig. 5C).
Strikingly, we found that both the total amount of ATG8 and the abundance of the ATG8-PE adduct in maize leaves were markedly affected by senescence. For example, when we compared the yellow and green sectors in an individual senescing leaf from a mature flowering plant, the yellow sectors had substantially more total ATG8 protein (Fig. 5D). Whereas the green sectors had a nearly equal ratio of lipidated ATG8 to free ATG8, similar to that found in the adjacent nonsenescing leaf positioned on the plant axis immediately above, the yellow sectors contained almost exclusively the ATG8-PE adduct, suggesting that high rates of autophagy were under way in these areas. This senescence-dependent effect could also be induced when the plants were grown hydroponically in a defined solution containing inorganic nutrients, a growth condition that accelerates the developmental senescence of leaves even though the plants appear well fertilized. As shown in Figure 5E for plants grown hydroponically for 10 DAG, the oldest leaf (L1) contained only traces of ATG8-PE, the next oldest leaf (L2) contained much higher levels, but almost all was lipidated, while younger leaves (L3) contained a mixture of the free and lipidated form, and the shoot apex contained almost exclusively the free form. Eventually, both forms of ATG8 disappeared along with most other proteins as the older leaves completed senescence (see L1 and L2 from 24-DAG seedlings in Fig. 5E).
Zm Atg Gene Expression and ATG8 Lipidation Increase during Nutritional Stress
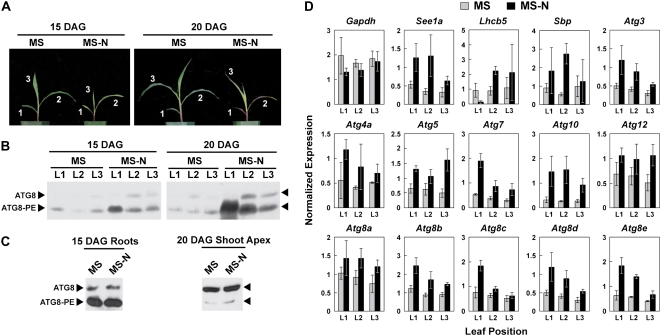
The age-dependent accumulation of the ATG8-PE adduct in the hydroponically grown seedling leaves (Fig. 5E) suggested that the availability of macronutrients and/or micronutrients regulates Zm Atg gene expression and the lipidation of the ATG8 protein. One obvious candidate is N, since its availability is often limiting to plant growth and protein synthesis. To test this possibility, we germinated and grew maize seedlings hydroponically for 15 d on deionized water, Murashige and Skoog (MS) medium lacking N (MS-N), or MS-N supplemented with 0.5, 1, or 2 times the normal concentration of nitrate (see “Materials and Methods”). As expected, the growth of seedlings treated with only water or MS-N was significantly retarded at 15 DAG compared with N-fertilized plants (Fig. 6). Concomitant with this growth reduction was an increase in the oldest leaves (L1) of both the amount of total ATG8 and the proportion converted to the lipidated form. As increasing amounts of N were added to the MS-N, the amounts of the ATG8-PE adduct and total ATG8 dropped in the L1 leaf (Fig. 6). Doubling the strength of N (2× N) did not significantly affect the levels of total and lipidated ATG8 when compared with single strength medium (1× N).
Figure 6.
N-dependent ATG8 lipidation. Maize seedlings were grown hydroponically on vermiculite for 15 DAG, supplied with deionized water (DW), MS-N alone (0 N), or MS-N supplemented with various strengths (0.5×, 1×, or 2× N) of nitrate (see “Materials and Methods” for details). Top, Typical growth of 15-DAG seedlings using the above conditions to show the effects of the N limitations. Bottom, Levels of lipidated and free ATG8 in the leaves of plants shown in the top panel. SDS-PAGE and immunoblot analysis were performed as in Figure 5. Each lane contains 20 μg of total protein.
To further investigate the effect of N limitation on ATG8 lipidation and Zm Atg gene expression, we collected leaf samples from 10-, 15-, and 20-DAG seedlings that were hydroponically grown with either MS or MS-N. At 10 DAG, no significant differences in seedling growth or in total and lipidated ATG8 levels were detected between leaf samples collected from MS- and MS-N-grown seedlings, suggesting that the plants had not yet experienced N deficiencies (data not shown). However, at 15 DAG, reductions in growth became evident for the seedlings fertilized with MS-N (Fig. 7A, left), indicating that they were now experiencing a N deficit. This deficiency became more obvious at 20 DAG, when the oldest leaves (L1) acquired the yellow/brown patches characteristic of strongly senescing leaves. In parallel with this N starvation was a substantial increase in the levels of the ATG8 protein. The 15- and 20-DAG plants grown on MS-N had higher levels of total and lipidated ATG8 than MS-grown seedlings, a difference that was especially apparent in the older (L1) leaves (Fig. 7B). In contrast to the effects on ATG8 levels in leaves, both the total amount of ATG8 and the ratio of lipidated ATG8 to the free form were unaffected by N limitation in the roots and shoot apex (Fig. 7C).
