Abstract
Objective
Unbalanced chromosomal aberrations are common in myelodysplastic syndromes, and have prognostic implications. An increased frequency of cytogenetic changes may reflect an inherent chromosomal instability due to failure of DNA repair. Therefore, it is likely that chromosomal defects in myelodysplastic syndromes may be more frequent than predicted by metaphase cytogenetics and new cryptic lesions may be revealed by precise analysis methods.
Methods
We used a novel high-resolution karyotyping technique, array-based comparative genomic hybridization, to investigate the frequency of cryptic chromosomal lesions in a cohort of 38 well-characterized myelodysplastic syndromes patients; results were confirmed by microsatellite quantitative PCR or single nucleotide polymorphism analysis.
Results
As compared to metaphase karyotyping, chromosomal abnormalities detected by array-based analysis were encountered more frequently and in a higher proportion of patients. For example, chromosomal defects were found in patients with a normal karyotype by traditional cytogenetics. In addition to verifying common abnormalities, previously cryptic defects were found in new regions of the genome. Cryptic changes often overlapped chromosomes and regions frequently identified as abnormal by metaphase cytogenetics.
Conclusion
The results underscore the instability of the myelodysplastic syndromes genome and highlight the utility of array-based karyotyping to study cryptic chromosomal changes which may provide new diagnostic information.
Keywords: Myelodysplastic syndromes, array-based comparative genomic hybridization, cytogenetic abnormalities, genomic instability
INTRODUCTION
In the myelodysplastic syndromes (MDS), clonal chromosomal defects convey important prognostic information and are likely responsible for the clinical behavior of the dysplastic hematopoietic clones1–4. There remains considerable clinical variability within MDS patients harboring identical non-random chromosome defects as well as in those with a normal karyotype. It is possible that, in addition to intrinsic factors such as immune function and clinical co-morbidities in the host, additional cryptic chromosome defects may shape the biological behavior of the clones. Such discrete lesions may defy current detection by metaphase cytogenetics, in agreement with the hypothesis that the presence of chromosomal defects is related to a generalized weakness in the DNA repair machinery. Between 40% and 60% of patients with MDS are said to have normal karyotypes, likely a large overestimate give the clonal nature of the disorder1. This pathogenetic mechanism could explain the frequent occurrence of complex karyotypes or the presence of multiple clones in MDS. Thus, the initial genomic defect may be the acquisition of multiple random lesions, while the subsequent process of selection leads to the establishment of the most permissive clone characterized by an individual or multiple defect(s) that provides the most favorable selection advantage.
Metaphase cytogenetics has genomic resolution limited to defects that produce visible changes in chromosome number or banding pattern, and requires live, dividing cells, precluding detection of submicroscopic chromosomal defects that may exist in a significant proportion of cases. Comparative genomic hybridization (CGH) allows for the direct comparison of normal and abnormal genomes for the identification of copy number changes5, but the level of resolution is limited by the platform on which it is performed; for example, metaphase-CGH can only detect lesions of 2 to 10 Mb6,7 when present in at least 50% of the cells analyzed8. To overcome the limitations of metaphase CGH, array-based CGH (A-CGH), utilizing well-defined genomic clones rather than metaphase chromosomes as a hybridization target, has recently been developed. The level of resolution in A-CGH is dependent on the size of the inserts and the genomic distance between the clones spotted on the array7 and theoretically can approach linearity. In addition, A-CGH does not require a live, dividing cell population and can be performed using DNA isolated from archived samples. Analysis of A-CGH is also objective, amenable to automation and can be performed without special training or equipment. Due to these advantages, A-CGH is anticipated to become a powerful and more widely used tool in molecular cytogenetics. In general, recently-developed array-based technologies have emerged, improving the resolution level and overcoming many of the technical limitations of traditional cytogenetics.
Since its introduction, A-CGH has been used primarily to study chromosomal abnormalities in solid tumors9–11. Fewer studies have focused on hematologic malignancies12–19, despite the fact that hematologic neoplasms are relatively easily sampled using blood or bone marrow, can readily be separated from contaminating normal cells using cell sorting techniques and often have less complex karyotypes than solid tumors.
We hypothesized that A-CGH would allow for the identification of previously cryptic chromosomal lesions in patients with MDS that would better define a phenotype of chromosomal instability. Detection of additional non-random lesions may lead to an improved classification of MDS cases, identification of unifying lesions and assignment of corresponding phenotypes.
MATERIALS AND METHODS
Patients and controls
Bone marrow samples were collected according to protocols approved by the Institutional Review Board of the Cleveland Clinic Foundation (Cleveland, OH) from 38 patients with MDS and 11 healthy controls (Table 1). Patients were classified according to FAB20 and WHO21 criteria as well as the IPSS scoring system1.
Table 1.
Patient characteristics.
| Diagnosis | Del 5q | −7 | +8 | Other | Complex | Normal/NG | |
|---|---|---|---|---|---|---|---|
| MDS (N=38) | RA/RARS (n=18) | 1 | 1 | 0 | 7 | 1 | 8 |
| RAEB/RAEB-t/AML (N=14) | 1 | 1 | 1 | 2 | 2 | 7 | |
| CMML (N=6) | 0 | 1 | 0 | 1 | 0 | 4 | |
| Normal (N=11) |
NG, no growth.
Cytogenetic analysis
Short term (24–48 hour) cultures were initiated in media with and without supplementation by GM-CSF or Conditioned medium III. Following harvest, metaphase preparations were G-banded (GTG) according to standard techniques. Clonal karyotypes were described according to ISCN(1995) 22. A minimum of twenty metaphases were analyzed whenever possible.
DNA extraction and quantification
DNA was extracted from whole bone marrow with the Puregene DNA Purification System Blood Kit (Gentra Systems, Minneapolis, MN). Red blood cell lysis solution was added to whole bone marrow at a 3:1 ratio and incubated with shaking for 10 minutes. The cells were pelleted and the DNA extracted as per the manufacturer’s instructions. The concentration of the DNA was obtained using a ND-1000 spectrophotometer (NanoDrop, Wilmington, DE) and the quality determined by gel electrophoresis.
Array-based comparative genomic hybridization analysis
The array platform used in these studies contained 2,632 BACs, containing large human genomic inserts, with an average coverage of 1 BAC per 1 Mb. Sample labeling and array hybridization was performed according to the manufacturer’s protocol. One μg of patient and reference male (Promega, Madison, WI) were labeled by random priming with either Cy3 or Cy5 (PerkinElmer Life Sciences, Boston, MA). To control for experimental error, a dye-swap protocol was used for all samples. The probes were applied to the arrays and hybridized overnight. After washing, the arrays were dried under a stream of compressed N2 and scanned using the GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA). Image analysis was performed using SpectralWare 2.2 (Spectral Genomics).
SNP microarray analysis
Karyotype changes as detected by A-CGH were verified by high-density microarray SNP analysis using the 50K Xba assay (GeneChip Mapping 100K Set, Affymetrix, Santa Clara, CA) as per manufacturer’s protocol. Briefly, 250 ng of patient genomic DNA was digested with XbaI and ligated with a Xba-specific adaptor. Ligated sequences were amplified with adaptor-specific primers, fragmented, labeled with a biotinylated deoxynucleotide and hybridized to the microarray. Hybridized probes were detected with streptavidin-conjugated phycoerythrin. The arrays were scanned and genotypes called as described previously23. Copy number analysis was performed using the Copy Number Analysis Tool (Affymetrix, Santa Clara, CA).
