Abstract
To better understand the mechanisms underlying the role of the immunoglobulin heavy chain gene (IgH) 3' enhancers on bcl-2 transcriptional deregulation in t(14;18) lymphoma, we characterized the physical interactions of the IgH 3' enhancer region with the bcl-2 promoters. Using the chromosome conformation capture (3C) technique, we found that the IgH 3' enhancers physically interact with the bcl-2 promoter region over a 350 kb genomic region in t(14;18) lymphoma cells. No interactions of the bcl-2 promoter region with sequences distant to the IgH enhancers were observed. The physical interactions of the IgH enhancers with the bcl-2 5' region are functionally involved in the transcriptional control of bcl-2. The histone deacetylase inhibitor, trichostatin A (TSA), repressed bcl-2 transcription and decreased the IgH enhancer-bcl-2 promoter region interactions. We showed by chromatin immunoprecipitation assay (ChIP) and siRNA transfection studies that the POU2 family transcription factor Oct-2 and its cofactor Bob-1 play a critical role in mediating the IgH enhancer-bcl-2 promoter region interactions. This study reveals a new aspect of the regulatory role of the IgH 3' enhancers on bcl-2 transcription in t(14;18) lymphomas.
Keywords: t(14;18) lymphoma, bcl-2, IgH enhancer, spatial interactions
INTRODUCTION
The t(14;18)(q32;q21) is the most common chromosomal translocation in human low grade lymphomas. More than 85% of follicular lymphomas and 25% of diffuse B-cell lymphomas possess this translocation. As a result of the translocation, one allele of the anti-apoptotic bcl-2 gene from chromosome 18q21 is juxtaposed to the immunoglobulin heavy chain (IgH) locus on chromosome 14q32. In t(14;18) lymphoma, the untranslocated bcl-2 allele is silent, and the translocated bcl-2 allele is aberrantly transactivated (Graninger et al., 1987). The prolonged cell survival due to increased bcl-2 expression from the translocated allele has been shown to contribute to the development of lymphomas and confer resistance to a variety of anticancer therapies (Desoize, 1994; Hockenberry et al., 1990; Reed et al., 1994; Schmitt and Lowe, 2001).
Two promoters mediate transcriptional control of the bcl-2 gene (Seto et al., 1988). The 5' promoter (P1) is located 1,386 to 1,423 bp upstream of the bcl-2 translational start site, and it is GC-rich with multiple Sp1 sites. The start sites of the 3' promoter (P2) are located 1.3 kb downstream of the P1 promoter. The P2 promoter has a classic TATA and CAAT box. The major positive cis-elements for P1 activity are a cAMP response element (CRE) and several Sp1 sites (Ji et al., 1996; Wilson et al., 1996). NF-κB family members bind to these sites and are essential for bcl-2 deregulation in t(14;18) cells (Heckman et al., 2002). The major positive regulatory site of P2 is a binding site for the homeodomain protein Cdx. A-Myb, a strong activator of P2 promoter activity, acts through the Cdx site (Heckman et al., 2000), as does (Heckman et al., 2003b). We also showed that Oct transcription factors mediate t(14;18) lymphoma cell survival by directly regulating bcl-2 expression (Heckman et al., 2006).
The 3' IgH enhancers consist of four DNase I-hypersensitive sites, HS1, 2, 3, and 4, which have been shown to function as a locus control region in B cells (Khamlichi et al., 2000; Madisen and Groudine, 1994; Mills et al., 1997). (HS sites 1 and 2 are located within 0.5 kb of each other and are usually assayed together as HS1, 2.) Several transcription factor-binding sites have been identified in the IgH 3' enhancer regions, including sites for NF-κB, Oct, Cdx, Pax5/Bsap (B-cell lineage-specific activator protein), Bach2/Maf, AP1, and Ets proteins. The conservation of the Oct, NF-κB, Cdx, AP1 and Ets sites across different species suggests that these sites may be critical for the function of the enhancers (Khamlichi et al., 2000; Mills et al., 1997).
Using in vitro and in vivo models, we have characterized the role that the IgH 3' enhancers play in the deregulation of bcl-2 transcription in t(14;18) lymphomas. Our initial transient transfection study showed that the murine IgH 3' enhancers HS1-4 increased bcl-2 transcription when they were linked to the bcl-2 promoter (Heckman et al., 2003a). To further characterize the role of the IgH 3' enhancers in bcl-2 transcriptional deregulation, we established an episomal reporter gene model system in which the bcl-2 promoter was linked to regions of the IgH 3' enhancers. This model system reproduces many aspects of bcl-2 deregulation in vivo, including the promoter shift and the increased histone H3 acetylation at the promoters (Duan et al., 2007). Our study of endogenous bcl-2 mRNA transcription in t(14;18) lymphoma cells using real-time reverse-transcriptase PCR revealed that the increased bcl-2 transcripts were derived from both the P1 and P2 promoters (Duan et al., 2007). Interestingly, the activation of the P2 promoter was more dramatic than that of the P1 promoter.
It is thus becoming increasingly clear that the IgH 3' enhancers are critically involved in aberrant transactivation of bcl-2 in t(14;18) lymphoma cells, but how the IgH enhancers mediate bcl-2 transactivation is not understood. Using the recently developed chromosome conformation capture (3C) technique to detect long-range interactions (Dekker et al., 2002), we demonstrate here that the IgH 3' enhancer regions are physically associated with the bcl-2 promoter region over a 350-kb genomic distance. These spatial interactions are functionally involved in the transcriptional deregulation of bcl-2 in t(14;18) lymphoma cells. Moreover, Oct-2 and its cofactor Bob-1 are important factors mediating these physical interactions.
