Abstract
Enterococcus faecium, an opportunistic pathogen that causes a significant number of hospital-acquired infections each year, presents a serious clinical challenge because an increasing number of infections are resistant to the so-called antibiotic of last resort, vancomycin. Vancomycin and other new glycopeptide derivatives target the bacterial cell wall, thereby perturbing its biosynthesis. To help determine the modes of action of glycopeptide antibiotics, we have developed a bottom-up mass spectrometry approach complemented by solid-state NMR to elucidate important structural characteristics of vancomycin-susceptible E. faecium peptidoglycan. Using accurate-mass measurements and integrating ion-current chromatographic peaks of digested peptidoglycan, we identified individual muropeptide species and approximated the relative amount of each. Even though the organism investigated is susceptible to vancomycin, only 3% of the digested peptidoglycan has the well-known D-Ala-D-Ala vancomycin-binding site. The data are consistent with a previously proposed template model of cell-wall biosynthesis where D-Ala-D-Ala stems that are not cross-linked are cleaved in mature peptidoglycan. Additionally, our mass-spectrometry approach allowed differentiation and quantification of muropeptide species seen as unresolved chromatographic peaks. Our method provides an estimate of the extent of muropeptides containing O-acetylation, amidation and hydroxylation, and the number of species forming cyclic imides. The varieties of muropeptides on which the modifications are detected suggest that significant processing occurs in mature peptidoglycan where several enzymes are active in editing cell-wall structure.
Keywords: carboxypeptidase, cell wall, cross-link, D-Ala-D-Ala, Enterococci, Enterococcus faecium, glycopeptide antibiotic, LC/MS, MS, peptidoglycan, solid-state NMR, vancomycin, VRE, VSE
Bacterial peptidoglycan is an attractive drug target because it has unique chemistry, and its integrity is crucial to protect cells from internal osmotic pressure. Vancomycin, a glycopeptide antibiotic targeting the peptidoglycan, has become increasingly important given its effectiveness against multiple drug-resistant bacteria. Alarmingly, some pathogens have developed resistance to even vancomycin, the so-called drug of last resort, leaving clinicians with limited treatment options. Among the most prevalent of these feared pathogens are vancomycin-resistant enterococci (VRE).
VRE infections, particularly E. faecium, have emerged as life-threatening pathogens in clinical settings worldwide. Since the first vancomycin-resistant E. faecium isolate was identified in 1986 (1), the number of VRE infections has increased dramatically, causing high mortality rates around the world. In some hospitals, over 90% of E. faecium isolates are vancomycin resistant (2). This presents a significant clinical challenge and manifests the urgency to find new therapeutic treatment possibilities.
Vancomycin inhibits peptidoglycan biosynthesis of the bacterial cell wall by forming complexes with the D-Ala-D-Ala carboxyl termini of peptidoglycan precursors outside the cytoplasmic membrane (3). Enterococcal resistance is generally thought to involve the modification of D-Ala-D-Ala binding sites to D-Ala-D-Lac (4). This change replaces a hydrogen bond between the nitrogen of the terminal D-Ala and the oxygen of vancomycin with an oxygen-oxygen repulsive interaction. The modified vancomycin-peptidoglycan stem complex has an in vitro binding constant over 1000 fold greater than that involving the D-Ala-D-Ala stem (5).
Recently, it was suggested that potent vancomycin-like glycopeptides, effective against VRE, recognize other peptidoglycan structural motifs in addition to D-Ala-D-Ala (6, 7). To understand the modes of action of these new glycopeptide analogues, as well as the action of vancomycin itself, atomic-level detail of the enterococcal peptidoglycan is required. To accomplish this objective, we have used bottom-up mass spectrometry to investigate the structure of vancomycin-susceptible E. faecium peptidoglycan. In particular, we are interested in the fine structure of E. faecium peptidoglycan as these structural details may be important to understanding cell-wall architecture and drug resistance.
Previously, mass spectrometric interrogations of enterococcal peptidoglycan did not address detailed muropeptide structural variations (8, 9). Those efforts, as well as other mass spectrometric analyses of bacterial peptidoglycan, relied on off-line MS identification (10–14) of HPLC peaks quantified by ultraviolet absorbance (15, 16). The strategy has proven to be powerful in characterizing the bacterial cell walls of many different organisms (14). Given the complexity of the biological system and difficulty of complete chromatographic separation, however, each HPLC peak may represent an indistinguishable mixture of muropeptides, some of which may not be assigned.
The approach we report here is to use LC/MS coupled with accurate-mass measurements of digested peptidoglycan to identify individual muropeptide species and, by integrating selected ion-current chromatographic peaks, estimate the relative amount of each. We verified the accurate-mass identifications with MS/MS and stable-isotope labeling experiments. After specifically enriching amino acids in the peptidoglycan stem with stable-isotope labels, we examined unique muropeptide mass shifts to check our assignments. The sensitivity of our method allowed us to assign nearly 90% of the peaks in those mass spectra occurring within the range of expected muropeptide retention times.
Experimental
Sample Preparation
Starting cultures of E. faecium (ATTC 49624) were prepared by inoculating brain heart infusion media with a single colony. Cultures were incubated overnight at 37 °C, but not aerated. Research samples were prepared by inoculating either brain-heart infusion or sterile enterococcal standard media (ESM) with the overnight starter cultures (1% final volume).
ESM was prepared as described before (17). The pH of ESM was adjusted to 7.0 prior to sterile filtration. Natural abundance amino acids in ESM were replaced by stable 13C and 15N isotope-enriched amino acids to incorporate specific labels into the samples for stable-isotope labeling experiments. All samples enriched with D-[1-13C]alanine were treated with 5 µg/mL of alaphosphin, an alanine racemase inhibitor, every 1.5 h of growth to prevent scrambling of the isotopic label from D-Ala to L-Ala (17).
Cells were harvested at the end of log phase, when the absorbance at 660 nm became approximately 1.0, by centrifugation at 10,000g for 25 min at 4 °C. Pellets were rinsed with 40 mM triethanolamine hydrochloride (pH = 7.0) three times followed by three rinses with H2O, centrifuging after each rinse. Cells were then resuspended in H2O, frozen, and lyophilized. Cell-wall isolates were prepared from lyophilized whole cells as detailed before (18). The isolated cell-walls were digested into muropeptides with lysozyme and mutanolysin by using a modified version of the protocol described by others (1, 8). All manipulations were performed at neutral pH.
LC-MS/MS
Liquid chromatography/MS and MS/MS were performed by using a PicoView PV-500 (New Objective, Wobum, MA) nanospray stage attached to either an LTQ-FT mass spectrometer or an LTQ-Orbitrap mass spectrometer (ThermoFisher, San Jose, CA).
Muropeptide samples were loaded into an uncoated 360/75 µm OD/ID fused-silica capillary column with a 15 µm picofritTM tip (New Objective, Wobum, MA), packed with C18 reverse-phase material (3 µm, 100 Å, Phenomenex, Torrance,CA). The column was eluted at a flow rate of 250 nL/min for 10 min with 0.1% (v/v) formic acid in water and subsequently with a 60-min linear acetonitrile gradient (0–40%) with 0.1% formic acid. The samples, as they emerged from the column, were sprayed into an LTQ-FT mass spectrometer. Full mass spectra were recorded in the FT component of the instrument at 100,000 resolving power.