Microsatellite analysis
Quantitative PCR for CA microsatellite was also used to verify A-CGH results. Microsatellites were identified using the Human Genome Browser (http://www.genome.ucsc.edu/cgi-bin/hgGateway). 40 ng of patient DNA was amplified as described24 in triplicate on the ABI 7500 Real Time PCR system (Applied Biosystems, Foster City, CA). Relative quantities of the microsatellites in patients as compared to a reference DNA (the same used in the A-CGH analysis) were determined using the Relative Quantification Study analysis of the 7500 System Software (Applied Biosystems, Foster City, CA).
RESULTS
Clinical features of patients with MDS
We utilized an array-based CGH approach to study the presence of cryptic chromosomal lesions undetectable by traditional karyotyping in patients with MDS. Large genomic defects have diagnostic and prognostic implications in MDS, but a considerable variability of clinical outcomes and features exist among patients with similar lesions or in those with a normal karyotype. We hypothesized that smaller chromosomal defects may be present in a much higher proportion of patients than estimated by karyotyping, implying an underlying chromosomal instability. Using A-CGH, we examined bone marrow samples from 38 patients with MDS (Table 1). The cohort included patients with low grade (RA/RARS, N=18) and high grade (RAEB/RAEB-t/AML, N=14) MDS as well as CMML (N=6).
Results of cytogenetic analysis
Results of cytogenetic analysis are summarized in Table 2. Nineteen patients (50%) had cytogenetic abnormalities as detected by traditional metaphase karyotyping; 19 patients (50%) had either a normal karyotype or were non-informative due to no growth. Monosomy 7 was found in 3 patients and del5q (q22q33 and q15q31) in 2 patients. Other karyotypic abnormalities included trisomy 8 (N=1), loss of chromosomes X and Y (N=2), gain of 11, 21 and Y, loss of material from 20q, gain of 12q and derivative chromosomes 3 and 16. Three patients had multiple karyotypic changes within a single clone.
Table 2.
A-CGH analysis of bone marrow from MDS patients.
| CGH results
|
|||||
|---|---|---|---|---|---|
| # | Cytogenetics | Gain | Loss | # changes | |
| Abnormal Metaphase Cytogenetics | 1 | 45,XY,t(1;4)(q32;q35),del(5) (q13q33), del(8)(p11.2-21)[20] | 17p12, 22q11, 22q13 | 5q21 | 4 |
| 3 | 46,XY,del(20)(q11.2q13.3)[19]/46,XY[1] | 2p22.3 | 3p14.1 | 2 | |
| 10 | 47,XX,+21[20] | 1p36.32 | NCD | 1 | |
| 12 | 46–47,XY,add(3)(q11.2),del(5) (q13q33),+8,-15,-16,+1-2 mar[cp20] | 3q13-3q25, 4q, 15q12-15q21, 16q12-qter | 4p, 8 | ND | |
| 14 | 46,XX,del(20)(q11.2q13.3)[7]/46,XX[13] | 2q22-q23, 21q22.3, | 20q | 3 | |
| 15 | 46,XY,del(7)(q22q34)[4]/45,X,-Y[2] | NCD | 7q21.1-31.3 | 21 | |
| 18 | 48-49,XY,+8,+9,+11[cp6]/46,XY[17] | 9p13.3, 11p12, 17q24 | 4q21.2 | 4 | |
| 23 | 47,XY,+Y[3]/47,XY,+8[3]/46,XY[16] | NCD | 2p14, 2q14.2, 2q23, 4p16.1, 4p15.1, 6p21.2, 6q12, 6q16.1, 6q23, 13q22.3, 15q21.1, 16p13.2 | 12 | |
| 26 | 46, XX,der(16)t(1;16) (q12;q11.2)[17]/46,XX[3] | 1p36, 1q arm, 22q13.33 | 8q24, 10q26; 16q arm | 83 | |
| 27 | 46,XY,del(5)(q15q31)[11]/46,XY[9] | NCD | 13q21.1, Xq27.3 | 2 | |
| 28 | 45,XX,-7[20] | 1p36.32, 1q31, 6q24 | whole 7, 8q24.2 | 49 | |
| 29 | 46,XY, del (5) (q22q33) [20] | 2p21 | 5q32 | 4 | |
| 32 | 47,XY,+Y[20] | Xq23, Yq11.2 | 8q24.2 | 4 | |
| 33 | 46,XY,del(20)(q11.2)[6]/46, idem,del(7)(p12)[2]/46,XY,ider(20) (q10)del(20)(q11.2)[8]/46,XY[4] | 20q13.33 | 12p13.2, 15q21.3, 20p11-13, 20q11-13 | 20 | |
| 34 | 47,XY,+11[2]/46,XY[18] | 1p36.3, 10q26.3, 10q13.33, 22q13.3 | 3q22 | 5 | |
| 36 | 47,XX,der (3) (3pter-3q13.1::3q21-3q21::3q24-3qter),+8[22] | NCD | 3q13, 3q21.3 | 6 | |
| 37 | 46,XY,del(7)(q11.2)[10]/46,XY,add(12)(q24.3)[7]/46,XY[3] | NCD | 11q14 | 1 | |
| 9 | 45,X,-X[5]/46,XX[15] | NCD | NCD | 0 | |
| 11 | 48,XY,+8,+8[6]/46,XY[14] | NCD | NCD | 0 | |
|
| |||||
| Normal Metaphase Cytogenetics | 2 | 46,XY[20] | 2q tel, 3p21.3, 5q33, 7p12, 7q11.23, 11ptel, 11p15.5, 17p12, 17q21, 22q11.2, 22q13.1 | 1q44, 2q23, 3p14, 3q26, 21q11.2, Xq12 | 20 |
| 7 | 46,XX[20] | NCD | 9q21.11 | 1 | |
| 13 | 46,XY[20] | 1p36.13, 1q22, 2p24, 3p22-3p25.3, 4p15.2, 4q32, 5q32, 6p21.1, 6q21, 14q12, 17p13.3, 17q24 | NCD | 15 | |
| 16 | 46,XX[20] | NCD | 1p21-1p22.1 (contiguous) | 2 | |
| 17 | 46,XY[20] | 1pter, 1p36.33, 2q35, 2q37.3, 4p, 5ptel, 6q24, 7ptel 8qtel, 9qtel, 10qtel, 13qtel, 17ptel, 17qtel, 19qtel 20qtel, 22qtel | NCD | 23 | |
| 19 | 46,XY[20] | TNTL | TNTL | 116 | |
| 20 | 46,XY[20] | 3q26 | 12q21.3 | 2 | |
| 21 | 46,XY[20] | 1p36.33, 9q34, 17p13.3 | 4q23, Xq21, Xq27.3, Xq28 | 8 | |
| 22 | 46,XY[20] | 7q11.2, 8q23, 9q21.2, 14q23.1, Xq21.1 | 1p35.1, 2q22, 4q32, 7p21, 9q21, 11p12, 12q12, 13q22.3, 15q23, 21p21.3 | 15 | |
| 24 | 46,XY[20] | 3p25, 5q32, 17p13.3 | NCD | 5 | |
| 25 | 46,XY[20] | 2q37.1, 5p15.33, 18p11.32, 19p13.2, Xp22.33 | NCD | 5 | |
| 35 | 46,XX[20] | 1q12 | 12q24.3 | 2 | |
| 38 | 46,XY | NCD | 2q32.3, 3p26.3, 8p12 | 3 | |
| 39 | 46,XY[20] | 10q26.3 (contiguous) | NCD | 2 | |
| 8 | 46,XY[6] | NCD | NCD | 0 | |
| 30 | 46,XY[20] | NCD | NCD | 0 | |
| 31 | 46,XY[20] | NCD | NCD | 0 | |
|
| |||||
| 5 | No growth | 10q26.3 | NCD | 1 | |
| 4 | No growth | NCD | NCD | 0 | |
Karyotype of patients as determined by traditional karyotyping and A-CGH are shown. NCD, no changes detected; ND, not determined; TNTL, too numerous to list.