Figure 7.
Maize seedlings respond to N limitation by increasing Atg gene expression and ATG8 protein abundance in a leaf age-dependent pattern. Seedlings were grown hydroponically in vermiculite subirrigated with MS medium or MS-N. A, Photographs of 15-DAG (left) and 20-DAG (right) seedlings grown on MS medium (left seedlings) and MS-N (right seedlings). Leaves at the various positions analyzed (L1 older to L3 younger) are indicated. B and C, Accumulation of free ATG8 and the ATG8-PE adduct in leaves (B) and in roots and the shoot apex (C). Crude extracts (20 μg of total protein) were subjected to SDS-PAGE and immunoblot analysis as in Figure 5. D, Levels of various Zm Atg mRNAs increase upon N limitation. Total RNA was isolated from the leaves shown in A and subjected to semiquantitative RT-PCR. Expression levels were normalized by dividing the amounts of PCR products by those produced from the Zm Ubc9 control transcript. Each bar represents the average of three biological replicates ± sd. Note that expression values among genes cannot be compared, due to the different amplification conditions used for each gene.
The increase in maize ATG8 protein levels during N starvation was consistent with previous data in Arabidopsis showing that the expression of the ATG system is up-regulated by nutrient stress (Contento et al., 2004; Yoshimoto et al., 2004; Buchanan-Wollaston et al., 2005; Sláviková et al., 2005; Thompson et al., 2005; Rose et al., 2006; van der Graaff et al., 2006; Osuna et al., 2007; Peng et al., 2007; Phillips et al., 2008). To test whether the suite of Zm Atg genes was affected similarly, we analyzed the transcript levels for each during N starvation along with several senescence-associated transcripts. Here, total RNA was isolated from maize leaves of various ages (L1–L3) from 15-DAG seedlings fertilized with either MS or MS-N, and the levels of individual mRNAs were determined by semiquantitative RT-PCR. Transcripts encoding the Ub-conjugating enzyme (Ubc9) and the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) were used for normalization and as an internal control, respectively (Czechowski et al., 2005). As can be seen in Figure 7D, the Gapdh/Ubc9 transcript ratio was not significantly affected by leaf age or N availability under these growth conditions. In contrast, the senescence-associated See1a transcript (Smart et al., 1995) was significantly up-regulated by the MS-N growth condition, confirming that premature senescence was induced in these N-limited leaves. Accelerated leaf senescence in N-limited leaves was also evident from RT-PCR analysis of the Lhcb5 transcript encoding one of the chlorophyll-binding proteins in the chloroplast (van der Graaff et al., 2006). The older senescing L1 leaves contained a very low level of Lhcb5 transcript. Unexpectedly, the second leaves (L2) of MS-N seedlings had higher levels of Lhcb5 transcript than MS-grown leaves. This is consistent with the RT-PCR data for sedoheptulose-bisphosphatase (Sbp) transcript, which encodes a Calvin cycle enzyme and shows a light-dependent expression pattern (Willingham et al., 1994). The Sbp transcript levels in N-limited L2 leaves were also higher than in L2 leaves fertilized with MS. The reason for this accumulation of these photosynthesis-related transcripts in N-limited middle leaves is not clear.
When the Zm Atg genes were analyzed similarly, we found that the transcript abundance of almost all were up-regulated by the absence of N and further up-regulated as the leaves aged (Fig. 7D). In most cases, the highest transcript abundance was found in the L1 leaves from MS-N-fertilized plants. This trend was particularly obvious for Atg3, Atg7, Atg10, and four of the five isoforms of Atg8 (Atg8b, -c, -d, and -e). Interestingly, in addition to Atg7, those genes encoding components required for ATG8 conjugation were more strongly affected by the combined action of N availability and leaf age (Atg3, Atg4b, and Atg8b to Atg8e) than those encoding components for ATG12 conjugation (Atg5, Atg10, and Atg12; Fig. 7D). This difference suggests that formation of the ATG8-PE adduct is more limiting to autophagic recycling during senescence.