Accurate mass product-ion spectra of muropeptides were acquired by introducing the samples by nanospray as they eluted from the LC to an LTQ-Orbitrap mass spectrometer. To obtain major components' product-ion spectra (i.e., species in scheme 2), cycles consisting of one full FT-scan mass spectrum and five ensuing data-dependent MS/MS scans acquired by the orbitrap (with a normalized collision energy setting of 35%) were repeated continuously throughout the elution with the following dynamic exclusion settings: repeat count, 3; repeat duration, 15 s; exclusion duration, 30 s. Sometimes, for minor components, an inclusion list containing the precursor ions' masses was applied.
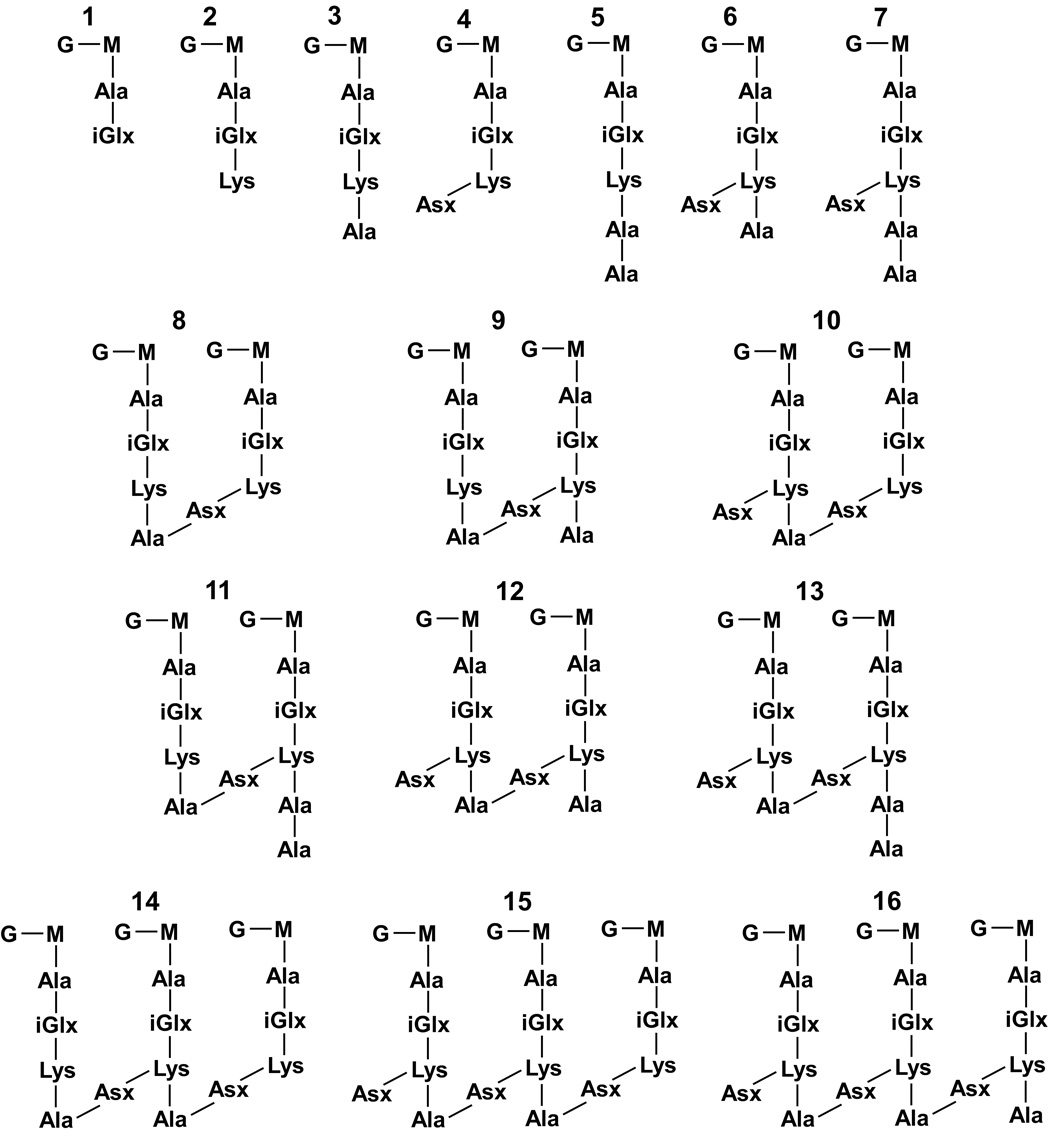
Scheme 2.
Structures of assigned muropeptides. The relative proportion of each structure is shown in Table 1.
Solid-State NMR
Experiments were performed using a four-frequency transmission-line probe (19) with a 14-mm long, 9-mm inside-diameter analytical coil, and a Chemagnetics/Varian magic-angle spinning ceramic stator. Lyophilized samples were spun in Chemagnetics/Varian 7.5-mm outside diameter zirconia rotors at 5.0 kHz under active control within +/− 2 Hz. Experiments were performed at room temperature with a Chemagnetics CMX-300 spectrometer operating at 75.453 MHz for carbon. Radio frequency pulses were produced by 1-kW Kalmus, ENI, and American Microwave Technology power amplifiers, each under active control (20); π pulse lengths were 10 µs for 15N. Cross-polarization transfer was at 50 kHz for 2 msec. Proton dipolar decoupling was at 105 kHz during data acquisition. The data were taken from a cell-wall sample with a mass of 150 mg, isolated from a 4 L preparation.