Validation of A-CGH results
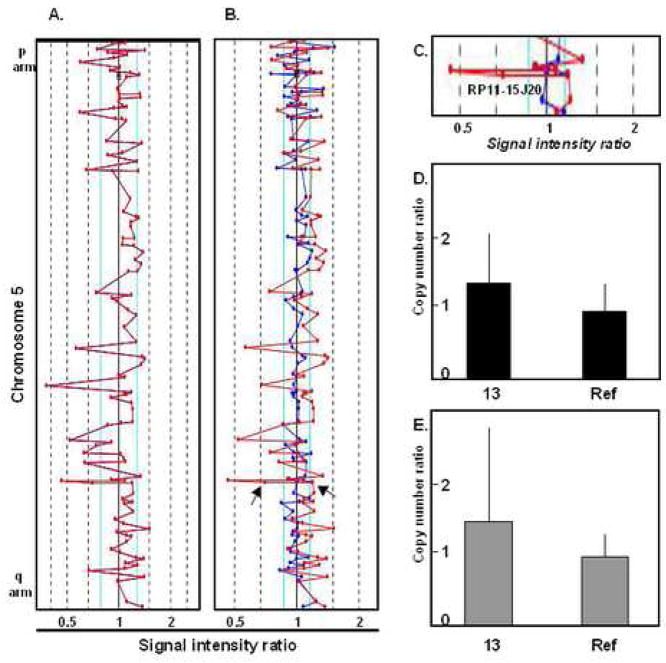
Traditional metaphase cytogenetic analysis has a level of resolution of approximately 5Mb. In contrast, the CGH arrays utilized in this study have an average coverage of 1 clone per Mb of genomic DNA, greatly increasing the level of resolution25. A dye-swap method, in which the test and reference DNA samples were reverse-labeled and hybridized to a second array, was used to control for experimental artifacts; this approach greatly reduced the number of potential false positive results (Figure 1A). For further analysis only concordant results (i.e. those found in both channels (Fig. 1B)) were used and validated by other methods (Fig. 1D, 1E).
Figure 1. Dye-swap technology as a method to control for experimental artifacts.
Microarray analysis and microsatellite quantitative PCR assay for chromosome 5 from patient #13 are shown. The dotted lines in the microarray analyses indicate the ratio of sample intensities between the test (patient bone marrow DNA) and reference (male genomic DNA) samples. Loci that fall outside of the solid lines have intensities that significantly differ from the reference and are scored as lesions by the SpectralWare Web version 2.2.40 analysis software. A. The signal intensity ratio results for the Cy3 channel are shown. If the Cy3 channel alone was analyzed multiple changes would be called along the length of chromosome 5. B. When both the Cy3 and Cy5 results are analyzed, only one loci (marked by an arrow) is significant in both channels. This change is scored as a gain of copy number. C. The locus expanded in #13 is identified as BAC RP11-15J20 at 5q31.3. If the gain identified is the duplication of BAC-specific sequences on one homologue, the copy number for that sequence would increase from 2 to 3, resulting in a copy number 1.5X that of the control, as seen here. D. The expansion within RP11-15J20 is verified using quantitative PCR for the CA microsatellite AF052687. E. Quantitative PCR for CA microsatellite A5S1979 also authenticates the expansion on 5q31.3 in #13.
Recently, it has become apparent that the human genome harbors a great deal of large-scale copy number polymorphisms25,26. If a clone from such a polymorphic region was included on the CGH array, a proportion of all samples tested would show changes at this locus. Chromosomal changes detected by A-CGH for each sample were compared to the Database of Genomic Variants25 (http://projects.tcag.ca/variation/) and those loci which showed copy number changes in healthy controls were excluded as they most likely fall within such polymorphic regions.
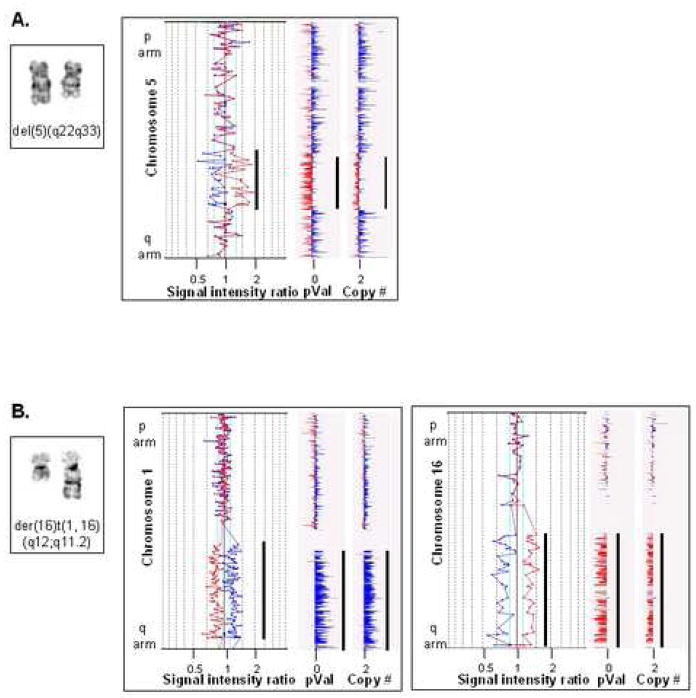
Although the dye-swap method negates many technical and experimental errors, the presence of abnormal chromosomal regions identified by A-CGH were confirmed by additional methods, including a 50K SNP array analysis (Fig. 2) and CA microsatellite-specific quantitative PCR (Fig. 3)24. Changes in small regions (Fig. 2A) as well as in large portions of chromosomal arms (Fig. 2B) detected by A-CGH were confirmed by copy number analysis using SNP arrays. Although the higher coverage of the genome by the SNP arrays unveils a finer structure of the genomic changes than the CGH array, CGH results were generally confirmed.
Figure 2. SNP chip validation of A-CGH results.