RESULTS
Experimental design of the chromosome conformation capture assay for the study of the physical interactions between the IgH enhancers and thebcl-2 promoter region
To determine whether the IgH 3' enhancers interact with the bcl-2 promoter region, 3C analysis was performed with primers near the bcl-2 transcriptional start sites on chromosome 18 and primers on chromosome 14 (105,051,517-105,351,516). The region of chromosome 14 is centered approximately between the two IgH 3' enhancer regions downstream of Cα1 and Cα2 (Figure 1a, b).
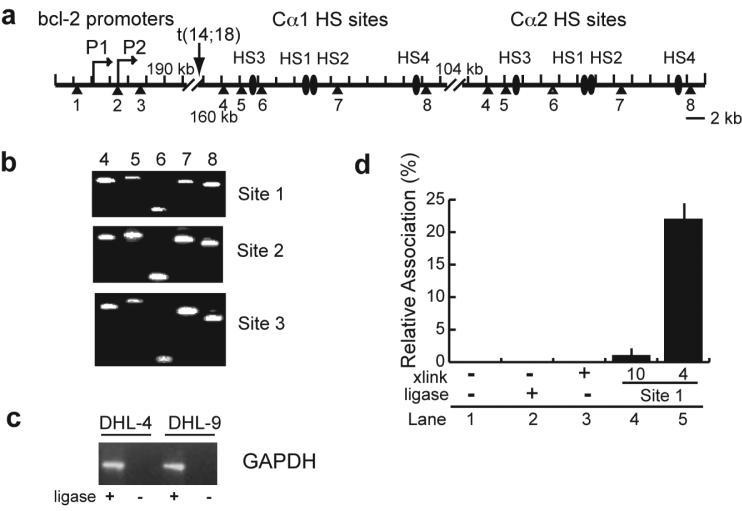
Figure 1. Diagram of the regions of human chromosomes 18 and 14 utilized in the 3C analysis of interactions between thebcl-2 promoter region and the IgH enhancers.
(a) 300 kb genomic region on chromosome 18 from 58,986,879 to 59,286,878 in the bcl-2 transcriptional orientation. The thick black line represents the genomic region. Gray lines above this line show the genes in the region with the arrows indicating the transcriptional orientation of each gene. Two vertical arrows represent the major transcriptional start sites of bcl-2 (P1 and P2). The lower lines indicate the span of BAC DNAs used to control for PCR efficiency in the 3C analysis. BamHI restriction sites are shown as small triangles under the BAC DNA.
(b) 300 kb genomic region on chromosome 14 from 105,051,517 to 105,351,516 in the IgH gene transcriptional orientation. The thick black line represents the genomic region, and the IgH genes are shown as gray rectangles. The black ovals following Cα1 and Cα2 represent the hypersensitive sites (HS3, HS1, HS2, and HS4). BAC DNAs and BamHI restriction sites are also shown.
The 3C assay measures the proximity and frequency of interaction between two points in chromatin. For the 3C assay, cells are treated with formaldehyde to cross-link proteins to other nearby proteins and DNA. After digestion with an appropriate restriction enzyme, ligation is performed under conditions of low DNA concentration to promote intramolecular ligation of the interacting cross-linked DNA fragments. The cross-links are reversed, and the ligation products are quantified by PCR. 3C analysis using BAC DNA was performed as a control for the real-time PCR efficiency of each primer/probe set (Hagege et al., 2007). To compare the 3C analysis between DHL-4 and DHL-9 cells, the interactions of two separate BamHI sites that are 11.5 kb away from each other in the GAPDH locus on chromosome 12 were determined. Similar interactions were observed in 3C samples at the GAPDH locus from DHL-4 and DHL-9 cells. (DHL-4 cells have the t(14;18) translocation, and DHL-9 cells lack this translocation.)
A higher resolution of the linear arrangement of the bcl-2 promoter region (BamHI sites 1-3) and the IgH enhancers (BamHI sites 4-8) in the t(14;18) translocation is shown in Figure 2a. Due to the conserved sequence in the enhancer regions 3' of Cα1 and Cα2, the primers for the two sets of IgH enhancers are the same. Examples of the PCR products with bcl-2 promoter region BamHI sites 1, 2, and 3 primers and primers at BamHI sites 4 though 8 of the IgH enhancer region are shown in Figure 2b. The PCR products with the primers at the GAPDH locus in DHL-4 and DHL-9 cells are shown in Figure 2c. As shown in Figure 2d (lanes 1-3), no PCR product was observed with DNA that was not cross-linked and ligated. When PCR was performed with cross-linked and ligated 3C DNA using bcl-2 promoter region BamHI site 1 primer and a primer at -90 (BamHI site 10) relative to the IgH enhancer site 4, a very low level of product was observed (Figure 2c, lane 4). In contrast, the yield of the PCR product was higher when bcl-2 promoter region BamHI site 1 primer was used with the primer at IgH enhancer BamHI site 4 (Figure 2c, lane 5).
Figure 2. Schematic of the BamHI sites at thebcl-2 promoter region and IgH enhancers, and controls for 3C analysis.
(a) Diagram of the bcl-2 promoters and the IgH 3' enhancers in t(14;18) lymphoma cells. The transcription start sites of bcl-2 (P1 and P2) and the IgH hypersensitive sites (HS3, HS1, HS2, HS4) following Cα1 and Cα2 are labeled. Numbers under each triangle show the BamHI sites in this region. Approximate distances in kilobases are shown at each of the gaps in the diagram.
(b) Agarose gels (2%) showing the PCR products of the 3C analysis of interactions of bcl-2 promoter region BamHI sites 1, 2, and 3 with IgH BamHI enhancer sites 4 through 8 in DHL-4 cells.