Results and Discussion
Muropeptide Identification
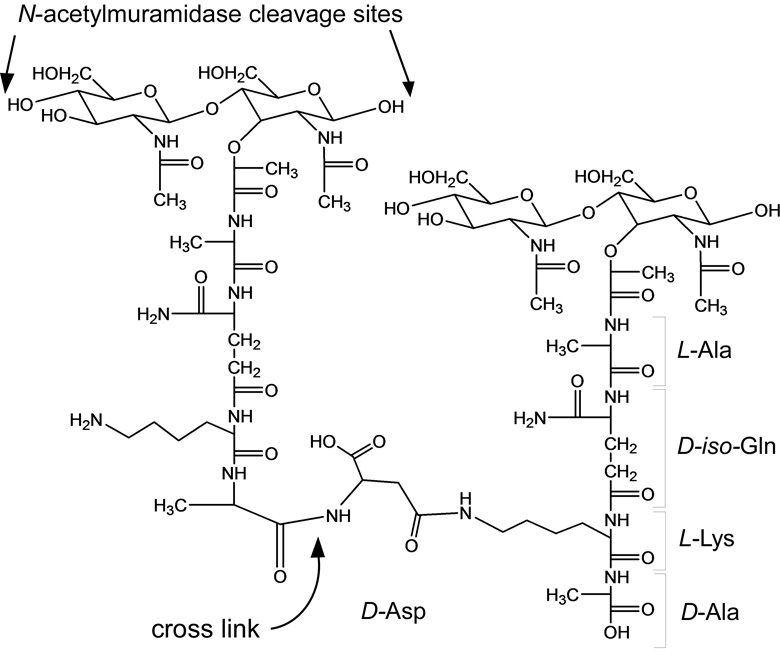
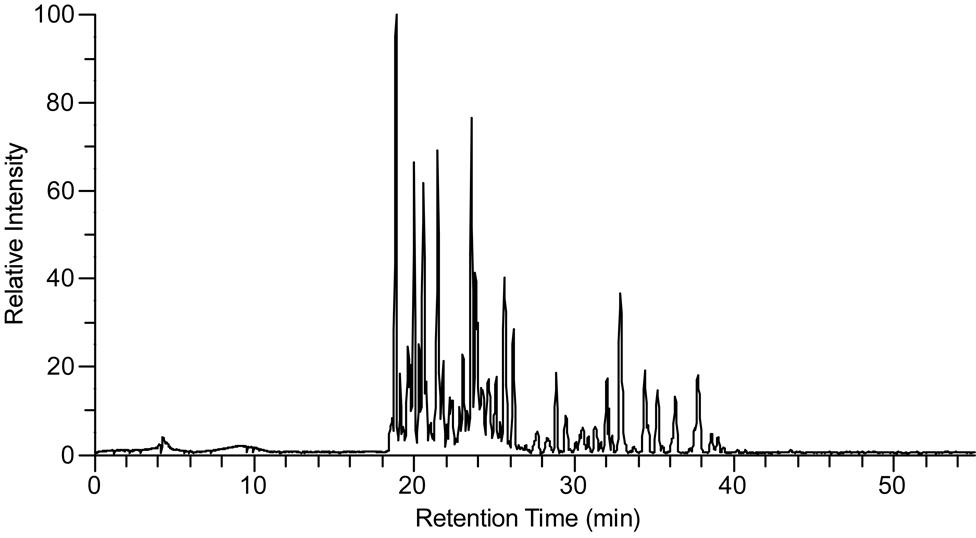
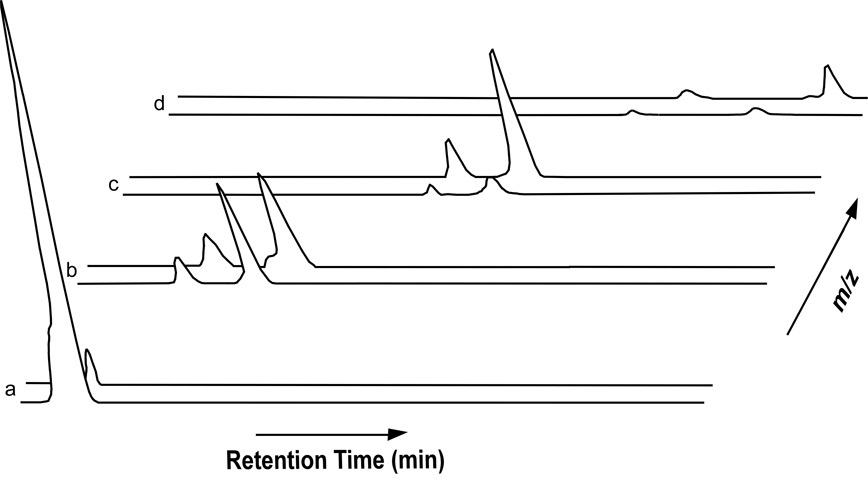
Enterococcal peptidoglycan-repeat units (see Scheme 1) are interconnected by cross-linked D-aspartic acid or D-asparagine (D-Asx) bridges and via the polymerization of the disaccharides. We used N-acetylmuramidase enzymes to catalyze selectively the hydrolysis of the β-1,4 linkages between N-acetylmuramic acid and N-acetylglucosamine, leaving the cross-links intact. As expected, on the basis of previous studies (8, 9), most of the muropeptide peaks in the ion chromatogram (Figure 1) were variations of monomers, dimers, and a few trimers.
Scheme 1.
Representative chemical structure of E. faecium peptidoglycan. The β-1,4 linkages between N-acetylmuramic acid and N-acetylglucosamine are cleaved by N-acetylmuramidase as indicated for the repeat unit on the left. The cross-links remain intact. D-iso-glutamine and D-aspartic acid may be substituted by either D-iso-glutamic acid or D-asparagine, respectively.
Figure 1.
Total ion chromatogram for digested E. faecium muropeptides. The relative intensity is represented by total ion current as a function of retention time.
All of the assigned muropeptide species eluted with a retention time between 15 and 35 min. The mass spectra, acquired so that accurate masses were taken for each scan of the ion chromatogram, serve as a means for evaluating muropeptide contributions. We considered first accurate-masses that are consistent with traditional muropeptide structures in a sample prepared with natural abundance isotopes only (no enrichment). These structural assignments were confirmed with MS/MS and stable-isotope labeling experiments. For example, an ion of m/z 824.4 corresponds to a species with a tripeptide stem without a bridge (see Scheme 2, structure 2). When E. faecium is grown in ESM with double-labeled aspartic acid, this mass is unaffected. When the sample is grown with L-[1-13C]lysine and D-[15N]alanine, the mass of the species increases by that consistent with the incorporation of one 13C.
Most of the other masses within 15–35 min retention time could be recognized as a variation of one of the traditional muropeptide structures. In fact, we were able to assign components represented by approximately 90% of the peaks in the mass spectra as muropeptide structures and verify them by MS/MS and stable-isotope labeling experiments. It should be emphasized that in the isolation of the muropeptides, the peptidoglycan was not subjected to any acid or base treatments. Thus, we associate nearly all of the identified muropeptide structures as representative of enterococcal peptidoglycan. The few exceptions found were not detected consistently in different sample preparations, and consequently ignored in the analysis.
Structural Quantification
The quantification of each muropeptide species was estimated using integrals of selected ion chromatograms. For species having structural isomers, the integral was taken as the sum over the entire selected ion chromatogram. Multiply charged species were also included in the integrals, and the distributions obtained by considering multiply charged ions were comparable to that of singly charged ions. The chromatographic separation has provided sufficient resolution such that the overlap of major and minor muropeptide components is minimal.
The monomers, dimers, and trimers represent 47, 43, and 8% of the muropeptides, respectively. The identified muropeptide structures (Scheme 2) and their relative abundances are listed in Table 1. The number of peptidoglycan stems terminating in D-Ala-D-Ala is only 3%. This quantification is reported as a percentage normalized to the total number of peptidoglycan stems. That is, dimer integrals are counted twice and trimer integrals are counted three times in determining the total number of peptidoglycan stems. This result is consistent with that determined in solid-state NMR studies of the same vancomycin-susceptible E. faecium organism, where the number of D-Ala-D-Ala terminating stems was found to be 7% in whole cells (17). While peptidoglycan precursors and lipid II may contribute to the difference in measurements for the concentration of D-Ala-D-Ala stems, the determinations are still within experimental error (see Validity of MS Quantification: Comparison with Solid-State NMR Results section); and we are unwilling to attribute any significance to the discrepancy.