Partial karyotypes, microarray analysis and 50K SNP chip analysis are shown for 2 patients. A. By traditional karyotyping patient #29 was scored as 46,XY, del(5)(q22q33). The deletion was detected by A-CGH analysis (middle panel, black bar) and verified by 50K SNP chip analysis (right panel, black bars). B. The karyotype of patient #26 was determined to be 46,XX, der(16)t(1,16)(q12;q11.2). A-CGH and SNP analysis of chromosome 1 is shown in the middle panel; both methods detected duplication of material on the q arm of chromosome 1. The right panel is the analysis and validation of loss of chromosome 16 q arm material.
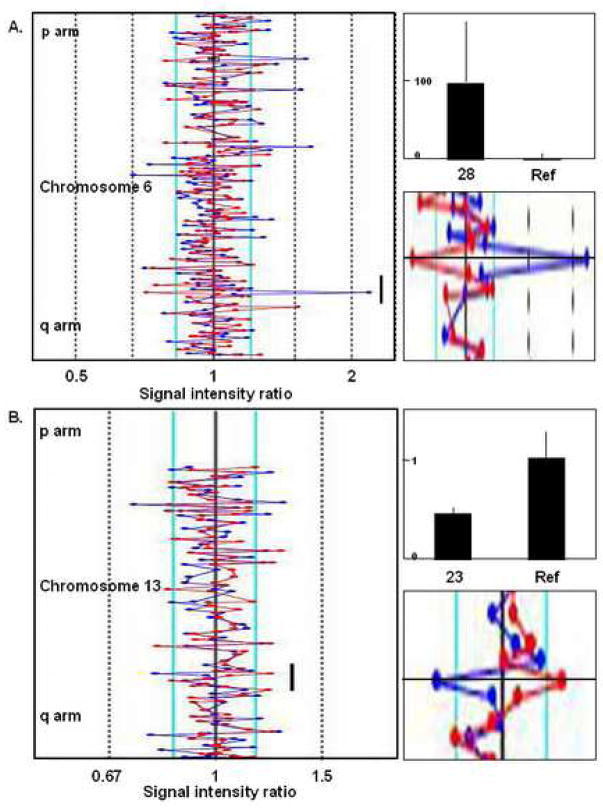
Figure 3. CA microsatellite validation of A-CGH results.
Cryptic chromosomal abnormalities undetectable by traditional cytogenetics and identified by A-CGH were also verified using a quantitative CA microsatellite PCR assay. A. A-CGH identified a gain of RP1-225E12 sequences on chromosome 6q in patient #28 (left panel, black bar). Quantitative microsatellite PCR for CA microsatellite repeat D6S1699 also detected the duplication (right top panel). The signal intensity ratios for both fluorescent channels are shown in detail for this region (right bottom panel). The lines have been traced for ease of viewing. B. #23 was determined to have a loss of RP11-753M10 sequences on chromosome 13q by A-CGH (left panel, bottom right panel). The deletion was verified by quantitative PCR analysis of CA microsatellite D13S1279 copy number (right top panel).
The copy number of microsatellites within the affected regions in the patient samples were determined relative to that of the same male genomic DNA sample used in the A-CGH experiments. Microsatellites on chromosomes not usually affected in MDS (chromosomes 2 and 12) were chosen as endogenous controls. We were able to verify the duplication and deletion of small cryptic chromosomal regions seen on A-CGH arrays that were undetectable by standard metaphase cytogenetics (for examples see Fig. 3).
A-CGH results from MDS patients and normal controls
The number of abnormalities in the genome without apparent phenotypic effect increases with age27–29. Since MDS is primarily a disease of older adults, a portion of the karyotypic lesions seen in MDS may reflect silent age-related changes. To identify the background rate of change in age-matched healthy controls, bone marrow samples from 11 healthy control individuals were studied (average 50 years old; range 35 to 66). One (N=2), 4 (N=1) and 5 (N=1) lesions (average 1) were found in the healthy controls, while the remaining 7 controls had no changes (data not shown). Additionally, no sharing of the altered loci was found.
By A-CGH analysis, cryptic chromosomal lesions were found in 32 patients (84.2% of all patients), including patients that were normal (N=14) or non-informative (N=1) by traditional metaphase karyotyping (Table 2). Consequently we were able to detect chromosomal abnormalities in 82.4% of patients with a normal cytogenetics and in 50% with an uninformative karyotype exam. Among the patients in whom karyotypic abnormalities were identified in metaphase analysis, A-CGH confirmed the results of cytogenetics in 10 patients (50%) and identified additional novel chromosomal changes in 17 samples (89.5%) known to contain karyotypic lesions. A normal karyotype by A-CGH was found in 6 patients by A-CGH: 2 patients had clonal chromosomal abnormalities by metaphase cytogenetics and 1 patients was non-informative.
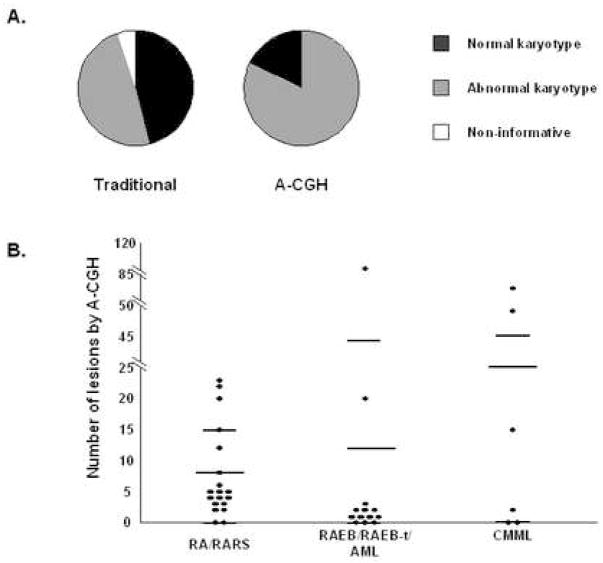
As a potential measure of chromosomal instability, we analyzed the total number of lesions. In the cohort of MDS patients studied, the average number of A-CGH abnormalities per patient was 14.82 (range 1–116, S.D. 26.27), indicating that A-CGH allows for a better detection of complex genotypes than metaphase cytogenetics. The genomes of several patients (#19, 26) contained large numbers (116 and 83 respectively) of defects. One sample (#17) demonstrated gains of multiple subtelomeric sequences. These findings demonstrate the ability of A-CGH to detect more chromosomal abnormalities than traditional cytogenetic techniques (Fig. 4A). As a consequence, A-CGH reduced the number of patients with a normal karyotype by over half. In addition, because A-CGH analysis does not require a dividing cell population, 2 cases formerly uninformative due to lack of growth of the culture could be analyzed.
Figure 4. Cryptic chromosomal abnormalities can be detected by A-CGH in patients with abnormal as well as normal karyotypes.
A. By metaphase karyotype analysis approximately 50% of patients had detectable chromosomal abnormalities. In addition 2 patients were non-informative due to a lack of growth of the culture. By A-CGH the number of patients with chromosomal abnormalities increased to 32; the non-informative cases were resolved. B. The number of lesions detected by A-CGH for patients with low- and high-grade MDS, as well as CMML is shown. Longer lines marks the average number of lesions while the shorter lines mark one standard deviation.
