Abstract
In Xenopus embryos, maternal cyclins drive the first 12 cell divisions after which several cyclins are terminally degraded, including cyclin B2. Cyclin B2 disappearance is due to transcription-mediated mRNA deadenylation at the midblastula transition, when transcription initiates and the cell cycle lengthens. To further define the mechanism, we characterized proteins capable of binding cyclin B2 3′UTR. We show that ElrA and AUF1 compete for binding to regions containing cytoplasmic polyadenylation elements (CPEs), with AUF1 binding increasing at the midblastula transition. Deletion of both CPEs abrogates polyadenylation but has no effect on deadenylation or binding of ElrA or AUF1. Overexpression of ElrA or AUF1 does not alter cyclin B2 mRNA stability. These results show that ElrA and AUF1 bind to cyclin B2 mRNA independent of CPEs and function by binding other elements.
Keywords: AUF1, CPE, Cyclin B2, ElrA, RNA, RBP, Xenopus
Introduction
Xenopus laevis is a classic model for post-transcriptional control of the cell cycle via regulation of mRNA adenylation. Early development occurs in the absence of transcription and the presence of stable maternal mRNAs. Global transcription begins at the midblastula transition (MBT), about 6 hours after fertilization [1, 2]. At this time previously deadenylated maternal mRNAs are destabilized and the embryonic cell cycle is remodeled from a rapid alternation between DNA synthesis and mitosis to a cell cycle containing gap-phases and checkpoint controls. Cell cycle remodeling in Xenopus is characterized by changes in maternal cell cycle regulators [3-6]. Among these, cyclin B2 is terminally degraded, due to deadenylation of its mRNA [7]. Deadenylation begins at the MBT and requires transcription. A 60-nucleotide (nt) region in the 3′-untranslated region (3′UTR) of cyclin B2 mRNA is required for but not capable of mediating deadenylation, demonstrating that other sequence is needed. This study was undertaken to further define the mechanism of cyclin B2 deadenylation. Regulatory elements in the 3′UTR generally function by associating with RNA binding proteins (RBPs). We thus set out to identify RBPs for cyclin B2 3′UTR.
Materials and Methods
Embryos
Embryos were obtained following standard procedures [4]. For immunoblots, 5 embryos were collected per timepoint and protein extracted [4]. 2-cell embryos (1.5 hours post-fertilization, hpf) were injected with 18.4 nl of in vitro transcribed, radiolabeled capped mRNA (1 fmol/μl), 1 ng of mRNA for myc-tagged protein, or 46 ng α-amanitin. Extracts were prepared from embryos as described [8].
Constructs
Primers used are listed in supplementary Table 1. GbB2 and GbA1Δ384 contain the T7 promoter, the globin 5′-UTR and ORF, and either cyclin B2 3′UTR or cyclin A1 3′UTR missing the last 384-nt [9]. B2 and A1Δ384 were constructed by deleting the globin sequence via digestion and recircularization. A1ΔARE was prepared by deleting two AREs from A1Δ384. B2 was used for PCR amplification of the constructs B2-2, 3 and B2-2-5 with the T7 promoter sequence. PCR products were used for RNA synthesis. B2-2,3,6 was prepared from GbB2Δ1 (missing the first 30-nt of cyclin B2 3′UTR) by first deleting the globin sequence and then deleting regions 4-5 (3′UTR nt 91-150) by site-directed mutagenesis. CPEs were deleted from cyclin B2 3′UTR in the context of GbB2-poly(A)65 by site-directed mutagenesis. Plasmids were linearized with EcoRV for poly(A)65 mRNA, or BamHI for poly(A)- mRNA. pCS2-MT-ElrA was used to create pCS2-MT-ElrAΔ by deleting the hinge region and third RRM [10]. To create pCS2-MT-AUF1, the AUF1 ORF was produced by PCR from pGEM-Teasy-AUF1 and inserted into pCS2-MT. Linearized plasmids were used for in vitro transcription of mRNAs.
Cloning of Xenopus AUF1
A nucleotide-nucleotide blast with human AUF1 (NM_031369) and the Xenopus EST-database showed a 519-nt region (human sequence 576-1095) conserved with 80% homology (EST BI348076). After alignment and translation there was 95% amino acid homology between the Xenopus EST and human p37AUF1. A second nucleotide-nucleotide blast using BI348076 found ESTs representing the 5′ (BJ031274 and BI477470) and the 3′ ends (AW638522). The entire ORF (943-nt) was amplified from Xenopus embryo cDNA, cloned into pGEM-Teasy and sequenced (AY726540). The ORF was cloned into pGEX to produce GST-AUF1.