Table 1.
Distribution of Structures in E. faecium Peptidoglycan
| Species | Theoretical Massa | Measured Mass | Percentage [%]b,c | Percentage [%]c,d |
|---|---|---|---|---|
| Monomers | 47 | 30 | ||
| 1 | 695.2856 | 695.2855 | <1 | <1 |
| 2 | 823.3805 | 823.3802 | 22 | 13 |
| 3 | 894.4177 | 894.4174 | 5 | 3 |
| 4 | 938.4075 | 938.4071 | 13 | 8 |
| 5 | 965.4548 | 965.4555 | <1 | <1 |
| 6 | 1009.4446 | 1009.4446 | 6 | 4 |
| 7 | 1080.4817 | 1080.4825 | <1 | <1 |
| Dimers | 43 | 51 | ||
| 8 | 1814.8151 | 1814.8150 | 8 | 10 |
| 9 | 1885.8522 | 1885.8510 | 3 | 4 |
| 10 | 1929.8421 | 1929.8420 | 23 | 27 |
| 11 | 1956.8894 | 1956.8899 | 2 | 2 |
| 12 | 2000.8792 | 2000.8784 | 7 | 8 |
| 13 | 2071.9163 | 2071.9158 | <1 | <1 |
| Trimers | 8 | 15 | ||
| 14 | 2806.2497 | 2806.2450 | 2 | 3 |
| 15 | 2921.2767 | 2921.2767 | 5 | 10 |
| 16 | 2992.3138 | 2992.3140 | 1 | 2 |
| Teramers | 2 | 4 | ||
Theoretical masses were calculated with Xcalibur™ 2.0
Percentages with respect to monomers, dimers, trimers, and tetramers
Both percentages include structural variations of the species (see Table 2). Under optimized digestion conditions, for data from triplicate runs of four different sample preparations, each species contribution was within +/− 3% of the values listed here.
Percentages with respect to total number of peptidoglycan stems
O-acetylated Muropeptides
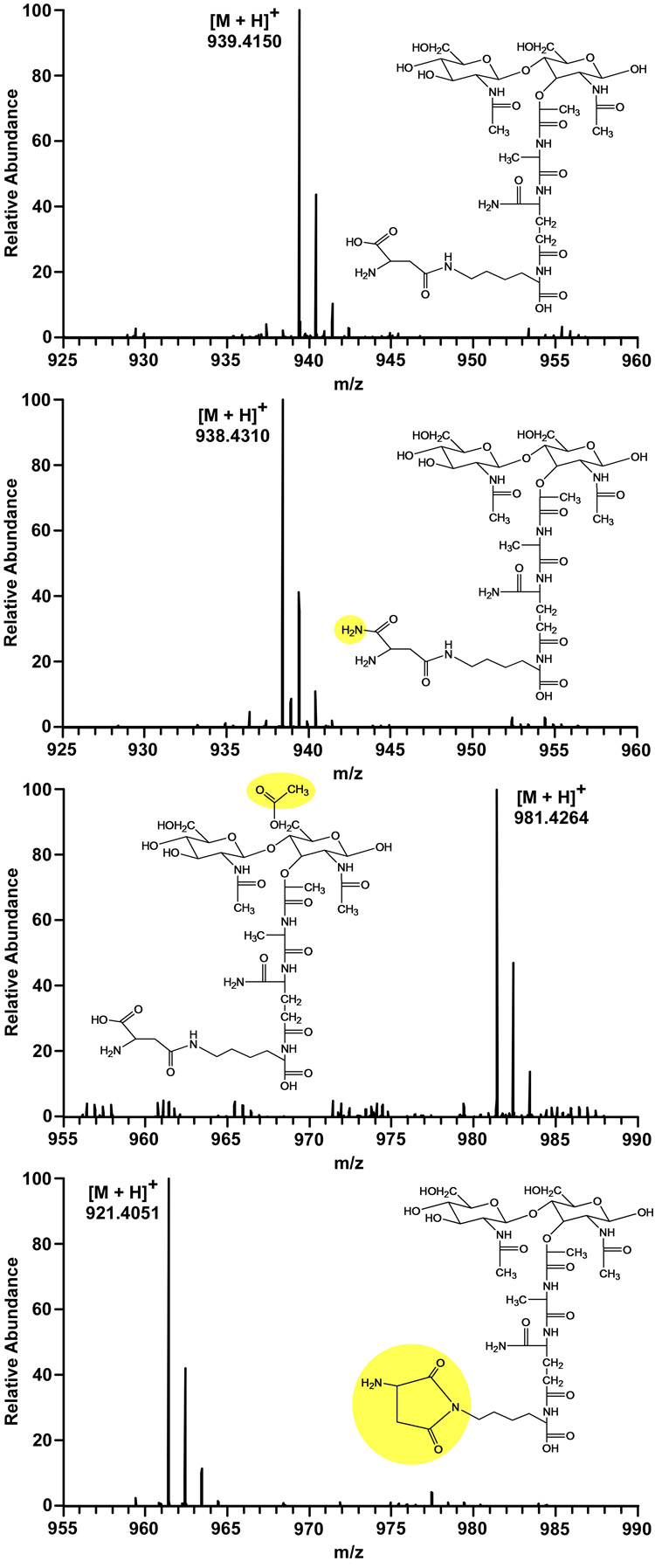
We observed several structural variations of each identified muropeptide species, one of which is consistent with O-acetylation of N-acetylmuramic acid corresponding to a mass increase of 42.011 (Figure 2, second from bottom). O-Acetylation of peptidoglycan has been well studied since it was first reported in 1958 (21, 22). Thus far, all analyses demonstrate that O-acetylation only occurs at C6 of N-acetylmuramic acid (23). The product-ion spectra (MS/MS) of structures with and without O-acetylation show the same fragmentation masses of N-acetylglucosamine and the peptide stem. On the other hand, the mass of N-acetylmuramic acid-peptide stem fragments increases by 42.011 for species with O-acetylation. These observations provide strong support for the previous conclusions.
Figure 2.
Mass spectra and assigned structures for variations of the muropeptide of m/z = 939.4150. The most likely structure for an m/z of 939.4150 (top) is that with a D-aspartic acid bridge. An m/z of 938.4310 (second from top) corresponds to the muropeptide with double amidation, D-asparagine and D-iso-glutamine. A mass increase of 42.011 is consistent with O-acetylation of N-acetylmuramic acid (second from bottom). A mass decrease of 18.010 suggests the loss of water, and could be indicative of two types of cyclic imides (bottom). The structure shown is consistent with NMR data (see Figure 4).
The ester bond of O-linked acetate is sensitive to mild alkaline hydrolysis (24). As described above, the peptidoglycan was isolated at physiological pH, leaving O-acetylated peptidoglycan intact for MS experiments. Our results suggest that each muropeptide structure has a different likelihood of being O-acetylated (Table 2). We estimate that 5–15% of the total peptidoglycan is O-acetylated on average. The extent of O-acetylation is significantly lower than the 46–57% that was previously reported for E. faecium (25). As was shown for other species (26–28), the extent of O-acetylation may vary in different enterococcal strains (25) and may even be correlated with vancomycin susceptibility in E. faecium.