(c) Agarose gel showing the PCR products of the 3C analysis of interactions of two BamHI sites in the GAPDH locus in DHL-4 and DHL-9 cells.
(d) Negative controls for 3C analysis in DHL-4 cells. Real-time PCR was performed with bcl-2 promoter region BamHI site 1 primer and probe and IgH enhancer BamHI site 4 primer on non-cross-linked DNA (lanes 1 and 2) and cross-linked DNA before and after ligation (lanes 3 and 5). The primer/probe at bcl-2 promoter region BamHI site 1 was also used in real-time PCR with the IgH locus BamHI site 10 primer (lane 4). (Lanes 4 and 5 utilized cross-linked and ligated DNA.)
Interactions of the core regulatory regions of the IgH 3' enhancers and thebcl-2 promoter region in t(14;18) DHL-4 cells but not in DHL-9 cells
Specific interactions of the bcl-2 promoter region BamHI site 1 were observed at the five BamHI sites in the IgH enhancer region, but no significant interactions of BamHI site 1 were observed with regions 5' or 3' to the IgH enhancers (Figure 3a). The results with sequences 5' and 3' to the Cα1 enhancers are shown, and numbering of the IgH region is relative to BamHI site 4, which was set at 0. Similar results were obtained with sequences flanking the Cα2 enhancers (data not shown). Bcl-2 promoter region BamHI site 2 also interacted with the five BamHI sites at the IgH enhancers but not with sequences 5' or 3' of the IgH enhancers (Figure 3b). The 3C results demonstrate that interactions of the bcl-2 promoter region with the IgH locus peak at the core enhancer regions and dramatically decrease with increasing genomic distance from the enhancers. These studies suggest that the interactions of the bcl-2 promoter region with the IgH enhancers are specific and most likely functionally related.
Figure 3. Interactions between the IgH 3' enhancers andbcl-2 promoter region are observed in human t(14;18) DHL-4 follicular lymphoma cells but not in non-t(14;18) DHL-9 cells.
(a) 3C analysis was performed in DHL-4 cells with real-time PCR using the primer and probe at bcl-2 promoter region BamHI site 1 and the other set of primers located at the IgH enhancers (BamHI sites 4, 5, 6, 7, 8, labeled on the graph) and regions 5' and 3' of the enhancers. Analysis of 3C samples of BAC DNA RP11-495C15 and RP11-815P20 or RP11-495C15 and RP11-731F5 was utilized to optimize the PCR conditions, and the relative association was determined relative to the interactions at BamHI sites at the GAPDH locus. The x-axis represents the genomic distance from Cα1 #4 site, which was arbitrarily defined as 0. The flanking sequences are 5' and 3' of the Cα1 region. No interaction was observed with sequences 5' and 3' to the Cα2 region although these results are not shown. The relative association is the average of three independent 3C analyses.
(b) 3C analysis was performed in DHL-4 cells with real-time PCR using the primer and probe at bcl-2 promoter region BamHI site 2 and the other set of primers located at the IgH enhancers and regions 5' and 3' of the enhancers as described for (a).
(c) 3C analysis was performed in DHL-4 cells with real-time PCR using the sets of primers and probes anchored at the bcl-2 promoter region (BamHI sites 1, 2, and 3) and the other set of primers located at the IgH enhancers (BamHI sites 4, 5, 6, 7, 8). The BamHI sites 4 through 8 at the IgH enhancers are shown on the x-axis, and the location of the IgH enhancer HS sites is shown above the x-axis.
(d) 3C analysis was performed in DHL-9 cells with real-time PCR using the primers and probes at the bcl-2 promoter region (BamHI sites 1, 2, and 3) and the primers at the IgH enhancers (BamHI sites 4, 5, 6, 7, and 8) as described for (c).
We focused on the interactions of the bcl-2 promoter region BamHI sites 1, 2, and 3 with the five BamHI sites at the IgH enhancers as shown in Figure 3c. The BamHI sites 1 and 2 at the bcl-2 promoter region have stronger interactions with the IgH enhancer regions than site 3, suggesting that BamHI sites 1 and 2 at the bcl-2 promoter region are in closer proximity than site 3 with the IgH enhancer regions. Moreover, the interactions of the bcl-2 promoter region with BamHI site 8 are significantly higher than with the other sites in the IgH enhancer region, suggesting a closer proximity of HS4 with the bcl-2 promoter regions compared to HS3 and HS1, 2. No interactions of the IgH enhancers with the bcl-2 promoter region were detected in DHL-9 cells which lack the t(14;18) translocation (Figure 3d).
The physical association of the IgH enhancers with thebcl-2 promoter region is functionally involved in the transcriptional regulation ofbcl-2
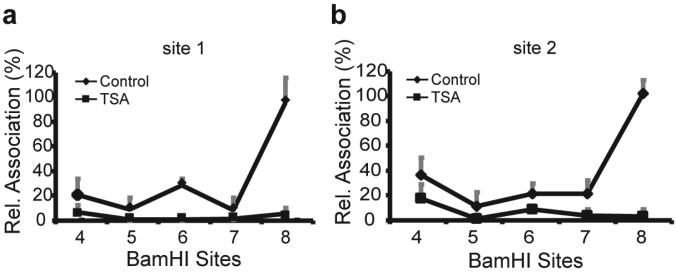
To demonstrate the functional involvement of the physical interactions of the IgH enhancer regions with the bcl-2 promoter region, we examined the effect of trichostatin A (TSA) on the physical association of the IgH enhancers with the bcl-2 promoter region. We have previously shown that TSA decreases the transcription of the translocated bcl-2 gene in t(14;18) lymphoma cells (Duan et al., 2005). DHL-4 cells were treated with 500 ng/ml TSA for 18 hours, and the relative association of the IgH enhancers and the bcl-2 promoter region was compared in cells treated with TSA or with solvent only. After normalization for PCR efficiency of the different primer/probe sets, the relative associations of the bcl-2 promoter region BamHI sites 1 and 2 and the IgH enhancers are shown in Figure 4a and b, respectively. TSA treatment dramatically decreased the IgH enhancer and bcl-2 promoter region interactions at both BamHI sites 1 and 2. We showed previously that treatment with 500 ng/ml TSA for 18 hours decreased bcl-2 transcription by 90% (Duan et al., 2005). The markedly decreased interaction of the IgH enhancers with the bcl-2 promoter region by treatment of the cells with TSA suggests that the association of the IgH enhancers with the bcl-2 promoter region correlates well with transcription.