UV-crosslink analysis
UV-crosslink analysis was carried out as described [11]. For competition assays, extract or recombinant protein was incubated with 1, 5 or 10 molar excess of unlabeled cyclin B2 3′UTR or cyclin A1-ΔARE for 15 min before addition of 32P-labeled RNAs and crosslinking. For immunoprecipitation, complexes resulting from crosslinking were incubated with 1 μg nonspecific IgG, anti-HuR, or anti-AUF1 followed by protein A/G-agarose. GST-ElrA and GST-p37AUF1 proteins were expressed in E.coli and purified as recommended (Amersham Biosciences).
Immunoblotting
One embryo per lane was resolved by SDS-PAGE, transferred onto nitrocellulose, and membranes incubated with anti-AUF1 (Upstate), anti-HuR to detect ElrA (Santa Cruz), or anti-myc (Cell Signaling Technology). For comparison of ElrA and AUF1 levels before biotin pulldowns, embryo extracts (20 μg) were blotted sequentially for ElrA, AUF1 or CPEB (Dr. J. Richter, Univ. Massachusetts). All Westerns were visualized by ECL (Pierce).
Northern analysis
Five embryos per timepoint were collected and total RNA extracted with TRIzol (Invitrogen). 10 μg RNA was electrophoresed on denaturing gels and transferred to nylon membrane. Membranes were probed with 32P-CTP-labeled cDNA for cyclin B2 and ODC (loading control) using Quikhyb (Stratagene) and analyzed by phosphorimaging.
Biotin pulldowns
4 and 8h embryo extracts (300 μg) or GST-ElrA and GST-AUF1 were incubated with 1μg biotinylated RNA for 30 min. For competition experiments, 0.05 μg GST-ElrA and an increasing amount of GST-AUF1 (0, 0.05, 0.25, 0.5, 1, 2 μg), or 0.2 μg GST-AUF1 and an increasing amount of GST-ElrA (0, 0.2, 0.5, 1, 2, 4 μg) were used. The RNA-protein complexes were isolated using pre-washed paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo). Proteins were eluted with SDS-sample buffer and analyzed by immunoblotting.
Adenylation analysis
Five embryos were collected upon completion of microinjections (2 hpf) and at the times shown. Total RNA was extracted and resolved by electrophoresis on 4% polyacrylamide-urea gels and analyzed by phosphorimaging.
Results
Deadenylated cyclin B2 mRNA accumulates during the MBT, by 8hpf [7]. To identify RBPs, extracts were prepared at 4 (pre-MBT) and 8hpf (post-MBT). UV-crosslink analysis was used to show similar sized proteins bound to the cyclin B2 3′UTR at 4 and 8hpf (Figure 1A, lanes 0). To determine the specificity, unlabeled cyclin B2 3′UTR and a partial cyclin A1 3′UTR (A1-ΔARE) were included as competitors in increasing concentration. At both times, an 37-kDa complex (arrow) was competed only by cyclin B2 3′UTR. In contrast, a 20-kDa complex was not competed (arrowhead), and a 45-50-kDa complex was competed by both (double arrowheads).
Figure 1. Endogenous ElrA and AUF1 bind cyclin B2 3′UTR.