Table 2.
Distribution of Selected Monomers
| Species | Acetylation [%] | ||||
|---|---|---|---|---|---|
| 2 | 19 | ||||
| 3 | 2 | Cyclic Imide [%] | Gln/Asn [%] | Gln/Asp (Glu/Asn) [%] | Glu/Asp [%] |
| 4 | 11 | 8 | 41 | 58 | 1 |
| 5 | 4 | 3 | 16 | 83 | 1 |
| 6 | 3 | 2 | 23 | 75 | 2 |
In digesting the peptidoglycan, we used two N-acetylmuramidase enzymes, mutanolysin and lysozyme. For many organisms, O-acetylation renders peptidoglycan resistant to the activity of lysozyme (28), whereas mutanolysin isolated from Streptomyces globisporus is not affected by this modification (29). Indeed, in the E. faecium preparations digested with only lysozyme, we observed no species with a mass increase consistent with O-acetylation for any muropeptide structures. Earlier studies showed large variations between different enterococcal strains in the lytic response of their cell walls to lysozyme (30, 31), indicating different degrees of O-acetylation (32). Thus, it is not surprising that the extent of O-acetylation in the strain of E. faecium we investigated varies from that of a different strain reported before.
We also observed muropeptide structures that were N-deacetylated, corresponding to a mass decrease of 42.011. The loss of an acetyl residue from N-acetylglucosamine or N-acetylmuramic acid is infrequent in E. faecium, detected in less than 0.2% of the peptidoglycan stems. We, therefore, ignored these structures in determining the extent of O-acetylation as it is highly unlikely that significant numbers of muropeptides are simultaneously O-acetylated and N-deacetylated.
Amidation of D-iso-Glu and D-Asp
The occurrence of D-iso-glutamic acid residues in muropeptide components is hypothesized to affect cross-linking (33) and vancomycin resistance in Staphylococcus aureus (34). Non-amidated peptidoglycan stems may be inefficient substrates for transpeptidase (35) and have higher affinity for vancomycin (34), thereby consuming the antibiotic in the cell wall before it can inhibit cell-wall biosynthesis at lipid II. E. faecium peptidoglycan has two potential sites for amidation, D-iso-Glx and D-Asx (see Scheme 1). Thus, for E. faecium peptidoglycan stems with bridges, there are three potential ions corresponding to double amidation, amidation/hydroxylation, and double hydroxylation. Because species with D-iso-glutamine/D-aspartic acid and D-iso-glutamic acid/D-asparagine have the same accurate mass, MS identification of each is difficult. There are, however, relatively few muropeptide structures without bridges that have a mass corresponding to hydroxylation. The singly charged muropeptide with an m/z of 824.4 (see Scheme 2, structure 2) does not have a bridge and contains D-iso-glutamine. The peak area for the ion of m/z 825.4, representing an accurate mass increase of 0.984, corresponding to hydroxylation, is relatively small compared to that of 824.4 (Figure 3a). This is consistent with there being very few D-iso-glutamic acid-containing peptidoglycan stems, as was suggested for other enterococcal organisms (9). Additionally, it is rare that muropeptides with bridges have a mass corresponding to double hydroxylation, supporting that few stems have D-iso-glutamic acid. Thus, we conclude that most muropeptide species with a mass increase corresponding to a single hydroxylation represent D-iso-glutamine/D-aspartic acid stems. Approximately 60% of peptidoglycan bridges are D-aspartic acid, and 2% of all peptidoglycan stems contain D-iso-glutamic acid (Table 2).
Figure 3.
Selected ion chromatograms for singly charged species with m/z of (a) 824.4 and 825.4; (b) 938.4 and 939.4; (c) 1009.5 and 1010.5; and (d) 1080.5 and 1081.5. The relative intensities of 1080.5 and 1081.5 are small, consistent with there being few D-Ala-D-Ala stems. Muropeptides with bridges elute as two distinct peaks with different retention times in the chromatogram. The selected ion chromatogram for m/z 824.4 shows only one peak.
Structural Bridge Isomers
Selected ion chromatograms of muropeptides with bridges show two peaks representing components with different retention times, suggesting that these muropeptides exist as two structural isomers associated with unique bridge conformations (Figure 3). The deamidation of D-asparagine is known to produce a mixture of α- and β-D-aspartic acid residues (36–38). Recently, however, it was suggested that D-asparagine bridges in E. faecium exclusively originate from the amidation of D-aspartic acid after its incorporation into the peptidoglycan (39). Further, the incorporation is thought to proceed specifically by way of a β-aspartyl-phosphate intermediate (40, 41), making α- and β-bridge isomers unlikely. Considering that β-D-aspartic acid is incorporated into peptidoglycan precursors, and that the 1 and 4 aspartyl-carbons are similar, we cannot rule out the possibility that either carbon could form a peptide bond with the 6-N-lysine, thereby creating two unique bridge conformations.
Cyclic Imides
Interestingly, the muropeptide monomers we assigned to have aspartic acid bridges show a relatively small amount of species with an accurate mass decrease of 18.010 consistent with the loss of water (Figure 2, bottom). MS/MS experiments indicate that the water loss is not from the disaccharides or the first amino acid of the peptide stem, L-alanine. Moreover, tripeptide monomers with bridges (see Scheme 2, structure 4) also undergo water loss prior to mass spectrometry analysis. We hypothesize that the water loss results in the formation of a cyclic imide structure, for which there are at least two possibilities. Aspartic acid could form a succinimide with 6-N-lysine (see Figure 2, bottom) or undergo cyclization with N-D-iso-glutamine.
Using the current mass spectrometric data, we are unable to differentiate between the two proposed varieties of cyclic imides. Monomeric muropeptides containing D-iso-glutamine and D-asparagine do not show mass decreases consistent with water loss, probably because there is no free hydroxyl to be protonated and become a leaving group. If water loss were to occur from stems containing both D-iso-glutamic acid and D-aspartic acid, it would suggest that the succinimide is the favored structure. Unfortunately, the infrequency of D-iso-glutamic acid containing muropeptide stems makes this analysis difficult. It should be noted that some structures with a missing water molecule, such as that with an m/z of 939.4, produce two peaks with different retention times in the ion chromatogram. This may be representative of both types of cyclic imide structures or a consequence of there being two bridge orientations as discussed above.