Figure 4. Treatment with TSA markedly decreases the interactions of the IgH enhancers with thebcl-2 promoter region.
(a) The effect of TSA treatment on the interactions of the IgH 3' enhancer BamHI sites 4 through 8 with the bcl-2 promoter region BamHI site 1 in DHL-4 cells was determined by 3C analysis. DHL-4 cells were treated with 500 ng/ml TSA for 18 hours. The PCR signal was normalized to the PCR efficiency derived from a parallel analysis of 3C samples of BAC DNA clones as well as to the association of the BamHI sites at the GAPDH locus in the untreated or treated samples.
(b) The effect of TSA on the interactions of the IgH 3' enhancer BamHI sites with the bcl-2 promoter region BamHI site 2 in DHL-4 cells was determined by 3C analysis as described for (a).
Oct-2 and its co-activator Bob-1 are involved in mediating the physical association of the IgH enhancers and thebcl-2 promoter region
Previously, we determined whether the level of expression of several transcription factors involved in the regulation of bcl-2 (CREB, CBP, Sp1, C/EBP, p53) and/or their binding to the bcl-2 promoter changed following treatment of DHL-4 cells with TSA (Duan et al., 2005). To extend these studies, we examined the level of expression of Oct family members in DHL-4 cells with and without TSA treatment. As seen in Figure 5a and b, TSA decreased both Oct-1 and Oct-2 protein levels in whole cell lysates and in nuclear extracts. A dose-dependent decrease of Oct-2 mRNA was also observed by real-time RT-PCR analysis (data not shown). TSA did not have a major effect on Bob-1 expression.
Figure 5. Treatment with TSA decreases the expression of Oct-1 and Oct-2 in DHL-4 cells.
(a) TSA decreases Oct-1 and Oct-2 expression in whole cell lysates in DHL-4 cells. Cells were incubated with or without TSA for 18 hours. The expression of Oct-1, Oct-2, and Bob-1 was determined by Western blot analysis, and β-actin was used as loading control.
(b) TSA decreases Oct-1 and Oct-2 expression in the nuclear extracts of DHL-4 cells.
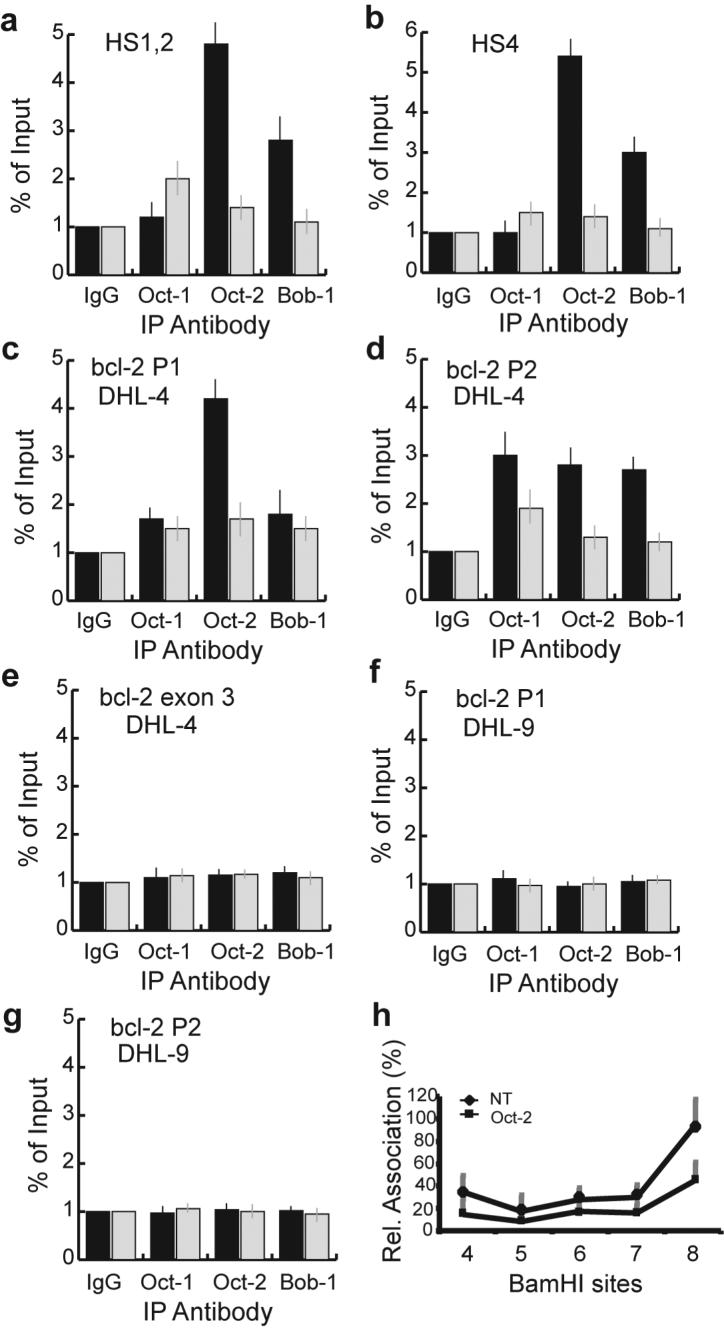
To further characterize the influence of TSA on Oct family members in the regulation of bcl-2 transcription, quantitative ChIP assays were performed to measure the binding of Oct-1, Oct-2, and Bob-1 to the consensus octamer sites in the IgH enhancer HS1, 2 and HS4 regions. As shown in Figure 6a and b, Oct-2 and Bob-1 bound to the IgH enhancer HS1, 2 and HS4 regions in DHL-4 cells. The binding of Oct-1 to HS1, 2 and HS4 was not above the background level. No significant binding of these factors to the HS3 region was observed (data not shown), which is consistent with the lack of an octamer site in HS3. Treatment with 500 ng/ml of TSA for 18 h dramatically decreased the binding of Oct-2 and Bob-1 to HS1, 2 and HS4 and had no significant effect on Oct-1 binding (Figure 6a and b).