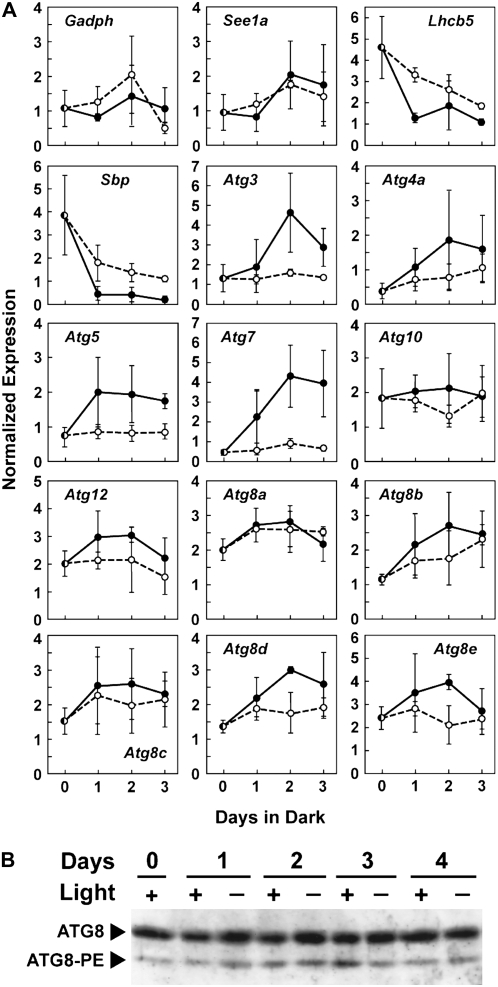
Fixed-C limitation has also been shown to increase the abundance of various ATG transcripts in Arabidopsis using either the withdrawal of Suc from the growth medium of cell cultures (Contento et al., 2004; Rose et al., 2006) or the placement of whole plants in extended darkness to deplete the availability of soluble sugars (Sláviková et al., 2005; Thompson et al., 2005; Phillips et al., 2008). To test the importance of fixed-C availability, we wrapped a 7.5-cm band of aluminum foil around individual maize leaf blades from plants grown in light to subject the region to extended darkness (Weaver and Amasino, 2001; van der Graaff et al., 2006). At various times, the darkened sectors and the corresponding sectors from light-exposed control leaves of the same developmental age were harvested and total RNA was isolated. The abundance of individual transcripts in each RNA pool was then determined by semiquantitative RT-PCR as described above using the signal from the Ubc9 mRNA for normalization. While short treatments (1–3 d) did not detectably affect the color of the darkened sectors, prolonged treatments up to 10 d induced obvious senescence, as indicated by leaf sector yellowing/browning.
Transcript analysis of Gapdh, See1a, Lhcb5, and Sbp demonstrated that this covering affected the photosynthetic machinery well before senescence became apparent. While the mRNA levels for Gapdh were unaffected and those for the See1a transcript were only modestly up-regulated in the darkened and presumably fixed-C-limited sectors relative to illuminated sectors, the levels of both Lhcb5 and Sbp dropped dramatically, consistent with their light requirement for transcription (Willingham et al., 1994; van der Graaff et al., 2006). As with N limitation, almost all of the Atg transcripts were up-regulated in the darkened sectors, with the maximal levels appearing after 2 d in the dark (Fig. 8A). Atg7 displayed the strongest up-regulation, with an approximately 8-fold increase in transcript levels in the darkened sectors. Only the abundance of the Atg8a mRNA appeared to be unaffected by darkness, with Atg8c and Atg10 exhibiting only small increases (Fig. 8A). As with the N-limiting growth condition (Fig. 7C), the genes associated with ATG8 conjugation were in general more strongly affected compared with those involved with ATG12 conjugation.
Figure 8.
Response of maize seedling leaves to dark-induced fixed-C limitation. A basal part of the second leaf blade (L2) of 15-DAG plants was wrapped with aluminum foil (75 mm in length). The leaf blade was harvested immediately or at 1, 2, or 3 d after wrapping and used for semiquantitative RT-PCR (A) or immunoblot analysis of the ATG8 protein (B) as described for Figure 7. The corresponding regions of light-exposed leaves of the same developmental age were used as controls. Black and white circles in A represent means of three normalized expression values from dark-treated and illuminated leaves, respectively.
Despite the increases in Atg mRNA abundance by darkening the leaf, the levels of the free ATG8 and the ATG8-PE adduct were not markedly altered. The steady-state levels of the free and conjugated forms were similar in the darkened and illuminated sectors even after 4 d of treatment (Fig. 8B). This lack of effect on ATG8 levels during fixed-C limitation as opposed to the strong effect induced by N limitation (Fig. 7B) could indicate that autophagy is more sensitive to N starvation. However, a more plausible interpretation is that our experiments with N starvation likely caused the entire plant to experience N deficits, while those testing fixed-C limitation subjected only part of the leaf to fixed-C deficits. Consequently, it is possible that the neighboring illuminated areas became a source of soluble sugars to the darkened areas, thus mitigating the drop in fixed-C in the absence of photosynthesis.