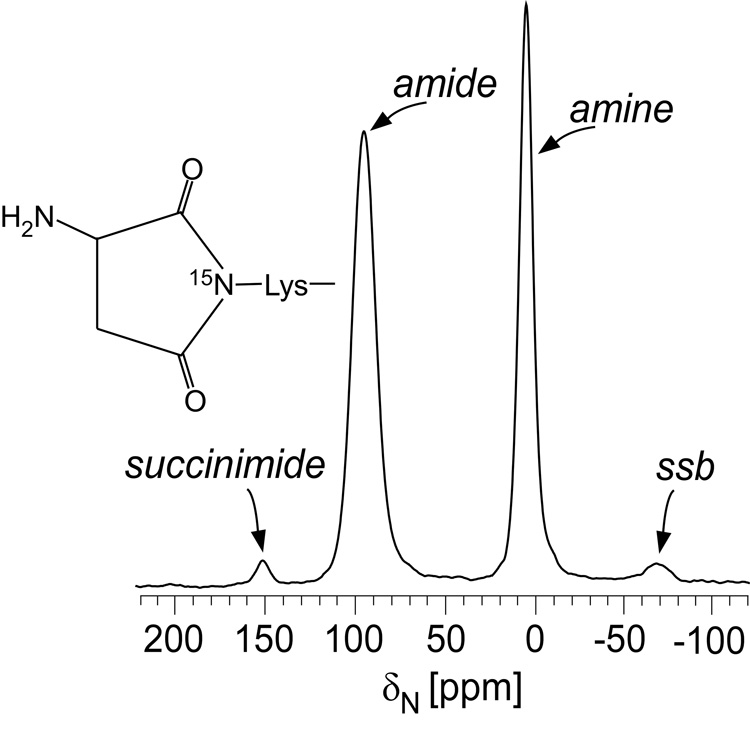
Solid-state NMR experiments performed on intact cell-wall isolates specifically labeled with L-[15N]lysine provide insight in the identification. An 15N cross-polarization magic-angle spinning (CPMAS) echo spectrum is consistent with a succinimide structure (Figure 4). The peak at 5 ppm is due to lysyl amines and the peak at 95 ppm is for lysyl amides. We suggest that the small peak at 151 ppm is from lysyl succinimides, representing a small percentage of peptidoglycan bridges. Although succinimides were proposed as intermediates (36), to our knowledge, this is the first detection of such structures in E. faecium peptidoglycan.
Figure 4.
15N CPMAS echo-spectrum of E. faecium cell-wall isolates enriched with L-[6-15N]lysine. The amide peak at 95 ppm occurs when lysine has a D-Asx bridge attached, and the amine peak at 5 ppm occurs when lysine does not have an attached bridge. We assign the 151 ppm peak to lysyl succinimides.
Muropeptides without Asx Bridges
The solid-state NMR 15N CPMAS echo spectrum of E. faecium cell-wall isolates shows, from the ratio of the amide to amine peak, that a significant percentage of peptidoglycan stems do not have bridges (17). This result contrasts with those from previous mass spectrometric studies in which it was concluded that over 95% of the enterococcal peptidoglycan contains bridges (8). Results from our MS experiments are consistent with solid-state NMR data, showing nearly 30% of peptidoglycan stems do not have bridges. The significant number of muropeptide stems without bridges is shown by the relative intensity of m/z 824.4 (amidated species 2 of Scheme 2) compared to other species (see Figure 3). The discrepancy in bridge-links between our data and those given in previous MS studies may be related to the specific strain of E. faecium investigated, or, it may result from excessive mutanolysin digestion of muropeptides as hypothesized here. The cell walls we digested with only lysozyme showed significant proportions of lysyl amine structures, but no stuctures consistent with O-acetylation. In cell walls digested with high concentrations of mutanolysin and lysozyme for 24 h, lysyl amine muropeptides decreased, representing less than 5% of the peptidoglycan. Additionally, the ion chromatogram profile changed. For example, the ratio of structures with an m/z of 824.4 to that of 825.4 (hydroxylated species 2 of Scheme 2) changed by a factor of six times, indicating that high concentrations of mutanolysin may further degrade some muropeptides after having cleaved the β-1,4 glycan linkages. To optimize the digestion, the concentration and digestion times were reduced such that the treatment produced significant proportions of lysyl amine muropeptides consistent with solid-state NMR measurements made on cell-wall isolates before digestion while still solubilizing a substantial amount of peptidoglycan and cleaving O-acetyl-substituted oligomers.
Validity of MS Quantification: Comparison with Solid-State NMR Results
The MS quantifications in this report are based on integrals obtained from ion chromatograms, and given that there are likely to be differences in ionization efficiencies between muropeptide structures and potential digestion sampling problems, we view them as approximations. Nonetheless, the investigation has revealed several new structural details of E. faecium peptidoglycan, the relative proportion of each, and the types of stems on which they occur. As a method to judge the quantitative integrity of the MS results, the percentage of total peptidoglycan stems cross-linked can be calculated and compared to that directly measured by solid-state NMR. Cross-linking is equal to: (dimers + 2·trimer + 3·tetramers) / (monomers + 2·dimers + 3·trimers + 4·tetramers). From the MS data, approximately 40% of E. faecium peptidoglycan is cross-linked. This result is in good agreement with the 47% obtained from solid-state NMR (17).
Glycan Chain Lengths
The absence of cross-linked chains longer than tetramers contrasts with the peptidoglycan from Staphylococcus aureus, another Gram-positive pathogen, where cross-linked chains greater than 9-mers were observed after N-acetylmuramidase digestion (42). In comparison to S. aureus, we propose that E. faecium compensates for the lack of peptidoglycan integrity resulting from short cross-linked fragments by extending the length of the glycan chains. No data have been reported for E. faecium glycan chain length, but we anticipate that it will be much longer than in S. aureus where the average was reported to be six disaccharides (43). Average glycan lengths as long as 140 disaccharides have been reported in some B. subtilis species (44).