Figure 6. Effect of TSA on Oct factor binding to the IgH enhancers andbcl-2 promoter region.
(a) TSA decreases Oct-2 and Bob-1 binding to the IgH HS1, 2 region as determined by quantitative ChIP assay. DHL-4 cells were untreated (black bars) or treated (gray bars) with 500 ng/ml of TSA for 18 h. Chromatin proteins and DNA were cross-linked by formaldehyde. The cross-linked chromatin was sheared and then fractionated using specific antibodies as indicated. Purified immunoprecipitated DNA was quantified using primer/probe sets corresponding to the IgH HS1, 2 region.
(b) TSA decreases Oct-2 and Bob-1 binding to the IgH HS4 region in DHL-4 cells as determined by quantitative ChIP assay.
(c) TSA decreases Oct-2 binding to the bcl-2 P1 promoter in DHL-4 cells as determined by quantitative ChIP assay.
(d) TSA decreases Oct-2 binding to the bcl-2 P2 promoter in DHL-4 cells as determined by quantitative ChIP assay.
(e) Oct-1, Oct-2, and Bob-1 do not bind to the bcl-2 exon 3 region in DHL-4 cells.
(f) Oct-1, Oct-2, and Bob-1 do not bind to the bcl-2 P1 promoter in DHL-9 cells.
(g) Oct-1, Oct-2, and Bob-1 do not bind to the bcl-2 P2 promoter in DHL-9 cells.
(h) siRNA targeting Oct-2 decreases the interactions of the IgH enhancers and bcl-2 promoter region as determined by 3C analysis. DHL-4 cells were transfected with siRNA targeting Oct-2 or a non-targeting siRNA. 24 hours after transfection, the cells were harvested, and dead cells were excluded by Ficoll purification. The relative associations of BamHI site 2 at the bcl-2 promoter region with the IgH enhancer BamHI sites were determined and quantified as described in Figure 4.
There are no octamer sites in either bcl-2 promoter region, but if Oct-2 and Bob-1 are involved in mediating the interactions of the bcl-2 promoter region with the IgH enhancers HS1, 2 and HS4, it may be possible to detect Oct-2 and Bob-1 binding at the bcl-2 promoter region. To determine whether Oct-2 and Bob-1 bound to the major active sites of the bcl-2 P1 and P2 promoters, ChIP assays were performed over these regions. Oct-2 bound to the bcl-2 P1 promoter region, and TSA decreased its binding to P1 by more than 50% (Figure 6c). Oct-1, Oct-2, and Bob-1 all bound to the bcl-2 P2 promoter, and TSA treatment decreased the binding of all three proteins to the P2 promoter by 40 to 55% (Figure 6d). As there are no consensus octamer motifs at the bcl-2 promoter regions, their binding to the promoter regions may derive from the IgH enhancers, which are physically associated with the bcl-2 promoter regions in t(14;18) cells. As controls, we examined the binding of Oct-1, Oct-2 and Bob-1 to a region in exon 3 of the bcl-2 gene. No octamer binding sites exist in this region. As shown in Figure 6e, no binding above background was detected in exon 3 and there was no change with TSA treatment. We wished to determine whether the binding of Oct-2 to the bcl-2 promoter was dependent on the presence of the IgH enhancers. We performed ChIP assays at the P1 and P2 promoters in the DHL-9 cell line which lacks the t(14;18) translocation. As shown in Figure 6f and 6g, there was no binding of Oct-1, Oct-2 or Bob-1 to either the P1 or P2 bcl-2 promoter in DHL-9 cells.
To further investigate the role of Oct-2 in mediating the interaction of the bcl-2 promoter region with the IgH enhancers, we used siRNA to decrease Oct-2 expression and then performed 3C analysis. Oct-2 siRNA transfection resulted in an 80% reduction of Oct-2 protein expression in the transfected DHL-4 cells, similar to our previous report (Heckman et al., 2006). Decreased interactions of the IgH enhancers with bcl-2 promoter region BamHI site 2 were observed by 3C analysis in cells transfected with siRNA to Oct-2 (Figure 6h). The interactions of IgH enhancer BamHI sites 4, 5, 6, 7, 8 with bcl-2 promoter region BamHI site 2 were decreased by 63%, 58%, 42%, 53% and 54%, respectively, relative to the results with the non-targeting siRNA. We also observed a decrease in the interactions of the IgH enhancers with bcl-2 promoter region BamHI site 1 in the cells transfected with siRNA to Oct-2 (data not shown). These results demonstrate that Oct-2 is involved in the physical association of the IgH enhancers with the bcl-2 promoter region. The effect of the Oct-2 siRNA on disruption of the interactions of the IgH enhancer with the bcl-2 promoter region was less than the effect of TSA, suggesting that additional transcription factors are also involved in mediating this association.