Genome-wide distribution of chromosomal lesions in MDS
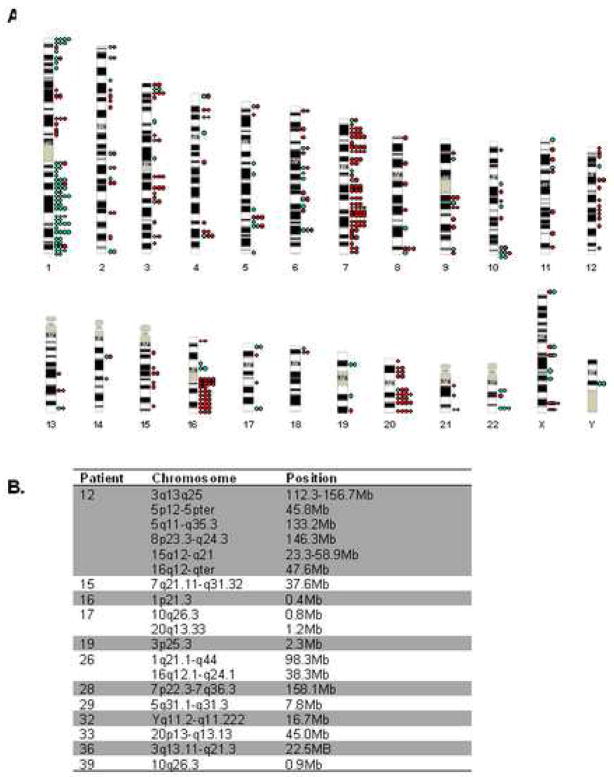
Examining the genome-wide pattern of karyotypic changes as detected by A-CGH may allow for the identification of chromosomes and/or chromosomal regions that play a role in the pathophysiology of MDS. It appears that within our cohort of MDS patients, the overall pattern of chromosomal changes is not entirely random (Fig. 5A). Multiple additional alterations were found on chromosomes, such as chromosomes 7 and 20, known to be frequently affected in patients with MDS. Additionally, chromosomes 1 and 16 harbored a large number of changes. In comparison, similarly sized chromosomes, such as chromosome 2 or chromosome 19, had fewer lesions.
Figure 5. Genome-wide view of chromosomal lesion as detected by A-CGH in patients with MDS.
A. Each dot represents a single change in a single BAC in a single patient. Gains are indicated by green and loss red. Due to the level of resolution of the chromosome ideograms lesions are grouped by band and not BAC. B. Patients that were found to harbor large changes in contiguous BACs are shown. Although many were previously identified by traditional karyotyping techniques, novel changes were found.
Within individual patients, we identified large changes in contiguous sequences (Fig. 5B). For 8 patients (#12, 15, 26, 28, 29, 32, 33, 36), these were known abnormalities; novel changes were identified for 4 patients. These included alterations on the p arm of chromosome 1 (#16), 10q and 20q (#17, Fig. 6A), 3p (#19) and the q arm of chromosome 10 (#39). Because the bacterial artificial chromosome (BAC) clones spotted on the array are on average spaced at 1Mb intervals across the genome, the presented sizes actually represent the minimum possible size of the lesion. Depending upon the coverage of that region, the changes may in fact be much larger than detected by the CGH array.
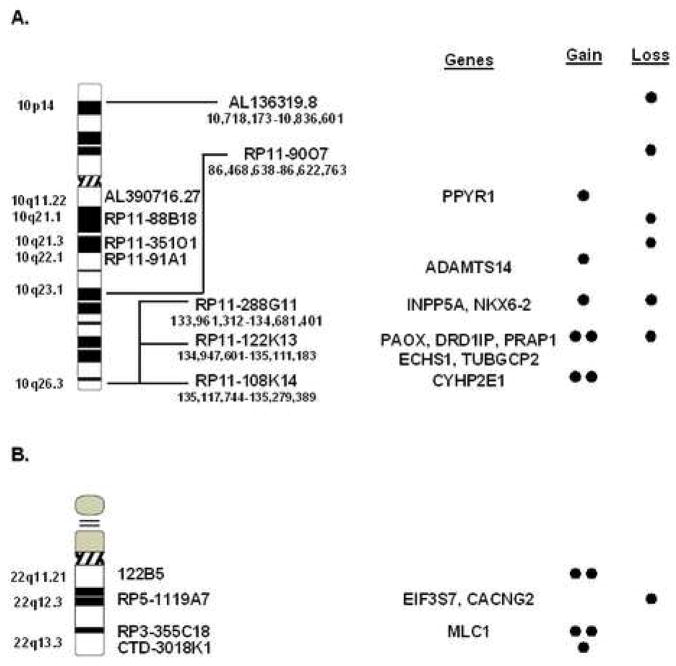
Figure 6. Regions that harbor chromosomal lesions identified by A-CGH harbor genes with a potential pathogenetic role in the etiology of MDS.
At the left the identity, cytogenetic and physical location of altered BACs are shown. Genes present on the BAC, along with whether the BACs were duplicated or deleted is indicated at the right. A. Chromosome 10. B. Chromosome 22.
Clinical implications of lesions detected by A-CGH
Three of the contiguous changes (1p21.3, 3p25, 10q26.3) identified were at chromosomal loci that are not commonly recognized as sites for non-random abnormalities in MDS. To cross reference our results, we searched the Mitelman Database of Chromosome Aberrations in Cancer30 (http://cgap.nci.nih.gov/Chromosomes/Mitelman). Patient #16, with a diagnosis of CMML, harbored cryptic loss of 1p21.3 material. Other patients with advanced forms of MDS and this particular lesion have been previously reported31,32. This region is clearly independent of the region of allelic loss of material on 1p32-p36.3 associated with progression from MDS to AML33. A-CGH also detected loss of 3p25 sequences in a patient with RAEB-t, #19. This deletion has been previously identified in two patients with MDS, including one also diagnosed as RAEBT 34,35. Two patients with RA, #17 and 39, had loss of 10q26 sequences, a finding which has been reported in one additional patient with RA36. Abnormalities of these regions have been identified in a number of malignancies, mainly solid tumors.
We were interested to learn whether small genomic changes identified by A-CGH in chromosomes that are monosomic or trisomic in MDS might pinpoint minimum common segments as targets for further study. We identified patients with A-CGH changes on chromosomes 5, 7, 8, 11 and 20 that were not detected by standard chromosomal analysis. Additionally, we identified 6 patients who had segmental gains mapping to 1p36.3; 5/6 patients were diagnosed with MDS/MPD overlap syndrome (including CMML and MPD/MDS-U), and 4/6 had a good IPSS score, that correlated with an absence of transformation to AML after 2 years of follow-up.