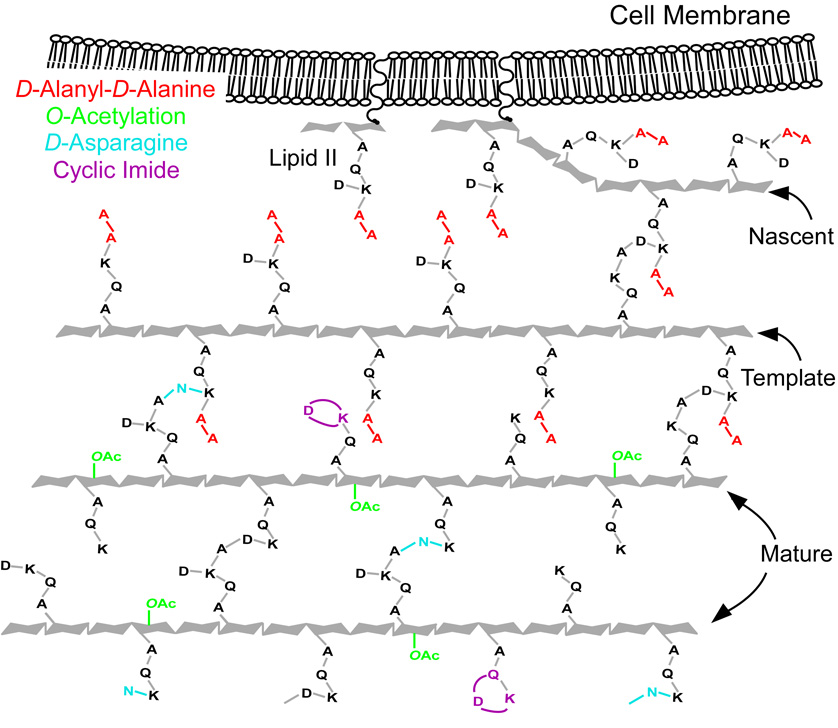
Template Model for Cell-wall Biosynthesis
Both MS and solid-state NMR results show that the number of peptidoglycan stems terminating in D-Ala-D-Ala is small despite this organism’s susceptibility to vancomycin. Nevertheless, these binding sites are crucial to cell-wall biosynthesis, and their small number is fundamentally consistent with the template model proposed for S. aureus (6, 7) and recently extended to E. faecium peptidoglycan assembly (17). In this single-strand addition model, the orientation of nascent peptidoglycan is signaled by the stem conformation of the template, the last completed glycan chain (Figure 5). For the template to form cross-links with the extending nascent peptidoglycan, it must have D-Ala-D-Ala stems. These few stems represent the 3% of all D-Ala-D-Ala peptidoglycan observed in MS experiments. Modification of the few D-Ala-D-Ala stems in nascent and template peptidoglycan prevents the binding of vancomycin and its ensuing inhibition of the enzymatic processes crucial for cell-wall assembly. We suspect that there is an active L,D-carboxypeptidase that cleaves D-Ala-D-Ala stems that are not cross-linked in mature peptidoglycan. It was suggested that L,D-carboxypeptidase activity is involved in peptidoglycan maturation for other organisms (45–48). In addition to L,D-carboxypeptidase activity, we propose that other enzymatic modifications also occur in mature peptidoglycan. Examples include O-acetylation, amidation of aspartic acid, and formation of cyclic imides, which occur less frequently on muropeptides associated with nascent and template peptidoglycan, namely those with D-Ala-D-Ala stems (see Table 2). The results suggest that significant processing occurs in mature peptidoglycan where several enzymes are active in editing cell-wall structure. Although the modified peptidoglycan precursors may produce inefficient substrates for transglycosylase and transpeptidase, modified mature peptidoglycan may play an important role in biosynthetic regulation and prove important in virulence, allowing bacterial infections to evade host-immune system response.
Figure 5.
Two-dimensional schematic representation of the template model of peptidoglycan biosynthesis for E. faecium. Chain extension occurs from right to left and is synchronized with cross-linking at lipid II. The few peptidoglycan stems terminating in D-Ala-D-Ala (red) occur in nascent and template peptidoglycan where they are cross-linked. We suspect that L,D-carboxypeptidase acts on stems in mature peptidoglycan that are not cross linked. We propose that other cell-wall editing enzymatic processes occur in mature peptidoglycan as well, creating muropeptides with O-acetylation (green), D-asparagine (blue), and cyclic imides (purple) as shown.
Conclusions
Mass spectrometry, particularly in LC/MS mode in which accurate masses and product-ion spectra are acquired, can provide new structural detail pertaining to variations of peptidoglycan stems. These results may prove important in understanding glycopeptide-antibiotic binding. One source of success of our bottom-up approach is the gentle treatments for digestion under neutral pH that allow the fine structure of the peptidoglycan to be preserved. Using this gentle digestion, we implemented an on-line LC/MS analysis that gives results suitable for comparison with those from solid-state NMR, a method with excellent quantitative sensitivity and reliability. The comparison verifies that, despite being susceptible to vancomycin, vancomycin-susceptible Enterococcus faecium have very few D-Ala-D-Ala binding sites. The finding is consistent with a template model for cell-wall biosynthesis and suggests that L,D-carboxypeptidase activity exists in E. faecium peptidoglycan. Although D-Ala-D-Ala terminating stems occur infrequently, their importance as the binding site for one of the most powerful antibiotics clinically available underscores the need for characterizing other fine peptidoglycan structural details such as were accomplished by this research.
Acknowledgements
This research was supported by the National Centers for Research Resources of National Institutes of Health (Grant No. P41RR000954) and the National Institutes of Health (Grant No. EB002058).
Abbreviations
- CPMAS
cross-polarization magic-angle spinning
- D-Asx
D-asparagine or D-aspartic acid
- D-iso-Glx
D-iso-glutamine or D-iso-glutamic acid
- LC/MS
liquid chromatography-mass spectrometry
- Lipid II
N-acetylglucosamine-N-acetyl-muramyl-pentapeptide-pyrophosphoryl-undecaprenol
- VRE
vancomycin-resistant enterococci
- VSE
vancomycin-susceptible enterococci
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Uttley AH, Collins CH, Naidoo J, George RC. Vancomycin-resistant enterococci. Lancet. 1988;1:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 2.Kak VAC, Joseph W. Acquired antibiotic resistances in enterococci. Washington, DC: ASM Press; 2002. [Google Scholar]
- 3.Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 2008;32:386–408. doi: 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- 4.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42. 2006;Suppl 1:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 5.Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 6.Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KSE, Parr TR, Far AR, Schaefer J. Mode of action of oritavancin in Staphylococcus aureus by solid-state NMR. J. Mol. Biol. 2007;377:281–293. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. Vancomycin derivative with damaged D-Ala-D-Ala binding cleft binds to cross-linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry. 2008;47:3822–3831. doi: 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billot-Klein D, Shlaes D, Bryant D, Bell D, van Heijenoort J, Gutmann L. Peptidoglycan structure of Enterococcus faecium expressing vancomycin resistance of the VanB type. Biochem. J. 1996;313(Pt 3):711–715. doi: 10.1042/bj3130711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jonge BL, Gage D, Handwerger S. Peptidoglycan composition of vancomycin-resistant Enterococcus faecium. Microb. Drug Resist. 1996;2:225–229. doi: 10.1089/mdr.1996.2.225. [DOI] [PubMed] [Google Scholar]
- 10.Singh PD, Johnson JH. Muraceins--muramyl peptides produced by Nocardia orientalis as angiotensin-converting enzyme inhibitors. II. Isolation and structure determination. J Antibiot (Tokyo) 1984;37:336–343. doi: 10.7164/antibiotics.37.336. [DOI] [PubMed] [Google Scholar]
- 11.Pittenauer E, S. E. R., Allmaier G, Pfanzagl B, Loffelhardt W, Quintela C, de Pedro MA, Stanek W. Structural characterization of the cyanelle peptidoglycan of Cyanophora paradoxa by californium-252 plasma desorption mass spectrometry and fast atom bombardment/tandem mass spectrometry. Biol. Mass Spectrom. 1993;22:524–536. doi: 10.1002/bms.1200220906. [DOI] [PubMed] [Google Scholar]
- 12.Pfanzagl B, Zenker A, Pittenauer E, Allmaier G, Martinez-Torrecuadrada J, Schmid ER, De Pedro MA, Loffelhardt W. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J. Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenker APB, Loffelhardt W, Allmaier G. Negative and positive ion matrix-assisted laser desorption ionization mass spectrometry of peptidoglycan fragments after size fractionation and reversed-phase high-performance liquid chromatography. J. Microbiol. Methods. 1998;32:237–246. [Google Scholar]