DISCUSSION
Gene expression is regulated in part by chromatin accessibility for transcription factors. It is also clear that the spatial structure of the chromatin is critical in the regulation of gene transcription. The β-globin genes and the locus control region, which is located over 50 kb away, form a loop structure during transcription, and the spatial interactions correlate with transcription of the β-globin genes (Palstra et al., 2003; Tolhuis et al., 2002). 3C analysis in murine plasmacytomas cells revealed mutual interactions over 22 kb at the transcriptionally active Igκ locus among the Vκ promoters and three Igκ enhancers (Liu and Garrard, 2005). Interactions between the IgH variable region and the IgH enhancers have also been reported recently (Ju et al., 2007).
It is widely believed that the IgH enhancers are responsible for the increased expression of bcl-2 in lymphomas with the t(14;18) translocation, but it is not known how this effect is mediated over a distance of 350 kb. Using chromosome conformation capture, we showed that the IgH enhancers physically associated with the bcl-2 promoter region. When we used the two BamHI sites flanking the bcl-2 promoter region as anchors, we found that the peak interactions of the bcl-2 promoter region were centered at the IgH enhancer HS sites, and they decreased rapidly with increasing genomic distance both 5'and 3' to the enhancers. It is likely that the intervening sequences between the bcl-2 promoter region and the IgH enhancers and the sequences between the two enhancer regions loop out, thus allowing the association of the bcl-2 promoter region with the IgH enhancers. Further evidence provides support for the hypothesis that these physical interactions are functionally involved in the transcriptional regulation of bcl-2 by the IgH enhancers.
We found that TSA, a transcriptional repressor of bcl-2, decreased the physical association of the IgH enhancers with the bcl-2 promoter region. Our previous study showed that histone H3 localized at the bcl-2 promoter region is hyperacetylated in t(14;18) DHL-4 cells compared to non-t(14;18) DHL-9 cells (Duan et al., 2007). TSA treatment decreased bcl-2 transcription and decreased histone H3 acetylation at the bcl-2 promoters (Duan et al., 2005). These results suggested that the IgH enhancers increased bcl-2 transcription through increasing bcl-2 promoter histone acetylation in t(14;18) cells. In the current study, we showed that TSA nearly abrogated the physical association of the IgH enhancers with the bcl-2 promoter region. The physical interactions of the IgH enhancers and the bcl-2 promoter region likely serve as the structural basis for the IgH enhancer-mediated bcl-2 promoter histone acetylation and increased transcription.
It is reasonable to assume that several transcription factors are involved in the physical association of the IgH enhancers with the bcl-2 promoter region. We investigated whether the expression levels of any of the transcription factors that are known to interact with the IgH enhancers were decreased by TSA treatment. TSA decreased the expression of both Oct-1 and Oct-2 at the protein level. We reasoned that decreased levels of Octamer proteins might be involved in the dramatic reduction in the interactions between the IgH enhancers and the bcl-2 promoter region with TSA treatment. Quantitative ChIP assays revealed that the binding of Oct-2 and Bob-1 at HS1, 2 and HS4 was decreased to baseline levels with TSA treatment.
We showed previously that levels of Oct-2 are increased in lymphoma cells with the t(14;18) translocation, and the Oct transcription factors mediate t(14;18) lymphoma cell survival by directly upregulating bcl-2 expression (Heckman et al., 2006). We previously observed binding of Oct-2 to the bcl-2 P2 promoter region despite the fact that there are no octamer sequences in this region. We have now shown that Oct-2 also binds to the bcl-2 P1 promoter. TSA decreased the binding of Oct family members, particularly Oct-2, to the bcl-2 promoters. Since no canonical octamer motifs exist at the bcl-2 promoters, the Oct family members at the bcl-2 promoter region are likely bound at the octamer sites in the IgH enhancers, which are then physically associated with the bcl-2 promoters. Decreased expression of Oct-2 by siRNA transfection results in decreased association of the IgH enhancers with the bcl-2 promoter region, providing further support for the critical involvement of Oct-2 in mediating the IgH enhancer and bcl-2 promoter region interactions.
In summary, our study provides solid evidence for the functional interactions of the IgH enhancers with the bcl-2 locus in t(14;18) lymphomas. Oct-2 is one of the transcription factors that bridges the IgH enhancers and the bcl-2 promoter region, and the Oct factors play a role in mediating the transcriptional deregulatory effect of the IgH enhancers on the bcl-2 promoters. The study of bcl-2 deregulation in t(14;18) lymphoma not only provides insight into the pathogenesis of the disease with the potential for identification of new targeted therapies, but additionally, the translocation itself serves as an ideal and unique model system in the study of the transcriptional regulation of a gene by remote enhancers. Furthermore, these results may be relevant to the molecular mechanisms involved in the deregulation of other oncogenes in translocations involving the IgH locus.
MATERIALS AND METHODS
Cell lines, drug treatment and siRNA transfection
The human t(14;18) lymphoma cell line DHL-4 and the DHL-9 lymphoma cell line that lacks a t(14;18) have been described previously (Ji et al., 1996). They were maintained in RPMI medium supplemented with 10% fetal calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM L-glutamine.
Where indicated, cells were treated with trichostatin A (TSA) (Sigma) for 18 h before further analysis. SiRNA targeting Oct-2 and control non-targeting siRNA were obtained from Dharmacon. Transfection of DHL-4 cells with siRNA molecules using Amaxa's Nucleofector device (Amaxa Biosystems) was performed as previously described (Heckman et al., 2006). The transfected cells were purifed by Ficoll separation to separate the dead cells before further analysis.