A genome-wide analysis of chromosomal lesions may allow for the identification of chromosomes and/or regions that play a role in the pathophysiology of MDS and, conversely, exclude certain chromosomes as uninvolved. Identification of smaller shared regions, such as individual BAC clones, would greatly aid in pinpointing genes with a potential role in the phenotype of MDS for further investigation. We identified 41 shared, or common, single BAC clones altered in two or more patients with MDS (for example, see Fig. 6). Several chromosomes (11, 12, 15, 16, 17, 18, 19 and 21) did not contain shared lesions. Approximately half of the shared lesions overlapped known genes or portions of genes. One example was gamma-tubulin complex component 2 (TUBGCP2) on chromosome 10, which is necessary for centrosome nucleation and may play a role in chromosome stability during mitosis (Fig. 6A)37,38. Also affected was eukaryotic translation initiation factor 3 (EIF3S7) on chromosome 22 which binds to the 40S ribosome and may play a role in the increased transcription rates seen in T cells during activation (Fig. 6B)39.
DISCUSSION
Large chromosomal lesions are frequently identified in MDS; however, approximately half of patients tested have a normal karyotype by metaphase cytogenetics. Phenotypic heterogeneity exists even between patients with the same karyotypic abnormality, strongly suggesting that genomic changes that are cryptic by traditional cytogenetic techniques, are common in MDS. For the first time and in a systematic fashion, we have utilized A-CGH to investigate the frequency and location of these cryptic changes in a large cohort of well-characterized patients with MDS to define the chromosomal instability phenotype that underlies the pathogenesis of the syndrome. Our study demonstrates the potential clinical applicability of array-based techniques, such as A-CGH, that allow for a more precise evaluation of chromosomal defects.
Although there was a high concordance between metaphase karyotyping and A-CGH results, genomic abnormalities were more frequently identified in patients using A-CGH than by cytogenetic analysis. In addition, we identified additional, cryptic lesions in patients with known cytogenetic abnormalities that most likely modify the phenotype. A-CGH identified genomic abnormalities in two cases in which cytogenetics was unsuccessful due to no growth.
As compared to healthy controls, patients with MDS had a higher number of changes overall. That the majority of changes involved single BAC clones further supports the suggestion that there is an underlying phenotype of chromosomal instability in MDS. Several of the affected BAC clones were on chromosomes or in chromosomal regions frequently identified as abnormal in MDS by traditional cytogenetics, including loci on 5q, 7 and 8. The comparison of clinical outcomes between patients with circumscribed lesions and much larger changes may allow for the definition of a minimal critical region(s) responsible for the phenotype.
We identified lesions within regions defined by changes in the copy number of contiguous clones that escaped detection by traditional karyotyping. Of note is that involvement of one BAC versus several consecutive BACs cannot be used to determine the size of the lesion. Due to the clone coverage within any given region and the genomic map distance between the BACs, a larger chromosomal region may still be involved if it is only represented by one BAC clone. However, the alteration of several contiguous clones more reliably defines a larger segmental change that may have greater biological significance. As with single BAC changes, these contiguous lesions most likely modify the effect of other chromosomal changes. For example, we identified a novel gain of material at 1p36.3 that appears to have a good prognosis, in stark contrast to the loss of 1p36.3 that is associated with poor prognosis in hematologic33 and solid40 malignancies.
Unlike in hematologic malignancies, A-CGH has been more often used to study chromosomal abnormalities in solid tumors41, including medulloblastoma9, breast10 and gastric11 cancers. By high-resolution karyotypic analysis, breast carcinoma karyotypes have been refined42 and novel lesions have been identified in non-Hodgkin lymphoma43. Copy number abnormalities detected by A-CGH can aid in the differential diagnosis of renal cell cancer44 and can correlate with disease stage and patient survival in lung and breast cancer45,46. High-resolution karyotypic analysis can also identify potential targets of new therapies in malignant histiocytomas47. In our studies, A-CGH results suggest a higher level of chromosomal instability in MDS, analogous to cancer using metaphase CGH46. Additionally, we detected novel chromosomal lesions involving single and multiple clones, similar to what has been seen in pediatric medulloblastoma9, breast cancer10 and natural killer cell lymphoma/leukemia14. In our cohort A-CGH identified novel karyotypic abnormalities in 3/5 patients with additional copies of chromosome 8. Similarly, in a recent report, 9/10 MDS patients with trisomy 8 harbored cryptic chromosomal lesions by A-CGH, some of the changes occurring in 2 or more patients12.
A-CGH has many advantages over traditional metaphase karyotyping, including a higher level of resolution and the ability to perform retrospective studies using DNA isolated from archived material. In addition, a much larger number of cells can be analyzed at one time. The fraction of abnormal cells in the sample from which DNA is extracted can have a critical impact on the detection of abnormalities by A-CGH. In one study, although a clone accounting for 12.5% of the total cell population was undetectable using A-CGH12, copy number changes could be identified in a sample consisting of 30% tumor cells and 70% normal tissue48. In MDS, several dysplastic clones may initially contribute to a largely oligoclonal stem cell pool. In this circumstance the pathogenetically relevant chromosomal abnormalities would remain undetected by A-CGH. Similarly, because abnormalities are scored within single mitotic cells in conventional cytogenetics, rather than within all cells in the sample as in A-CGH, abnormalities that are more representative of the dividing cellular population may be better identified with cytogenetics. Unlike traditional cytogenetics, A-CGH cannot detect balanced chromosomal abnormalities. However, as a majority of chromosomal lesions in MDS are unbalanced, this limitation would have little effect on our study. Such mechanisms may account for some of the discrepancies observed between A-CGH and traditional chromosomal analysis.
Additionally, some changes may be reflective of as of yet unidentified genomic copy number polymorphisms. We took several measures to reduce the number of false positives generated by A-CGH. We used a dye-swap technique to reduce the number of false calls that could arise from technical artifacts. Any clones that fell within regions of genomic copy number polymorphisms were excluded from further analysis. We used both quantitative PCR for microsatellites and SNP array analysis to confirm our A-CGH findings. For quantitative PCR, we used the same male reference DNA used in A-CGH as the calibrator sample. SNP array analysis was performed for lesions that did not overlap a CA microsatellite and allowed us to more finely delineate the boundaries of the lesions.
Although our cohort of healthy controls consisted primarily of older individuals, the age distribution is younger than the patient cohort. Therefore, it is possible that a proportion of the lesions detected in the patients reflect normal age-related chromosomal changes. However, it is difficult to obtain putatively “normal” samples from older individuals, as most receive bone marrow biopsies on the suspicion of hematologic disease. Therefore, the number of healthy individuals studied was limited.
The application of A-CGH to study karyotypic abnormalities in MDS has many implications. From an investigative standpoint it may be possible to define a minimal shared region within the large genomic alterations such as monosomy or trisomy that play a role in MDS. Additionally, cryptic lesions that modify the expression of the MDS phenotype can be identified. The rate of general chromosomal instability, which may predispose to MDS or have prognostic significance, can be measured and quantified in a large number of patients. It may also be possible to determine the a pathogenetic sequence of genetic abnormalities, which in turn may help define whether the instability phenotype is due to e.g. inefficiency of the DNA repair pathways or spindle formation defects. Clinically, A-CGH may help to refine the prognosis for known lesions (i.e. trisomy 8, monosomy 7) according to what smaller lesions are present in the clone. New lesions identified by high-resolution karyotyping methods may have clinical significance, or they may prove to be targets of novel therapies.
Table 3.