- 14.Bacher G, Korner R, Atrih A, Foster SJ, Roepstorff P, Allmaier G. Negative and positive ion matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and positive ion nano-electrospray ionization quadrupole ion trap mass spectrometry of peptidoglycan fragments isolated from various Bacillus species. J Mass Spectrom. 2001;36:124–139. doi: 10.1002/jms.109. [DOI] [PubMed] [Google Scholar]
- 15.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 16.Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J. Biol. Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 17.Patti GJ, Kim Sung Joon, Schaefer J. Characterization of the peptidoglycan of vancomycin-susceptible Enterococcus faecium. Biochemistry Submitted. 2008 doi: 10.1021/bi8008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong G, Pan Y, Dong H, Pryor R, Wilson GE, Schaefer Jacob. Structure and dynamics of pentaglycyl bridges in the cell walls of Staphylococcus aureus by 13C-15N REDOR NMR. Biochemistry. 1997;36:9859–9866. doi: 10.1021/bi970495d. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer J, McKay RA. Patent, U. S. 1999
- 20.Guillion TSJ. Detection of Weak Heteronuclear Dipolar Coupling by Rotational-Echo Double-Resonance Nuclear Magnetic Resonance. Adv. Magn. Reson. 1989;13:57–83. [Google Scholar]
- 21.Abrams A. O-acetyl groups in the cell wall of Streptococcus faecalis. J. Biol. Chem. 1958;230:949–959. [PubMed] [Google Scholar]
- 22.Brumfitt W, Wardlaw AC, Park JT. Development of lysozyme-resistance in Micrococcus lysodiekticus and its association with an increased O-acetyl content of the cell wall. Nature. 1958;181:1783–1784. doi: 10.1038/1811783a0. [DOI] [PubMed] [Google Scholar]
- 23.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 24.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer JM, Strating H, Weadge JT, Clarke AJ. Peptidoglycan O acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J. Bacteriol. 2006;188:902–908. doi: 10.1128/JB.188.3.902-908.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenthal RS, Folkening WJ, Miller DR, Swim SC. Resistance of O-acetylated gonococcal peptidoglycan to human peptidoglycan-degrading enzymes. Infect. Immun. 1983;40:903–911. doi: 10.1128/iai.40.3.903-911.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swim SC, Gfell MA, Wilde CE, 3rd, Rosenthal RS. Strain distribution in extents of lysozyme resistance and O-acetylation of gonococcal peptidoglycan determined by high-performance liquid chromatography. Infect. Immun. 1983;42:446–452. doi: 10.1128/iai.42.2.446-452.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke AJ, Dupont C. O-acetylated peptidoglycan: its occurrence, pathobiological significance, and biosynthesis. Can. J. Microbiol. 1992;38:85–91. doi: 10.1139/m92-014. [DOI] [PubMed] [Google Scholar]
- 29.Hamada S, Torii M, Kotani S, Masuda N, Ooshima T, Yokogawa K, Kawata S. Lysis of Streptococcus mutans cells with mutanolysin, a lytic enzyme prepared from a culture liquor of Streptomyces globisporus 1829. Arch. Oral Biol. 1978;23:543–549. doi: 10.1016/0003-9969(78)90268-6. [DOI] [PubMed] [Google Scholar]
- 30.Chesbro WR. Lysozymes and the Production of osomotic fragility in enterococci. Can. J. Microbiol. 1961:952–955. [Google Scholar]
- 31.Toennies G, Iszard L, Rogers NB, Shockman GD. Cell multiplication studied with an electronic particle counter. J. Bacteriol. 1961;82:857–866. doi: 10.1128/jb.82.6.857-866.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metcalf RH, Deibel RH. Growth of Streptococcus faecium in the presence of lysozyme. Infect. Immun. 1972;6:178–183. doi: 10.1128/iai.6.2.178-183.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stranden AM, Roos M, Berger-Bachi B. Glutamine synthetase and heteroresistance in methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 1996;2:201–207. doi: 10.1089/mdr.1996.2.201. [DOI] [PubMed] [Google Scholar]
- 34.Hanaki H, Labischinski H, Inaba Y, Kondo N, Murakami H, Hiramatsu K. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 1998;42:315–320. doi: 10.1093/jac/42.3.315. [DOI] [PubMed] [Google Scholar]
- 35.Nakel M, Ghuysen JM, Kandler O. Wall peptidoglycan in Aerococcus viridans strains 201 Evans and ATCC 11563 and in Gaffkya homari strain ATCC 10400. Biochemistry. 1971;10:2170–2175. doi: 10.1021/bi00787a033. [DOI] [PubMed] [Google Scholar]
- 36.Ghuysen JM. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol. Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- 37.Ikawa M. The Configuration of aspartic acid in cell walls of lactic acid bacteria and factors affecting the racemization of aspartic acid. Biochemistry. 1964;3:594–597. doi: 10.1021/bi00892a021. [DOI] [PubMed] [Google Scholar]
- 38.Suginaka H, Kotani S, Kato K, Kashiba S, Amano T. Action of a staphylolytic enzyme (ALE) of a strain of Staphylococcus epidermidis. Biken's J. 1968:13–24. [Google Scholar]
- 39.Bellais S, Arthur M, Dubost L, Hugonnet JE, Gutmann L, van Heijenoort J, Legrand R, Brouard JP, Rice L, Mainardi JL. Aslfm, the D-aspartate ligase responsible for the addition of D-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J. Biol. Chem. 2006;281:11586–11594. doi: 10.1074/jbc.M600114200. [DOI] [PubMed] [Google Scholar]
- 40.Staudenbauer W, Strominger JL. Activation of D-aspartic acid for incorporation into peptidoglycan. J. Biol. Chem. 1972;247:5095–5102. [PubMed] [Google Scholar]
- 41.Staudenbauer W, Willoughby E, Strominger JL. Further studies of the D-aspartic acid-activating enzyme of Streptococcus faecalis and its attachment to the membrane. J. Biol. Chem. 1972;247:5289–5296. [PubMed] [Google Scholar]
- 42.de Jonge BL, Chang YS, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 43.Boneca IG, Huang ZH, Gage DA, Tomasz A. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J. Biol. Chem. 2000;275:9910–9918. doi: 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- 44.Archibald AR, Hancock IC, Harwood CR. Cell wall structure, synthesis, and turnover. Washington, D.C.: ASM Press; 1993. [Google Scholar]
- 45.Courtin P, Miranda G, Guillot A, Wessner F, Mezange C, Domakova E, Kulakauskas S, Chapot-Chartier MP. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L,D-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 2006;188:5293–5298. doi: 10.1128/JB.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korza HJ, Bochtler M. Pseudomonas aeruginosa LD-carboxypeptidase, a serine peptidase with a Ser-His-Glu triad and a nucleophilic elbow. J. Biol. Chem. 2005;280:40802–40812. doi: 10.1074/jbc.M506328200. [DOI] [PubMed] [Google Scholar]
- 47.Templin MF, Ursinus A, Holtje JV. A defect in cell wall recycling triggers autolysis during stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ursinus A, Steinhaus H, Holtje JV. Purification of a nocardicin A-sensitive LD-carboxypeptidase from Escherichia coli by affinity chromatography. J. Bacteriol. 1992;174:441–446. doi: 10.1128/jb.174.2.441-446.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]