DISCUSSION
A number of genetic studies with the ATG8/12 conjugation cascades in Arabidopsis have demonstrated the importance of autophagy to nutrient recycling and survival of plants under N/C-limiting growth conditions (see above). In an effort to determine the importance of autophagy to crop plants, we completed a molecular description of an orthologous ATG system in the cereals rice and maize. We then assayed in maize their expression patterns and the accumulation of the lipidated ATG8 adduct during senescence and in response to N or fixed-C limitation. All components required for ATG8/12 conjugation in yeast and Arabidopsis (Ohsumi, 2001; Thompson and Vierstra, 2005) were identified in both rice and maize, indicating that the pathway is highly conserved among plant species. As in Arabidopsis, four maize components are encoded by single genes (Atg3, Atg5, Atg7, and Atg10), whereas two are encoded by small gene families (two for Atg4 and five for Atg8). This increased diversity for the ATG8 protein tag and the ATG4 processing protease required for ATG8 maturation suggests both subfunctionalization of the ATG8 isoforms and distinct roles for the two ATG4 isoforms in processing the ATG8 precursor and/or recycling ATG8 modified with PE. As reported for the Arabidopsis ATG8 gene family (Sláviková et al., 2005; Thompson et al., 2005), individuals in the five-member maize Atg8 family display subtle differences in their tissue expression patterns and varying responses to N and fixed-C limitations, indicating that the individual ATG8 isoforms in maize have cell/tissue-specific and/or stress-specific roles.
Our description of the maize Atg genes is one of the earliest comprehensive genomic analyses on a specific metabolic pathway since the release of the draft genome sequence (http://maizesequence.org). Despite difficulties in determining the exact number of genes encoding each Zm ATG protein, given the incomplete annotation of the sequence and the large number of discontinuous segments still remaining, we are certain about the gene identifications described in Figure 1 and Table II for the following reasons. First, we focused on genes involved in the ATG8/12 conjugation cascades. They appear to be the most conserved members in the Atg system, which in turn simplified the identification of the rice and then the maize orthologs. For example, like its structural relative Ub, ATG8 is arguably one of the most conserved proteins in eukaryotes (Fig. 2; Supplemental Fig. S1; Doelling et al., 2002; Xie and Klionsky, 2007). Second, by exploiting the deep EST database for maize (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html) in combination with the B73 reference BAC sequences, we could better identify true functional loci. For instance, we initially found numerous maize cDNA and genomic sequences related to various rice Atg loci. However, only a smaller number was confirmed as real by exact matching with the corresponding BAC sequences. The less-related sequences likely reflect pseudogenes, misassemblies, sequence polymorphism, and/or sequencing errors.
Although we are confident in most of the rice and maize ATG gene models, several await full-length cDNAs and/or more refined chromosomal sequence data before final confirmation of their gene organizations. For example, we could not assign a BAC sequence to the Zm Atg8e transcript, although there are two candidate BACs (AC203867 and AC203875) predicted to encode very similar transcripts (Table II). In addition, the precise gene models for rice Atg3b and Atg10b await full-length cDNAs. cDNAs encoding partial polypeptides are available for both, indicating that these loci are transcribed. However, based on the predicted high expression of the paralogous Os Atg3a and Os Atg10a loci (based on the number of ESTs [Supplemental Table S1]), it is likely that the rice Atg3b and Atg10b loci are minor contributors to the pools of each enzyme. And lastly, the rice Atg8e gene is currently without any EST or cDNA data, thus leaving its gene model and transcriptional activity unsupported (Table I).
While almost all of the rice and maize ATG proteins are structurally orthologous to their Arabidopsis and yeast counterparts, ATG7 is notably distinct. The rice and maize polypeptides contained an approximately 325-amino acid duplication of their C-terminal half that replicates the region encompassing the active-site Cys and the nucleotide-binding pocket necessary for ATG8/12 activation. While the nucleotide-binding pocket sequence is evident in both repeats, the more N-terminal repeat had its active-site Cys replaced with a Ser. Despite this duplication, maize ATG7 remains functional, as judged by the ability of the full-length protein to interact by Y2H analysis with ATG8 and ATG12 and by our detection of its ATG8-PE product in planta. At present, it is unclear how the catalytic regions of rice and maize ATG7s are organized and whether the more N-terminal predicted nucleotide-binding pockets can actually participate in ATG8/12 activation.