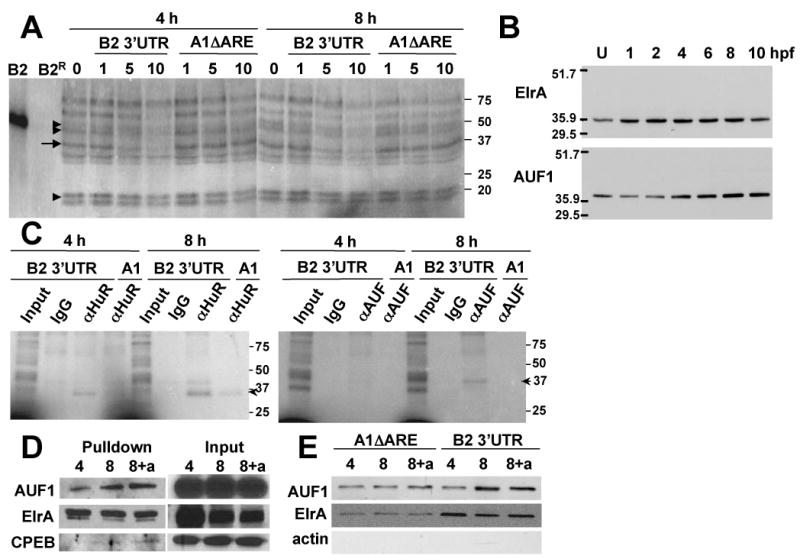
A, UV-crosslink analysis of 4 and 8h embryos with radiolabeled cyclin B2 3′UTR in the presence of 0, 1, 5, or 10 molar excess unlabeled cyclin B2 3′UTR or cyclin A1ΔARE. Arrow, 37-kDa protein competed by B2 3′UTR. Arrowhead, proteins not competed. Double arrowheads, proteins competed by both. B2, cyclin B2 3′UTR; B2R, cyclin B2 3′UTR+RNase. B, Immunoblot of unfertilized eggs (U) through 10 hpf embryos sequentially probed with ElrA and AUF1 antibodies. Top numbers, hours postfertilization. C, UV-crosslink of 4 and 8h embryos with radiolabeled cyclin B2 3′UTR or A1ΔARE (A1) followed by immunoprecipitation with IgG, αElrA or αAUF1. Input, UV-crosslink before immunoprecipitation. Arrowheads, immunoprecipitated protein. D, RNA-pulldown assay of 4 and 8h embryos with biotinylated cyclin B2 3′UTR; followed by immunoblotting for AUF1, ElrA, and CPEB (Pulldown). Input, immunoblot of embryos used for the pulldown. 8+a, embryos injected with α-amanitin. E, RNA-pulldown of 4 and 8h embryos with biotinylated cyclin B2 3′UTR or A1ΔARE; followed by immunoblotting for AUF1, ElrA, and actin. Markers are in kDa for A-C; n=3 for A, C-E; n=5 for B.
We focused on the 37-kDa complex, which was similar in size to ElrA and the 37-kDa isoform of AUF1, both proteins that bind AU-rich elements (AREs). ElrA is the Xenopus homolog of human HuR and a member of the ELAV family of stabilizing RBPs [10]. AUF1 destabilizes mRNAs [12], with the 37-kDa isoform having the highest destabilizing activity for mRNAs containing noncanonical AREs [13]. Cyclin B2 3′UTR contains noncanonical AREs, known as cytoplasmic polyadenylation elements (CPEs). Since ElrA and AUF1 were present (Figure 1B), we tested if they bound cyclin B2 3′UTR. 4 and 8h extracts were incubated with radiolabeled cyclin B2 3′UTR or A1-ΔARE. After crosslinking, the complexes were immunoprecipitated with normal IgG, HuR, or AUF1 antibodies. The HuR antibody immunoprecipitates ElrA better than an ElrA antibody, and ElrA doesn't bind A1-ΔARE [14]. Immunoprecipitated complexes were visualized by SDS-PAGE and phosphorimaging (Figure 1C). ElrA from both 4 and 8h embryos associated with cyclin B2 3′UTR (αHuR), and surprisingly, with A1ΔARE (A1). In contrast, AUF1 bound only to cyclin B2 3′UTR at 8h (αAUF).
To confirm this, we performed RNA-pulldowns followed by immunoblotting for ElrA and AUF1. 4 and 8h extracts were incubated with biotinylated cyclin B2 3′UTR and RNA-protein complexes isolated using paramagnetic streptavidin-conjugated Dynabeads. Proteins were eluted using SDS sample buffer and analyzed by immunoblotting. As shown in Figure 1D, Pulldown, ElrA and AUF1 from 4 and 8h extracts bound cyclin B2 3′UTR. The amount of ElrA bound was similar at 4 and 8h while AUF1 binding increased at 8h. As a control, we assessed CPE binding protein (CPEB), which was low. Deadenylation of cyclin B2 mRNA requires transcription. To determine if increased AUF1 binding at 8h required transcription, we injected transcription inhibitor α-amanitin into embryos. Blocking transcription had no effect on AUF1 binding (8+a). Immunoblotting for ElrA, AUF1 and CPEB in the extracts used for pulldowns (Figure 1D, Input) showed that they were present. To determine if binding was specific, we repeated pulldowns with A1ΔARE (Figure 1E). Similar to Figure 1C and D, more AUF1 associated with cyclin B2 3′UTR at 8 versus 4 h, even when transcription was inhibited. Low amounts of ElrA and AUF1 were also pulled down with A1ΔARE at 4 and 8h. This may be due to more nonspecificity in pulldowns, which require more starting protein than UV-crosslinks.