Shared genomic regions of change as detected by A-CGH.
| Clone | Cytogenetic localization | Sample and change | Genes |
|---|---|---|---|
| RP11-421C4 | 1p36.3 | 17 G, 34G | MRPL20 |
| RP4-703E10 | 1p36.32 | 10 G, 26 G, 28 G | |
| RP1-62I8 | 1p36.33 | 17 G, 21 G | |
| RP11-433J22 | 1q21.1 | 19 L, 26 G | ACP6, GJA5 |
| RP11-79M15 | 1q23.3 | 13 G, 19 L | CD48, SLAMF7, LY9* |
| RP11-2l82G6 | 2p24.3 | 13 G, 19 L | |
| RP11-30M1 | 2q32.3 | 19 L, 38 L | |
| RP11-155G3 | 3p25.3 | 13 G, 19 L | |
| RP11-63O1 | 3p26.3 | 19 L, 38 L | CNTN4 |
| RP11-745L2 | 3q13.13 | 19 L, 36 L | |
| RP11-81H11 | 4p15.1 | 13 G, 19 L | |
| RP11-6F19 | 4q32.3 | 13 G, 19 L | TLL1 |
| AC008406.7 | 5q31.1 | 19 L, 29 L | CATSPER3, PITX1, |
| RP11-15J20 | 5q31.3 | 13 G, 19 L, 24 G | DIAPH1, HDAC3*, FCHSD1, CENTD3 |
| RP11-81F7 | 6p21.1 | 13 G, 19 L | |
| RP1-129L7 | 6q12 | 19 G, 23 | |
| RP11-91B17 | 6q21 | 13 G, 19 L | SEC63, OSTM1 |
| RP1-225E12 | 6q24.1 | 17 G, 28 G | HECA |
| RP11-89B15 | 7p21.1 | 22 L, 28 L | MEOX2 |
| RP11-88D24 | 7q21.11 | 15 L, 28 L | |
| RP11-46O13 | 7q21.13 | 15 L, 28 L | |
| RP11-79O7 | 7q21.3 | 15 L, 28 L | |
| AC005064.3 | 7q22.1 | 15 L, 28 L | PRES, RELN |
| RP11-80P24 | 7q22.1 | 15 L, 28 L | CUTL1* |
| RP11-72J24 | 7q22.3 | 15 L, 28 L | MLL5*, SRPK2 |
| RP11-77E2 | 7q31.1 | 15 L, 19 L | DLD, LAMB1, LAMB4 |
| RP11-110C11 | 7q31.2 | 15 L, 28 L | |
| RP11-51M22 | 7q31.2 | 15 L, 28 L | TES |
| RP11-112P4 | 7q31.32 | 15 L, 28 L | PTPRZ1 |
| RP11-140O21 | 7q31.31 | 15 L, 19 L | |
| RP11-17M8 | 8q24.23 | 26 L, 28 L, 32 L | |
| RP11-203L2 | 9q13 | 7 L, 19 L | PIP5K1B |
| RP11-108K14 | 10q26.3 | 5 G, 39 G | GTP, CYP2E1 |
| RP11-122K13 | 10q26.3 | 17 G, 19 L, 34 G | D26579, TUBGCP2*, ZNF511, DRD1IP, PRAP1, ECHS1, PAOX, GTP |
| RP11-288G11 | 10q26.3 | 17 G, 26 L | INPP5A |
| RP11-753M10 | 13q31.3 | 22 L, 23 L | |
| RP11-557O15 | 14q13.1 | 13 G, 19 L | NPAS3 |
| RP5-824J5 | 20q12 | 14 L, 33 L | |
| AL118506.27 | 20q13.3 | 17 G, 33 G, 34 G | TPD52L2, DNAJC5, UCKL1, SAMD10 |
| 122B5 | 22q11.21 | 17 G, 19 G | |
| RP3-355C18 | 22q13.33 | 17 G, 34 G | MLC1 |
The identities of clones that are altered in 2 or more samples are shown. Cytogenetic localization and genes within the clones were identified using the UCSC Human Genome Browser (http://www.genome.ucsc.edu/cgi-bin/hgGateway). Genes marked with * play a role in hematopoiesis, chromosomal stability and gene expression. G, gain; L, loss.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- 1.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 2.Morel P, Hebbar M, Lai JL, et al. Cytogenetic analysis has strong independent prognostic value in de novo myelodysplastic syndromes and can be incorporated in a new scoring system: a report on 408 cases. Leukemia. 1993;7:1315–1323. [PubMed] [Google Scholar]
- 3.Pfeilstocker M, Reisner R, Nosslinger T, et al. Cross-validation of prognostic scores in myelodysplastic syndromes on 386 patients from a single institution confirms importance of cytogenetics. Br J Haematol. 1999;106:455–463. doi: 10.1046/j.1365-2141.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- 4.Sole F, Luno E, Sanzo C, et al. Identification of novel cytogenetic markers with prognostic significance in a series of 968 patients with primary myelodysplastic syndromes. Haematologica. 2005;90:1168–1178. [PubMed] [Google Scholar]
- 5.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 6.Inazawa J, Inoue J, Imoto I. Comparative genomic hybridization (CGH)-arrays pave the way for identification of novel cancer-related genes. Cancer Sci. 2004;95:559–563. doi: 10.1111/j.1349-7006.2004.tb02486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantripragada KK, Buckley PG, de Stahl TD, Dumanski JP. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Lapierre JM, Tachdjian G. Detection of chromosomal abnormalities by comparative genomic hybridization. Curr Opin Obstet Gynecol. 2005;17:171–177. doi: 10.1097/01.gco.0000162188.99219.04. [DOI] [PubMed] [Google Scholar]
- 9.Rossi MR, Conroy J, McQuaid D, et al. Array CGH analysis of pediatric medulloblastomas. Genes Chromosomes Cancer. 2006;45:290–303. doi: 10.1002/gcc.20292. [DOI] [PubMed] [Google Scholar]
- 10.Naylor TL, Greshock J, Wang Y, et al. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–R1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss MM, Kuipers EJ, Postma C, et al. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004;26:307–317. doi: 10.1155/2004/454238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsson K, Heidenblad M, Strombeck B, et al. High-resolution genome-wide array-based comparative genome hybridization reveals cryptic chromosome changes in AML and MDS cases with trisomy 8 as the sole cytogenetic aberration. Leukemia. 2006 doi: 10.1038/sj.leu.2404145. [DOI] [PubMed] [Google Scholar]
- 13.Tyybakinoja A, Saarinen-Pihkala U, Elonen E, Knuutila S. Amplified, lost, and fused genes in 11q23–25 amplicon in acute myeloid leukemia, an array-CGH study. Genes Chromosomes Cancer. 2006;45:257–264. doi: 10.1002/gcc.20288. [DOI] [PubMed] [Google Scholar]
- 14.Nakashima Y, Tagawa H, Suzuki R, et al. Genome-wide array-based comparative genomic hybridization of natural killer cell lymphoma/leukemia: different genomic alteration patterns of aggressive NK-cell leukemia and extranodal Nk/T-cell lymphoma, nasal type. Genes Chromosomes Cancer. 2005;44:247–255. doi: 10.1002/gcc.20245. [DOI] [PubMed] [Google Scholar]
- 15.Rubio-Moscardo F, Climent J, Siebert R, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 16.Babicz M, Kowalczyk JR, Winnicka D, et al. The effectiveness of high-resolution-comparative genomic hybridization in detecting the most common chromosomal abnormalities in pediatric myelodysplastic syndromes. Cancer Genet Cytogenet. 2005;158:49–54. doi: 10.1016/j.cancergencyto.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Kim MH, Stewart J, Devlin C, et al. The application of comparative genomic hybridization as an additional tool in the chromosome analysis of acute myeloid leukemia and myelodysplastic syndromes. Cancer Genet Cytogenet. 2001;126:26–33. doi: 10.1016/s0165-4608(00)00386-1. [DOI] [PubMed] [Google Scholar]