An interesting aspect of the cereal Atg gene models is that most of the genes are associated with alternative splicing of pre-mRNA. Current estimates of genes with multiple splice variants are approximately 21% and 22% for Arabidopsis and rice, respectively (Wang and Brendel, 2006). Although the number of maize genes subjected to alternative splicing is not yet available, it appears to be similar to that of rice (T. Chung and B.A. Larkins, unpublished data). Conversely, the numbers of genes affected by alternative splicing within the ATG8/12 conjugation cascades are notably larger, with splice variants evident for 41%, 79%, and 92% of the respective Arabidopsis, rice, and maize Atg loci examined here (Table III). Such increases imply that alternative splicing has a role in either diversifying the ATG protein catalog without increasing the gene number or providing a sophisticated means to control the abundance of functional Atg transcripts in response to various developmental and environmental cues. The former scenario could explain why rice and maize have fewer ATG8 isoforms than Arabidopsis despite having larger proteomes overall. The latter scenario is also reasonable given the importance of the ATG autophagic system to cell survival and its likely regulation by a number of internal and external signals. Curiously, all of the splice variants from the Arabidopsis ATG genes are predicted to encode proteins that retain their active site and/or other amino acids essential for function (Table III). On the contrary, many splice variants of the rice and maize Atg genes encode proteins lacking one or more essential residues and therefore are likely to be nonfunctional. The possibility that this splice variant distinction reflects unique methods employed by dicotyledonous and monocotyledonous plants to control the ATG system is intriguing.
As observed previously with Arabidopsis (Contento et al., 2004; Sláviková et al., 2005; Thompson et al., 2005; Phillips et al., 2008), the ATG8/12 conjugation cascades are ubiquitously expressed in most, if not all, tissues of maize, with a strong up-regulation of most genes during senescence and under N- and fixed-C-limiting growth conditions. For senescing leaves and leaves harvested from plants grown under N-limiting conditions, this up-regulation also led to an increase in the levels of the ATG8 protein. Analysis of the ATG8 protein pool in turn revealed that the steady-state ratio of the lipidated to free form of ATG8 is strongly affected in some tissues by their developmental and nutritional states. As examples, (1) the abundance of the ATG8-PE adduct relative to free ATG8 increased as the endosperm and embryo matured following pollination. This increase in the developing endosperm could reflect a role for autophagy in the programmed cell death process that begins in the central region of the starchy endosperm and then proceeds to outer regions of the endosperm as the seed matures (Young and Gallie, 2000). (2) In the germinating seedling, the scutellum had relatively high levels of lipidated ATG8, which is consistent with its role in absorbing nutrients from the endosperm for seedling development. (3) And finally, both natural and hydroponics-induced senescence and N starvation dramatically increased the relative abundance of the ATG8-PE adduct. This effect was particularly striking in senescing leaves, where the yellow sectors had high levels of total ATG8 with almost all of the pool converted to the lipidated form.
What drives this substantial increase in the steady-state levels of the ATG8-PE adduct during seed development, leaf senescence, and N starvation is not yet known. In yeast, this lipidation is controlled by a set of nutrient-sensing systems (e.g. target-of-rapamycin, Snf1, and cAMP-dependent protein kinases) that suppress the activity of the ATG1 kinase complex, whose action ultimately impacts the ATG8/12 conjugation machinery (Klionsky, 2007; Mizushima, 2007). Assuming that the ATG12-ATG5 adduct represents the E3 activity that transfers activated ATG8 from the ATG3 E2 to PE (Hanada et al., 2007; Fujioka et al., 2008), the assembly of this adduct in particular could be the rate-limiting step. Since the pool of the ATG8-PE adduct is also modulated by turnover, the steady-state increase could also reflect a decrease in delivery of the adduct along with the autophagosome to the vacuole, a decrease in the recycling of ATG8 by the ATG4 protease, and/or an increased half-life of the adduct once in the vacuole (Mizushima and Yoshimori, 2007). Clearly, reverse genetic analyses of the two conjugation cascades and upstream regulators and a detailed understanding of the intracellular dynamics of ATG8 will be needed to identify these control points in maize.