Since HuR and AUF1 concurrently and competitively bind to a subset of target mRNAs [15-18], we tested if ElrA and AUF1 compete for binding. We used GST- ElrA and GST-AUF1 for in vitro binding assays. GST-AUF1 eluted as a band of 63-kDa, while GST-ElrA eluted as a doublet of 62- and 55-kDa upon purification (not shown). UV-crosslink competition experiments (Figure 2A) showed that both GST-ElrA and GST-AUF1 bound cyclin B2 3′UTR (lanes labeled 0) and could be competed with unlabeled cyclin B2 3′UTR, but less well by A1ΔARE. RNA-pulldown competition experiments showed that ElrA and AUF1 compete for binding (Figure 2B). When cyclin B2 3′UTR was incubated with a constant amount of GST-ElrA or GST-AUF1, increasing the opposing protein decreased the amount of initial protein binding.
Figure 2. ElrA and AUF1 bind cyclin B2 3′UTR in vitro.
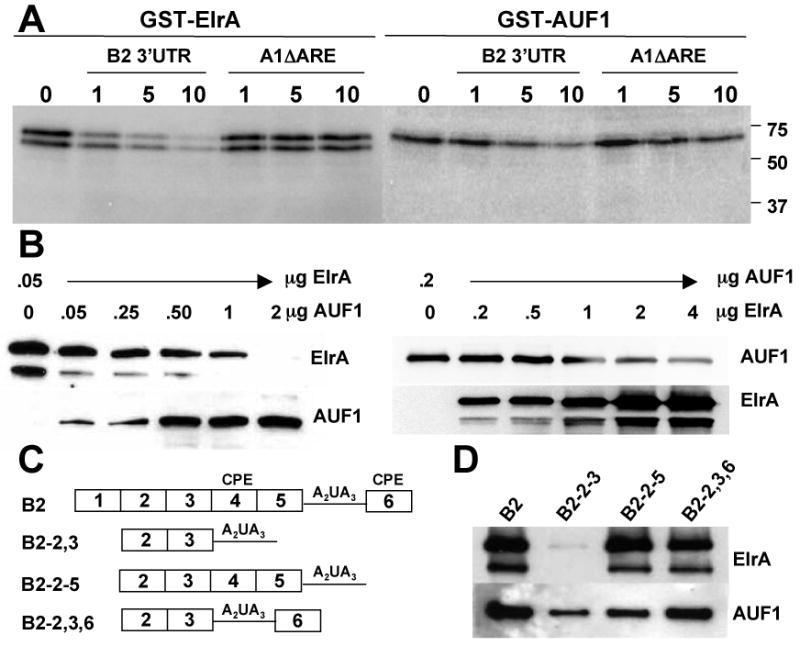
A, UV-crosslink analysis of GST-ElrA and GST-AUF1 with radiolabeled cyclin B2 3′UTR in the presence of 0, 1, 5, or 10 molar excess unlabeled cyclin B2 3′UTR or A1ΔARE. Markers are in kDa. B, RNA-pulldown of biotinylated cyclin B2 3′UTR with GST-ElrA and increasing GST-AUF1 (left panel), or GST-AUF1 and increasing GST-ElrA; followed by immunoblotting for ElrA and AUF1. C, Diagram of cyclin B2 3′UTR deletion mutants. Mutants contain the 30-nt regions shown (1-6) and the NPS (A2UA3). CPE, regions containing this element. D, RNA-pulldown with the biotinylated RNA indicated and 0.05 μg GST-ElrA or 0.2 μg GST-AUF1, followed by immunoblotting for ElrA and AUF1. n=3 for A and B; n=4 for D.