- 18.Castuma MV, Rao PH, Acevedo SH, Larripa IB. Comparative genomic hybridization study of de novo myeloid neoplasia. Acta Haematol. 2000;104:25–30. doi: 10.1159/000041065. [DOI] [PubMed] [Google Scholar]
- 19.Wilkens L, Burkhardt D, Tchinda J, et al. Cytogenetic aberrations in myelodysplastic syndrome detected by comparative genomic hybridization and fluorescence in situ hybridization. Diagn Mol Pathol. 1999;8:47–53. doi: 10.1097/00019606-199903000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Bennett JM, Catovsky D, Daniel MT, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982;51:189–199. [PubMed] [Google Scholar]
- 21.Harris NL, Jaffe ES, Diebold J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999;17:3835–3849. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 22.ISCN 1995. An International System for Human Cytogenetic Nomenclature. Basel: S. Karger; 1995. [Google Scholar]
- 23.Kennedy GC, Matsuzaki H, Dong S, et al. Large-scale genotyping of complex DNA. Nat Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 24.Nigro JM, Takahashi MA, Ginzinger DG, et al. Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am J Pathol. 2001;158:1253–1262. doi: 10.1016/S0002-9440(10)64076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 26.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 27.Guttenbach M, Koschorz B, Bernthaler U, Grimm T, Schmid M. Sex chromosome loss and aging: in situ hybridization studies on human interphase nuclei. Am J Hum Genet. 1995;57:1143–1150. [PMC free article] [PubMed] [Google Scholar]
- 28.Neurath P, DeRemer K, Bell B, Jarvik Kato T. Chromosome loss compared with chromosome size, age and sex of subjects. Nature. 1970;225:281–282. doi: 10.1038/225281a0. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee AB, Thomas S. A longitudinal study of human age-related chromosomal analysis in skin fibroblasts. Exp Cell Res. 1997;235:161–169. doi: 10.1006/excr.1997.3673. [DOI] [PubMed] [Google Scholar]
- 30.Mitelman Database of Chromosome Aberrations in Cancer. 2005.
- 31.Andersen MK, Christiansen DH, Pedersen-Bjergaard J. Centromeric breakage and highly rearranged chromosome derivatives associated with mutations of TP53 are common in therapy-related MDS and AML after therapy with alkylating agents: an M-FISH study. Genes Chromosomes Cancer. 2005;42:358–371. doi: 10.1002/gcc.20145. [DOI] [PubMed] [Google Scholar]
- 32.Tien HF, Wang CH, Chuang SM, et al. Cytogenetic studies, ras mutation, and clinical characteristics in primary myelodysplastic syndrome. A study on 68 Chinese patients in Taiwan. Cancer Genet Cytogenet. 1994;74:40–49. doi: 10.1016/0165-4608(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 33.Mori N, Morosetti R, Mizoguchi H, Koeffler HP. Progression of myelodysplastic syndrome: allelic loss on chromosomal arm 1p. Br J Haematol. 2003;122:226–230. doi: 10.1046/j.1365-2141.2003.04434.x. [DOI] [PubMed] [Google Scholar]
- 34.Horiike S, Taniwaki M, Misawa S, Abe T. Chromosome abnormalities and karyotypic evolution in 83 patients with myelodysplastic syndrome and predictive value for prognosis. Cancer. 1988;62:1129–1138. doi: 10.1002/1097-0142(19880915)62:6<1129::aid-cncr2820620616>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 35.Andreasson P, Hoglund M, Bekassy AN, et al. Cytogenetic and FISH studies of a single center consecutive series of 152 childhood acute lymphoblastic leukemias. Eur J Haematol. 2000;65:40–51. doi: 10.1034/j.1600-0609.2000.90190.x. [DOI] [PubMed] [Google Scholar]
- 36.Nowell P, Bergman G, Besa E, Wilmoth D, Emanuel B. Progressive preleukemia with a chromosomally abnormal clone in a kindred with the Estren-Dameshek variant of Fanconi’s anemia. Blood. 1984;64:1135–1138. [PubMed] [Google Scholar]
- 37.Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neben K, Tews B, Wrobel G, et al. Gene expression patterns in acute myeloid leukemia correlate with centrosome aberrations and numerical chromosome changes. Oncogene. 2004;23:2379–2384. doi: 10.1038/sj.onc.1207401. [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto S, Patel P, Hershey JW. Changes in ribosomal binding activity of eIF3 correlate with increased translation rates during activation of T lymphocytes. J Biol Chem. 2005;280:28251–28264. doi: 10.1074/jbc.M414129200. [DOI] [PubMed] [Google Scholar]
- 40.Caron H, van Sluis P, de Kraker J, et al. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–230. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- 41.Sanlaville D, Lapierre JM, Turleau C, et al. Molecular karyotyping in human constitutional cytogenetics. Eur J Med Genet. 2005;48:214–231. doi: 10.1016/j.ejmg.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 42.Albertson DG, Ylstra B, Segraves R, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 43.Wessendorf S, Schwaenen C, Kohlhammer H, et al. Hidden gene amplifications in aggressive B-cell non-Hodgkin lymphomas detected by microarray-based comparative genomic hybridization. Oncogene. 2003;22:1425–1429. doi: 10.1038/sj.onc.1206297. [DOI] [PubMed] [Google Scholar]
- 44.Wilhelm M, Veltman JA, Olshen AB, et al. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res. 2002;62:957–960. [PubMed] [Google Scholar]
- 45.Kristiansen G, Yu Y, Petersen S, et al. Overexpression of c-erbB2 protein correlates with disease-stage and chromosomal gain at the c-erbB2 locus in non-small cell lung cancer. Eur J Cancer. 2001;37:1089–1095. doi: 10.1016/s0959-8049(01)00096-x. [DOI] [PubMed] [Google Scholar]
- 46.Jain AN, Chin K, Borresen-Dale AL, et al. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc Natl Acad Sci U S A. 2001;98:7952–7957. doi: 10.1073/pnas.151241198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chibon F, Mariani O, Derre J, et al. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14-q15 and 6q23 amplifications. Genes Chromosomes Cancer. 2004;40:32–37. doi: 10.1002/gcc.20012. [DOI] [PubMed] [Google Scholar]
- 48.Garnis C, Coe BP, Lam SL, MacAulay C, Lam WL. High-resolution array CGH increases heterogeneity tolerance in the analysis of clinical samples. Genomics. 2005;85:790–793. doi: 10.1016/j.ygeno.2005.02.015. [DOI] [PubMed] [Google Scholar]