Whatever the mechanisms, the strong correlations between the steady-state abundance of the ATG8-PE adduct and leaf senescence and growth under N-limiting conditions suggest that the level of the adduct represents a sensitive indicator of both the amount of autophagic recycling and the nutrient status of the plant/organ/tissue. For specific tissues, it could also be a measure of their source/sink status. For example, in hydroponically grown plants, older leaves, which are expected to be important nutrient sources for the growing regions as they senesce, contained mostly the conjugated form of ATG8, while the shoot apex, composed of rapidly expanding young leaves that are strong nutrient sinks, contained mostly the free form. Taking this possibility one step further, our analysis of ATG8 lipidation also implies that autophagic recycling is regulated in a tissue/organ-dependent manner in maize and likely in other plants. In N-fertilized plants, the high ATG8-PE to free ATG8 ratio in roots relative to shoots and leaves would imply that roots are relatively more active in autophagy under N-rich growth conditions. When depleted of N, root autophagy appeared unaffected, as this ratio remained unchanged. Instead, a dramatic increase in the ATG8-PE adduct was observed in leaves, suggesting that autophagy in this tissue was specifically induced. This N starvation-induced increase further underscores the notion that senescing leaves are a major source of remobilized nutrients during nutrient starvation (Lim et al., 2007; Munné-Bosch, 2008). While more data are clearly needed to connect ATG8 lipidation to autophagic recycling rates and plant nutrition, our data suggest that the formation of the ATG8-PE adduct could be a simple indicator for the source/sink status of individual tissues and thus could provide a useful marker for the nutritional state of crops grown in the field.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All experiments were performed with the maize (Zea mays) inbred B73. Seeds were germinated in pots (125 cm3) with a 1:1:1 mixture of compost, peat moss, and vermiculite and were grown at 21°C in a growth chamber with a 16-h-light/8-h-dark photoperiod. For dark-induced fixed-C limitation, the basal part of the second leaf blade was wrapped with aluminum foil (75 mm in length) at 15 DAG. At various times, the wrapped part of the leaf blade was harvested along with a corresponding region from a light-exposed leaf blade of the same developmental age, which served as the control. For N limitations, seedlings were grown hydroponically in pots (450 cm3) with vermiculite. The pots were subirrigated with 10 mL of various liquid media every 5 d, starting at 1 DAG. The media included (1) MS medium (MS basal salts), (2) MS-N (MS micronutrient solution plus 3 mm CaCl2, 1.5 mm MgSO4, 1.25 mm KH2PO4, and 5 mm KCl [pH 5.7]), or (3) MS-N supplemented with 0.825 g L−1 NH4NO3 and 0.95 g L−1 KNO3 (MS-N + 0.5× N medium), 1.65 g L−1 NH4NO3 and 1.9 g L−1 KNO3 (MS-N + 1× N medium), or 3.3 g L−1 NH4NO3 and 3.8 g L−1 KNO3 (MS-N + 2× N medium). When needed, seedlings were grown to maturity in a greenhouse to allow for natural senescence and to obtain developing kernels following self-pollination. Extra lighting was provided to maintain a 14-h-light/10-h-dark photoperiod.
DNA and Protein Sequence Analyses
Maize Atg genes were identified by TBLASTN (Altschul et al., 1990) using the orthologous protein sequences from Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa subsp. japonica) as queries. The rice Atg sequences were first identified in the GenBank rice Non-redundant and The Institute for Genomic Research (TIGR) pseudomolecule (Ouyang et al., 2007) databases by TBLASTN using the yeast (Saccharomyces cerevisiae) and Arabidopsis orthologs as queries. Corresponding rice Atg genomic sequences were subsequently used as queries against rice Non-redundant and EST databases. Rice Atg gene models and potential splice variants were identified by manual inspection of the resulting sequence alignments. The Arabidopsis and rice ATG protein sequences were then used to search by TBLASTN the ZmGI EST contig (Lee et al., 2005) and TIGR AZM5 genomic assembly (Chan et al., 2006) databases (last searched January 2007). The identified ZmGI cDNA sequences were also used as BLASTN query sequences against the AZM5 genomic sequence database to help assign each cDNA to a genomic locus.
Sequence alignments between individual cDNAs and genomic sequences were manually inspected for consensus coding regions, intron splice sites, and potential splice variants. Sequence gaps were filled in by BLASTN of the resulting gene models to the maize HTGS BAC sequence database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Dot plots revealing local sequence similarities were generated by PLALIGN (Pearson, 2000). Full-length cDNAs for each Zm Atg gene were generated by RT-PCR of total RNA isolated from leaves, using primers that bracketed the translation start and stop codons (see Supplemental Table S2 for the primer sequences). The entire RT-PCR product populations were inserted into pENTR/D-TOPO (Invitrogen). Multiple inserts (between three and 10 depending on the locus), especially from those populations containing individuals with unique electrophoretic mobilities, were sequenced in their entirety and compared with the gene models to reveal novel splice variants. The current Zm Atg gene models, including splice variants and the GenBank accession numbers of the corresponding cDNAs, can be found in Supplemental Data Set S1.