To localize binding, cyclin B2 3′UTR deletion mutants were used for RNA-pulldowns with GST-ElrA and GST-AUF1 (Figure 2C and D). The 3′UTR was divided into six regions of about 30-nt and various regions deleted as described [7]. All mutants contained the nuclear polyadenylation sequence (NPS, A2UA3). Mutant B2-2,3 contained regions 2 and 3, the deadenylation element. Mutant B2-2-5 contained regions 2-5, with a maturation CPE in region 4; and mutant B2-2,3,6 contained regions 2, 3 and 6, with a maturation CPE in region 6. Maturation CPEs (U5AU) specify polyadenylation in oocytes; embryonic CPEs (poly(U)12-27) function in embryos, and both need the NPS for activity [19, 20]. ElrA bound CPE-containing B2-2-5 and B2-2,3,6 similarly to full length 3′UTR (B2) but didn't bind B2-2,3. AUF1 bound B2-2,3,6 and B2-2-5 best, and B2-2,3 weakly. The overlapping RNA-binding requirements of ElrA and AUF1 are in agreement with Figure 2B.
Since AUF1 and ElrA bound RNAs with CPEs best, we deleted the CPEs and assayed binding. Deletions were performed in GbB2, containing globin 5′-UTR and ORF and cyclin B2 3′UTR. Figure 3A shows UV-crosslinks of these mRNAs with GST-ElrA and GST-AUF1. As compared to GbB2 (1), deletion of one (2, 3) or both (4) of the CPEs had no effect on protein binding. RNA-pulldowns indicated that endogenous ElrA and AUF1 bound equally well to these RNAs (not shown). These results show that CPEs are not required for ElrA and AUF1 binding.
Figure 3. CPEs are not required for ElrA and AUF1 binding.
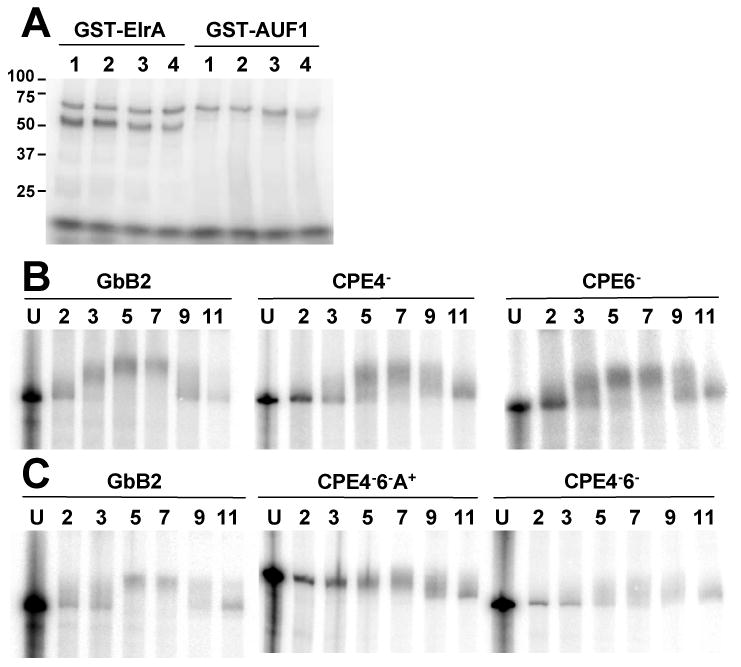
A, UV-crosslink analysis of GST-AUF1 or GST-ElrA with radiolabeled RNA: 1: GbB2, 2-4: GbB2 with region 4 (CPE4-), region 6 (CPE6-) or both CPEs deleted (CPE4-6-). Markers are in kDa. B and C, The indicated RNA was injected into 2-cell embryos, extracted at the indicated hpf (top numbers) and resolved on denaturing gels. U, RNA before injection. n=3.
GbB2 recapitulates the adenylation profile of endogenous cyclin B2 mRNA, being polyadenylated after injection through 7 hpf, after which it is deadenylated [7]. As the CPEs in the cyclin B2 mRNA had only been shown to function in egg extracts [19, 20], we tested if they mediated polyadenylation in embryos. The mRNAs were injected into 2-cell embryos and the first timepoint taken upon completion of injections (2 hpf). The size of GbB2 mRNA increased (polyadenylation) between 3-7 hpf and decreased (deadenylation) after 7 hpf (Figure 3B and C). Deletion of one CPE (CPE4- and CPE6-) didn't affect polyadenylation or deadenylation while deletion of both decreased polyadenylation dramatically (CPE4-6-). When CPE4-6- was transcribed with a poly(A)65 tail (CPE4-6-A+), it was deadenylated (Figure 3C). These experiments showed that only one CPE is necessary for polyadenylation but neither was necessary for deadenylation. As both ElrA and AUF1 bound mRNAs lacking CPEs, they also indicate that ElrA is not involved in maturation CPE function and leave open a possible deadenylation role for AUF1.