Chromatin conformation capture (3C) assay
Generation of 3C templates was performed following the standard protocol with minor modifications (Hagege et al., 2007; Miele et al., 2006). Briefly, approximately 2×107 cells were centrifuged and resuspended in 45 ml of fresh RPMI1640 medium. The suspended cells were cross-linked by 1% formaldehyde for 10 min at room temperature and then quenched by the addition of glycine to 0.125 M with incubation for 5 min at room temperature. After sitting on ice for 15 min, the cells were harvested and lysed in 2 ml ice-cold lysis buffer (10 mM Tris-Cl, pH 8.0, 10 mM NaCl, 0.2% NP-40) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstain A) for 15 min. Nuclei were harvested and washed once using ice-cold 1x BamHI buffer and then resuspended in 1x BamHI buffer containing 0.3% sodium dodecyl sulfate (SDS) and incubated at 65°C for 10 min. Triton X-100 was added to 1.8%. 5×106 cross-linked nuclei were digested with 400 units BamHI (New England Biolabs) at 37°C overnight with constant rotating to achieve >80% digestion. (BamHI was chosen for the 3C analysis because it cuts regularly several kilobases around the bcl-2 promoters and the IgH 3' enhancers. Several other BamHI sites located at a distance from the IgH enhancers were also chosen for the comparison of the relative associations of the core regulatory regions and the flanking regions with the bcl-2 promoters.) The restriction enzyme was inactivated by the addition of SDS to 1.6% with incubation at 65°C for 30 min. Triton X-100 was added to 1%, and 5×106 digested nuclei were subjected to ligation with 4000 Weiss units of T4 DNA ligase (New England Biolabs) in 7.5 ml of 1x ligation buffer and 1% BSA for 5 h at 16°C. 50 μl of 10 mg/ml proteinase K was added to the ligation complex followed by incubation at 65°C overnight to reverse the cross-links. The DNA was purified twice by standard phenol-chloroform extraction and ethanol precipitation. Purified DNA was dissolved in TE and digested with 10 μg/ml DNase free RNase A at 37°C for 15 min. 100 ng of DNA was used for each PCR analysis. The PCR primers were designed to hybridize as close as possible to the restriction sites.
A control template was utilized to optimize the PCR conditions and determine the minimal amount of ligation product that can be reliably quantified and to control for differences in amplification efficiency between primer sets. Generation of the control 3C template using BAC clone DNA was essentially the same as described above. BAC clone RP11-495C15 or CTD2270-P21 on chromosome 18 was utilized with RP11-815P20 or RP11-731F5 covering 105,051,517 to 105,351,516 on chromosome 14 (Invitrogen). 20 μg of one BAC clone or equal molar amounts of two BAC clones were used to generate 3C control templates which were then mixed with 100 ng of genomic DNA prior to PCR. To control for differences in the 3C efficiency in different samples, an internal standard was utilized, the association of two BamHI sites in the GAPDH locus (Hagege et al., 2007; Splinter et al., 2004). PCR was performed at 95°C for 15 min, cycles 2-45 of 95°C for 10 s, 60°C for 60 s. To confirm that the correct bcl-2/IgH hybrid molecules were present in the 3C analysis, the PCR products were electrophoresed on 2% agarose gels, and the gel-purified DNA was sequenced.
The following primers and probes were used for the analysis ofbcl-2 and IgH enhancer interactions by TaqMan primer/probe based real-time PCR (probes are double-dye 5'FAM and 3'TAMRA).
bcl-2 BamHI site primers and probes:
#1 CAAGATGCCACATAAGGAATCAGTC, CCTTTGCTCCCACAGAGCCTCACTCTATG
#2 TCATGTGTGTGGAGAGCGTCAAC, ATGACTGAGTACCTGAACCGGCACCTGCAC
#3 CTGGAAGAATTTGCTAAAGGGTGAAAAG,
TTGGGAATCTGGAAGTCCCAACCCCTTTAG
IgH enhancer BamHI site primers:
#4 CAAAGAAGCCTGCTCCTAAGAACTAC; #5 CACTGAGCCCTGGACCAGAC;
#6 GATGACTCTGAGCATCACGCTGTC; #7 GCTGCCCTCACCACCTGCTG;
#8 TCCAGGGAGGACTCAAGTTTGAG
IgH flanking BamHI site primers:
#9 CACTGAGGAGCTGAGGTTCTGGAGAG; #10 TAGAGTTGAGTGCCTGTGGCTTTTCC
#11 AAATGTATGCCCACCTGGAACCTCAG; #12 ATGGATGCAGTTTCTCCTCCTGCTG
#13 CACGCCTTGAGCTAGTCCATGTGC; #14 CTGTTCGCGGATGCCACAGCCATCTC
#15 ATGCTATCCTCCCACCTCAGTCTTCG;
#16 CTGTGGCTGGTATGTGAGCAGCCAGGT
All quantitative 3C assay results are presented as the average and standard deviation from at least three independent preparations of 3C DNA followed by duplicate real-time PCR analysis.
Quantitative ChIP assay
The ChIP assay was performed as outlined previously (Duan et al., 2005). Antibodies for Oct-1, Oct-2, Bob-1, and IgG were from Santa Cruz Biotechnology. Real-time PCR was performed to quantify the amount of immunoprecipitated DNA. The primer and probe sequences for the ChIP assay of the bcl-2 promoter P1 and P2 regions and exon 3 were described previously (Duan et al., 2005; Duan et al., 2007). The following primer and probe sequences were used for the analysis of IgH enhancers HS1, 2 and HS4 regions:
HS1, 2 forward primer: CATGCAAATGGTTGTTTGTTCCAC
HS1, 2 reverse primer: GTGGAGAATCGTGCAAGCTATTT
HS1, 2 probe: Probe: [6-FAM]TTTCTTGCCCTCTGAGGCTGTTTCCA[Tamra-Q]
HS4 forward primer: AGATGGCGATTTGCATTGG
HS4 reverse primer: GAATAGTCAGGAATCCTGCAAACC
HS4 probe: [6-FAM]AAGGCTGGCACCCAGGCAGCT[Tamra-Q]
All quantitative ChIP assay results are presented as the average and standard deviation from at least three independent immunoprecipitations followed by duplicate real-time PCR analysis.