RT-PCR Analyses
RNA was extracted from three biological replicates of various maize tissues using Trizol (Invitrogen). First-strand cDNA synthesis was achieved with SuperScript II (Invitrogen) reverse transcriptase and an oligo(dT)18 primer (Fermentas). The amount of RT product in each sample was standardized by a second round of PCR using primers against Zm Ubc9 (named because of its closest amino acid match to Arabidopsis UBC9; Czechowski et al., 2005). After appropriate dilutions, the RT products were subjected to semiquantitative PCR using Zm Atg gene-specific primers (Supplemental Table S2). The number of PCR cycles was adjusted beforehand to obtain a nearly linear amplification of each transcript. The amounts of PCR products were quantified using GelDoc imaging software (Bio-Rad Laboratories) following agarose gel electrophoresis and ethidium bromide staining of the product. Normalized expression values were generated using RT-PCR products generated from Zm Ubc9 as the standard.
Y2H Assays
Y2H assays were conducted with full-length cDNAs as described previously with minor modifications (Gingerich et al., 2007). Using the LR Clonase reaction (Invitrogen), the At ATG8a, At ATG12a, At ATG12b, Zm Atg8a, and Zm Atg12 cDNAs were recombined into the pDEST22 activation domain plasmid, whereas the cDNAs encoding At ATG7, At ATG7(C558A), and Zm ATG7 were recombined into the pDEST32 binding domain plasmid (Invitrogen). The plasmids were then introduced into the haploid yeast strains YPB2a and LB41α, respectively, and diploid yeast strains were then generated by mating. Protein-protein interactions were identified by testing for growth of the diploid strains on selection medium lacking His, Leu, and Trp and containing 5 mm 3-amino-1′,2′,4′-triazole. The At ATG7(C558A) mutant, in which the Cys-558 codon was replaced with that encoding Ala, was generated by the PCR-based QuickChange method (Stratagene) with appropriate mutagenic primers.
SDS-PAGE Analysis
In most cases, maize tissue was harvested, immediately frozen to liquid N temperatures, and homogenized in SDS-PAGE sample buffer. The extracts were clarified by centrifugation at 16,000g and then heated before use. Protein amounts were determined by spotting aliquots of the extracts onto Whatman 3MM filter paper (Fisher Scientific) and comparing the Coomassie Brilliant Blue staining intensity of the spots with bovine serum albumin standards (Ghosh et al., 1988). In most cases, equal amounts of total protein were subjected to SDS-PAGE with or without 6 m urea in the separating gel. Immunoblot analyses were performed with affinity-purified anti-At ATG8 antibodies (Thompson et al., 2005) or anti-5His antibodies (Qiagen) following electrophoretic transfer of the proteins onto Immobilon polyvinylidene difluoride membranes (Millipore). Cross-reactivity of anti-At ATG8 antibodies with Zm ATG8 protein isoforms was checked using recombinant 6His-tagged full-length versions expressed in Escherichia coli BL21(DE3) cells. The pDEST17 plasmids expressing each of the Zm ATG8a, -b, -d, and -e isoforms were generated by a series of LR Clonase reactions (Invitrogen) with appropriate pENTR/D-TOPO clones containing the full-length cDNAs. The pET28 plasmid expressing At ATG8a was previously described (Thompson et al., 2005).
To characterize the ATG8-PE adducts, various maize tissues were homogenized in TNPI buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm phenylmethanesulfonyl fluoride, and 10 mm iodoacetamide). The extract was filtered through four layers of cheesecloth and clarified by centrifugation at 2,000g for 5 min, and the supernatant was centrifuged at 100,000g for 1 h. The resulting 100-k pellet was resuspended in TNPI buffer, solubilized by adding Triton X-100 at a final concentration of 0.5%, and clarified at 16,000g for 10 min to obtain the solubilized membrane fraction. This membrane fraction was incubated at 37°C for 1 h with Streptomyces chromofuscus PLD (Biomol; 250 units mL−1 final concentration) or an equal volume of its buffer. The reactions were stopped by the addition of SDS-PAGE sample buffer and then subjected to SDS-PAGE on 12% polyacrylamide gels with 6 m urea. Free ATG8 and the ATG8-PE adduct were detected by immunoblot analysis with anti-At ATG8 antibodies.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ445019 and FJ444997 (Zm Atg3, splice variants 1 and 2), FJ444998 and FJ444999 (Zm Atg4a, splice variants 1 and 2), FJ445000 to FJ445004 (Zm Atg4b, splice variants 1, 3, 4, 5, and 6), FJ445005 (Zm Atg7), FJ445006 (Zm Atg8a, splice variant 1), FJ445007 (Zm Atg8b, splice variant 1), FJ445008 (Zm Atg8c, splice variant 1), FJ445009 and FJ445010 (Zm Atg8d, splice variants 1 and 4), FJ445011 (Zm Atg8e, splice variant 1), FJ445012 to FJ445014 (Zm Atg10, splice variants 1, 5, and 6), and FJ445015 and FJ445016 (Zm Atg12, splice variants 1 and 2), respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple amino acid sequence alignment for select ATG proteins from yeast, Arabidopsis, rice, and maize.