We next tested if ElrA could function in mRNA stabilization and AUF1 in destabilization. The constitutive presence of maternal ElrA and AUF1 in the Xenopus embryo made knockdown experiments unfeasible, so we overexpressed ElrA and AUF1 and assayed effects on cyclin B2 mRNA by Northern analysis. To block the function of endogenous ElrA, we also overexpressed ElrAΔ, a mutant missing the hinge region and RRM3 that binds but doesn't stabilize mRNA [21]. Figure 4A shows that mRNAs encoding myc-tagged AUF1, ElrA, and ElrAΔ were translated after microinjection into 2-cell embryos and the proteins persisted through 24 hpf.
Figure 4. Overexpression of ElrA and AUF1 does not affect cyclin B2 mRNA stability.
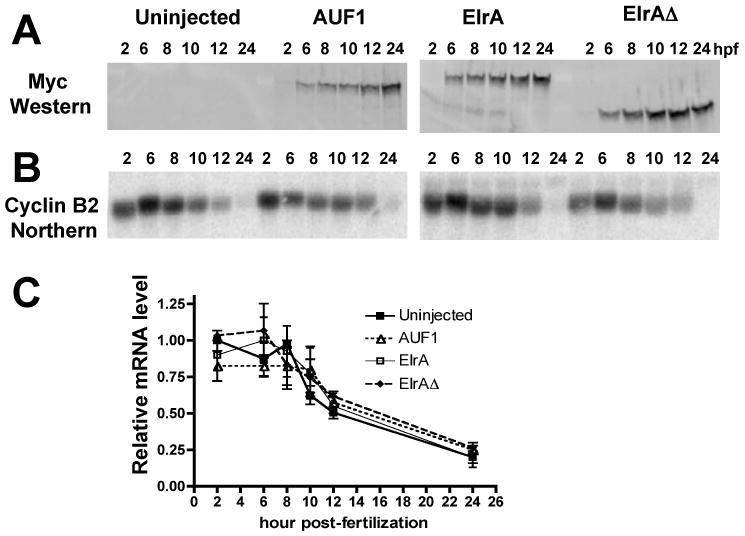
2-cell embryos remained uninjected or microinjected with mRNAs encoding myc-tagged AUF1, ElrA, ElrAΔ and processed at the hour indicated (hpf) for either A, myc immunoblotting or B, cyclin B2 Northern analysis. C, Relative amount of cyclin B2 mRNA +/- SEM as compared to 2 hpf uninjected embryos; n=4.
Figure 4B shows the decay profile for cyclin B2 mRNA in uninjected embryos and after expression of myc-ElrA, myc-AUF1 or myc-ElrAΔ. 4 repetitions of this experiment showed no statistical difference in the level of cyclin B2 mRNA in any experimental condition when compared to uninjected embryos (Figure 4C). Based on these results, there is no clear indication that ElrA or AUF1 determine the stability of cyclin B2 mRNA.
Discussion
Our aim was to identify RBPs interacting with the cyclin B2 mRNA in Xenopus embryos. Cyclin B2 mRNA is polyadenylated and translated during oocyte maturation, under control of two maturation CPEs in the 3′UTR [19, 20]. After fertilization, cyclin B2 mRNA is further polyadenylated and translated [7, 19] with protein peaking during mitosis and then degrading. At the MBT, cyclin B2 mRNA is deadenylated, leading to the terminal disappearance of the maternal protein.
We demonstrate that ElrA and AUF1 bind the cyclin B2 3′UTR. ElrA is the Xenopus homolog of HuR, a stabilizing RBP, while AUF1 is a destabilizing RBP. ElrA binds equally well pre- and post-MBT while AUF1 binding increases post-MBT, when cyclin B2 mRNA is deadenylated and destabilized. ElrA and AUF1 possess overlapping binding specificity to regions containing maturation CPEs and compete with each other for binding. Our results are consistent with others showing that HuR and AUF1 bind to a subset of common target mRNAs [15-18], including cyclin D1 mRNA, and expand the list of cyclin mRNAs they bind [22-24].