Western blot analysis
Generation of the whole cell lysates and nuclear extracts has been described previously (Duan et al., 2005). Cells were washed with ice-cold PBS and lysed in Triton X-100 extract buffer. Samples (50 μg) were electrophoresed in a 10% SDS-polyacrylamide gel. All antibodies were from Santa Cruz Biotechnology. Detection of β-actin expression was performed to ensure equivalent protein loading.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant CA56764.
We are grateful to Dr. Job Dekker (U Mass) for advice and for providing the 3C protocol, and to Drs. Tao Li, Jianqun Lin, and Xinwen Qiu in Dr. Andrew Hoffman's lab at Stanford University for expert assistance with the 3C analysis.
REFERENCES
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Desoize B. Anticancer drug resistance and inhibition of apoptosis. Anticancer Res. 1994;14:2291–2294. [PubMed] [Google Scholar]
- Duan H, Heckman CA, Boxer LM. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell. Biol. 2005;25:1608–1619. doi: 10.1128/MCB.25.5.1608-1619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Heckman CA, Boxer LM. The immunoglobulin heavy-chain gene 3' enhancers deregulate bcl-2 promoter usage in t(14;18) lymphoma cells. Oncogene. 2007;26:2635–2641. doi: 10.1038/sj.onc.1210061. [DOI] [PubMed] [Google Scholar]
- Graninger WB, Seto M, Boutain B, Goldman P, Korsmeyer SJ. Expression of bcl-2 and bcl-2-Ig fusion transcripts in normal and neoplastic cells. J. Clin. Invest. 1987;80:1512–1515. doi: 10.1172/JCI113235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nature Protocols. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Cao T, Somsouk L, Duan H, Mehew JW, Zhang C, et al. Critical elements of the immunoglobulin heavy chain gene enhancers for deregulated expression of bcl-2. Cancer Res. 2003a;63:6666–6673. [PubMed] [Google Scholar]
- Heckman CA, Duan H, Garcia PB, Boxer LM. Oct transcription factors mediate t(14;18) lymphoma cell survival by directly regulating bcl-2 expression. Oncogene. 2006;25:888–898. doi: 10.1038/sj.onc.1209127. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Mehew JW, Boxer LM. NF-κB activates bcl-2 expression in t(14;18) lymphoma cells. Oncogene. 2002;21:3898–3908. doi: 10.1038/sj.onc.1205483. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Mehew JW, Ying G-G, Introna M, Golay J, Boxer LM. A-Myb up-regulates Bcl-2 through a Cdx binding site in t(14;18) lymphoma cells. J. Biol. Chem. 2000;275:6499–6508. doi: 10.1074/jbc.275.9.6499. [DOI] [PubMed] [Google Scholar]
- Heckman CA, Wheeler MA, Boxer LM. Regulation of Bcl-2 expression by C/EBP in t(14;18) lymphoma cells. Oncogene. 2003b;22:7891–7899. doi: 10.1038/sj.onc.1206639. [DOI] [PubMed] [Google Scholar]
- Hockenberry D, Nunez G, Milliman C, Schreiber RD, Korsmeyer SJ. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348:334–336. doi: 10.1038/348334a0. [DOI] [PubMed] [Google Scholar]
- Ji L, Mochon E, Arcinas M, Boxer LM. CREB proteins function as positive regulators of the translocated bcl-2 allele in t(14;18) lymphomas. J. Biol. Chem. 1996;271:22687–22691. doi: 10.1074/jbc.271.37.22687. [DOI] [PubMed] [Google Scholar]
- Ju Z, Volpi SA, Hassan R, Martinez N, Giannini SL, Gold T, et al. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3' regulatory region. J. Biol. Chem. 2007;282:35169–35178. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogne M. The 3' IgH regulatory region: A complex structure in a search for a function. Adv. Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3' boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt's lymphoma cells. Gene & Devel. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by chromosome conformation capture (3C) In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al., editors. Current Protocols in Molecular Biology. John Wiley & Sons; New York: 2006. pp. 21.11.1–21.11.20. [DOI] [PubMed] [Google Scholar]
- Mills FC, Harindranath N, Mitchell M, Max EE. Enhancer complexes located downstream of both human immunoglobulin C-alpha genes. J. Exp. Med. 1997;186:845–858. doi: 10.1084/jem.186.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra R-J, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The β-globlin nuclear compartment in development and erythroid differentiation. Nature Genetics. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Reed JC, Kitada S, Takayama S, Miyashita T. Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin's lymphoma and lymphocytic leukemia cell lines. Ann. Oncol. 1994;5(Suppl 1):S61–S65. doi: 10.1093/annonc/5.suppl_1.s61. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Lowe SW. Bcl-2 mediates chemoresistance in matched pairs of primary Eμ-myc lymphomas in vivo. Blood Cells Mol. Dis. 2001;27:206–216. doi: 10.1006/bcmd.2000.0372. [DOI] [PubMed] [Google Scholar]
- Seto M, Jaeger U, Hockett RD, Graninger W, Bennett S, Goldman P, et al. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol. Cell. Biol. 1996;16:5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]