Supplemental Table S1. GenBank accession numbers associated with splice variants (SVs) of selected Arabidopsis (At), rice (Os), and maize (Zm) Atg genes.
Supplemental Table S2. Oligonucleotide primers used for RT-PCR in this study.
Supplemental Data Set S1. Current maize gene models for Atg3, Atg4a and Atg4b, Atg5, Atg7, Atg8a to Atg8e, Atg10, and Atg12.
Supplementary Material
Acknowledgments
We acknowledge the Maize Genome Sequencing Consortium for making the sequence available prior to publication. We thank Joseph Walker and Dr. Allison Phillips for helpful advice and Ashvini Damodaran for assistance with the Y2H analyses.
This work was supported by grants from the National Research Initiative Competitive Grants Program (grant nos. 2005–35301–15768 and 2008–02545) of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service to R.D.V. and by a Thailand Predoctoral Fellowship to A.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard D. Vierstra (vierstra@wisc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Bassham DC (2007) Plant autophagy: more than a starvation response. Curr Opin Plant Biol 10 587–593 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42 567–585 [DOI] [PubMed] [Google Scholar]
- Chan AP, Pertea G, Cheung F, Lee D, Zheng L, Whitelaw C, Pontaroli AC, SanMiguel P, Yuan Y, Bennetzen J, et al (2006) The TIGR maize database. Nucleic Acids Res 34 D771–D776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277 33105–33114 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F (2008) In vitro reconstitution of plant ATG8 and ATG12 conjugation systems essential for autophagy. J Biol Chem 283 1921–1928 [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Gepstein S, Heikkila JJ, Dumbroff EB (1988) Use of a scanning densitometer or an ELISA plate reader for measurement of nanogram amounts of protein in crude extracts from biological tissues. Anal Biochem 169 227–233 [DOI] [PubMed] [Google Scholar]
- Gingerich DJ, Hanada K, Shiu SH, Vierstra RD (2007) Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19 2329–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282 37298–37302 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M, Taylor C, Liu ZD, Ma FS, McNamara L, Wang TW, Thompson JE (2007) Regulation and execution of molecular disassembly and catabolism during senescence. New Phytol 175 201–214 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 47 1641–1652 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8 931–937 [DOI] [PubMed] [Google Scholar]
- Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J (2005) The TIGR gene indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58 115–136 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121 567–577 [DOI] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21 2861–2873 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3 542–545 [DOI] [PubMed] [Google Scholar]
- Moriyasu Y, Hattori M, Jauh GY, Rogers JC (2003) Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol 44 795–802 [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S (2008) Do perennials really senesce? Trends Plant Sci 13 216–220 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130 165–178 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2 211–216 [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, et al (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49 463–491 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR (2000) Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol 132 185–219 [DOI] [PubMed] [Google Scholar]
- Peng MS, Bi YM, Zhu T, Rothstein SJ (2007) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65 775–797 [DOI] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F (2006) Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell 98 53–67 [DOI] [PubMed] [Google Scholar]
- Sláviková S, Shy G, Yao YL, Giozman R, Levanony H, Pietrokovski S, Elazar Z, Galili G (2005) The autophagy-associated ATG8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot 56 2839–2849 [DOI] [PubMed] [Google Scholar]
- Smart CM, Hosken SE, Thomas H, Greaves JA, Blair BG, Schuch W (1995) The timing of maize leaf senescence and characterization of senescence-related cDNAs. Physiol Plant 93 673–682 [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E (2004) HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABA(A) receptor-associated protein-phospholipid conjugates. J Biol Chem 279 36268–36276 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8 165–173 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32 275–302 [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves but inhibited in whole darkened plants. Plant Physiol 127 876–886 [PMC free article] [PubMed] [Google Scholar]
- Willingham NM, Lloyd JC, Raines CA (1994) Molecular cloning of the Arabidopsis thaliana sedoheptulose-1,7-biphosphatase gene and expression studies in wheat and Arabidopsis thaliana. Plant Mol Biol 26 1191–1200 [DOI] [PubMed] [Google Scholar]
- Xie ZP, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Suzuki T, Moriyasu Y (2007) Constitutive autophagy in plant root cells. Autophagy 3 360–362 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR (2000) Programmed cell death during endosperm development. Plant Mol Biol 44 283–301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.