We showed previously that ElrA binds a noncanonical ARE and an embryonic CPE in the cyclin E1 mRNA, suggesting a function in polyadenylation and stabilization [14], as does its embryonic CPE-dependent binding to two other mRNAs [10]. Deletion of both maturation CPEs in the cyclin B2 3′UTR shows that ElrA and AUF1 bind sequence other than the CPEs, that ElrA binding is not sufficient for polyadenylation and that CPEs don't function in deadenylation. In Xenopus, AREs direct deadenylation immediately upon fertilization [25]. ElrA doesn't inhibit ARE-mediated deadenylation [26], but it does protect the body of deadenylated mRNA from degradation [24]. Despite this, ElrA overexpression didn't protect deadenylated cyclin B2 mRNA from degradation, and AUF1 overexpression didn't hasten decay. In addition, cyclin B2 protein level was not altered (not shown), arguing against a role in translation. One problem with overexpression studies is that the amount of ElrA or AUF1 is likely not limiting.
Other RBPs as well as microRNAs interact with AUF1, HuR or with AREs to influence function [27-31]. We recently found that a microRNA targets cyclin B2 deadenylation element (E. Lund et al., in preparation). We are exploring a link between ElrA/AUF1 and microRNA targeting of cyclin B2 mRNA to increase our understanding of mRNA deadenylation as an early cell cycle regulatory paradigm.
Supplementary Material
Acknowledgments
Supported by American Cancer Society (RSG-01-054-CCG) and National Institutes of Health (R01CA0958980) grants to RSH. Drs. P. Good (NIH), MD Sheets (Univ. Wisconsin), P. Krieg (Univ. Arizona) and JD Richter (Univ. Massachusetts) provided reagents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:673–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 2.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 3.Hartley RS, Sible JC, Lewellyn AL, Maller JL. A role for cyclin E/Cdk2 in the timing of the midblastula transition in Xenopus embryos. Dev Biol. 1997;188:312–321. doi: 10.1006/dbio.1997.8647. [DOI] [PubMed] [Google Scholar]
- 4.Hartley RS, Rempel RE, Maller JL. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- 5.Howe JA, Newport JW. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc Natl Acad Sci U S A. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howe JA, Howell M, Hunt T, Newport JW. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- 7.Audic Y, Anderson C, Bhatty R, Hartley RS. Zygotic regulation of maternal cyclin A1 and B2 mRNAs. Mol Cell Biol. 2001;21:1662–1671. doi: 10.1128/MCB.21.5.1662-1671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 9.Audic Y, Garbrecht M, Fritz B, Sheets MD, Hartley RS. Zygotic control of maternal cyclin A1 translation and mRNA stability. Dev Dyn. 2002;225:511–521. doi: 10.1002/dvdy.10191. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, Good PJ, Richter JD. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartley R, Le Meuth-Metzinger V, Osborne HB. Screening for sequence-specific RNA-BPs by comprehensive UV crosslinking. BMC Mol Biol. 2002;3:8. doi: 10.1186/1471-2199-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 13.Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol Cell Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slevin MK, Gourronc F, Hartley RS. ElrA binding to the 3′UTR of cyclin E1 mRNA requires polyadenylation elements. Nucleic Acids Res. 2007;35:2167–2176. doi: 10.1093/nar/gkm084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. Rna. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommer S, Cui Y, Brewer G, Fuqua SA. The c-Yes 3′-UTR contains adenine/uridine-rich elements that bind AUF1 and HuR involved in mRNA decay in breast cancer cells. J Steroid Biochem Mol Biol. 2005;97:219–229. doi: 10.1016/j.jsbmb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J Biol Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. Embo J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheets MD, Fox CA, Hunt T, Woude GV, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 20.Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. Embo J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 21.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. Embo J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. Embo J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo X, Hartley RS. HuR Contributes to Cyclin E1 Deregulation in MCF-7 Breast Cancer Cells. Cancer Res. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 24.Aoki K, Matsumoto K, Tsujimoto M. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J Biol Chem. 2003;278:48491–48497. doi: 10.1074/jbc.M308328200. [DOI] [PubMed] [Google Scholar]
- 25.Voeltz GK, Steitz JA. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol. 1998;18:7537–7545. doi: 10.1128/mcb.18.12.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 28.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–7913. [Google Scholar]
- 29.Wilson GM, Sutphen K, Bolikal S, Chuang KY, Brewer G. Thermodynamics and kinetics of Hsp70 association with A + U-rich mRNA-destabilizing sequences. J Biol Chem. 2001;276:44450–44456. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 31.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


