Abstract
Understanding the molecular underpinnings of cancer is of critical importance to developing targeted intervention strategies. Identification of such targets, however, is notoriously difficult and unpredictable. Malignant cell transformation requires the cooperation of a few oncogenic mutations that cause substantial reorganization of many cell features1 and induce complex changes in gene expression patterns2-6. Genes critical to this multi-faceted cellular phenotype thus only have been identified following signaling pathway analysis7-10 or on an ad hoc basis4, 11-14. Our observations that cell transformation by cooperating oncogenic lesions depends on synergistic modulation of downstream signaling circuitry15-17 suggest that malignant transformation is a highly cooperative process, involving synergy at multiple levels of regulation, including gene expression. Here we show that a large proportion of genes controlled synergistically by loss-of-function p53 and Ras activation are critical to the malignant state. Remarkably, 14 among 24 such ‘cooperation response genes’ (CRGs) were found to contribute to tumor formation in gene perturbation experiments. In contrast, only one in 14 perturbations of genes responding in a non-synergistic manner had a similar effect. Synergistic control of gene expression by oncogenic mutations thus emerges as an underlying key to malignancy and provides an attractive rationale for identifying intervention targets in gene networks downstream of oncogenic gain and loss-of-function mutations.
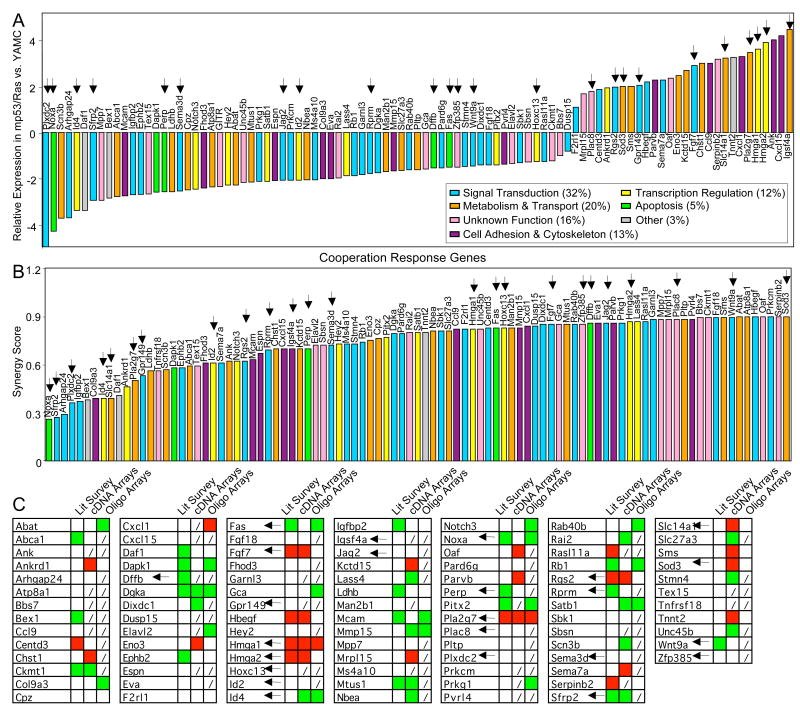
To identify genes regulated synergistically by cooperating oncogenic mutations at genomic scale, we compared mRNA expression profiles of young adult murine colon (YAMC) cells with those of YAMC cells expressing mutant p53175H (mp53), activated H-Ras12V (Ras) or both mutant proteins together (mp53/Ras)17 using Affymetrix microarrays. Using a step-wise procedure, we first identified 538 genes differentially expressed between mp53/Ras and YAMC control cells with a statistical cut off at p < 0.01 (N-test, Westfall-Young adjusted). A further subset of 95 annotated genes that respond synergistically (28 up/67 down) to the combination of mutant p53 and Ras proteins, termed ‘cooperation response genes’ (CRG) was then determined using a synergy score, as described in methods (Figure 1, Supplementary Table 1, Supplementary File 1). Expression values and synergy scores for the CRGs derived from TaqMan low-density QPCR array (TLDA) data showed strong positive correlation with the values for the same genes obtained from microarray analysis (Supplementary Figures 1 and 2, Supplementary Table 2 and Supplementary File 2). Thus CRG identification was confirmed by independent methods, with final CRG selection based on microarray data, due to higher sample replication in this data set.
Figure 1. Identification and characterization of cooperation response genes (CRGs).

Raw expression values (log2) of 538 differentially expressed genes (represented by 657 probe sets) for mp53, Ras and mp53/Ras cells, as compared to YAMC controls, are shown rank ordered according to synergy score. Red and green indicate relative gene expression in the cells indicated versus YAMC cells. Purple or blue indicate the synergy score for each gene plotted. A synergy score of 0.9 or less defines CRGs. The cut off is indicated by arrowheads or the threshold line (stippled).
CRGs encode proteins involved in the regulation of cell signaling, transcription, apoptosis, metabolism, transport or adhesion (Figure 2A, B and Supplementary Table 1), and in large proportion appear misexpressed in human cancer. For 47 of 75 CRGs tested co-regulation is found in primary human colon cancer and our murine colon cancer cell model (Figure 2C, Supplementary Figure 3). Furthermore, altered expression of 29 CRGs has been reported in a variety of human cancer types, consistent with the direction of the change in gene expression observed in our experiments (Figure 2C, Supplementary Table 1 and references therein). Thus, modulation of CRG expression has common features in malignant cell transformation of both murine and human cells.
Figure 2. Differential expression and synergy scores of CRGs in mp53/Ras cells and CRG co-regulation in human colon cancer.
Bar graphs ranking CRG expression measured by microarray in mp53/Ras vs. YAMC cells (A) and CRG synergy scores (B). Bars are color-coded for gene-associated biological processes according to Gene Ontology (GO) database. C) Table summarizing co-regulation of CRGs in mp53/Ras cells and human cancer based on literature survey for a variety of human cancers and two independent expression analyses of primary human colon cancers. Up- or down-regulation of CRG expression vs. controls is indicated by red or green, lack of CRG representation on arrays by (/). Arrows indicate genes perturbed in this study.
The relevance of differentially expressed genes for malignant cell transformation was assessed by genetic perturbation of a series of 24 CRGs and 14 genes responding to p53175H and/or activated H-Ras12V in a non-cooperative manner (non-CRGs). Perturbed genes were chosen across a broad range of biological functions, levels of differential expression and synergy scores (Figure 2, Supplementary Figure 4, Supplementary File 3). Gene perturbations were carried out in mp53/Ras cells with the goal to re-establish mRNA expression of the manipulated genes to levels relatively close to those found in YAMC control cells, and to monitor subsequent tumor formation following sub-cutaneous injection of these cells into immuno-compromised mice. Of the perturbed genes, 18 were up- and 20 down-regulated in mp53/Ras cells, relative to YAMC.
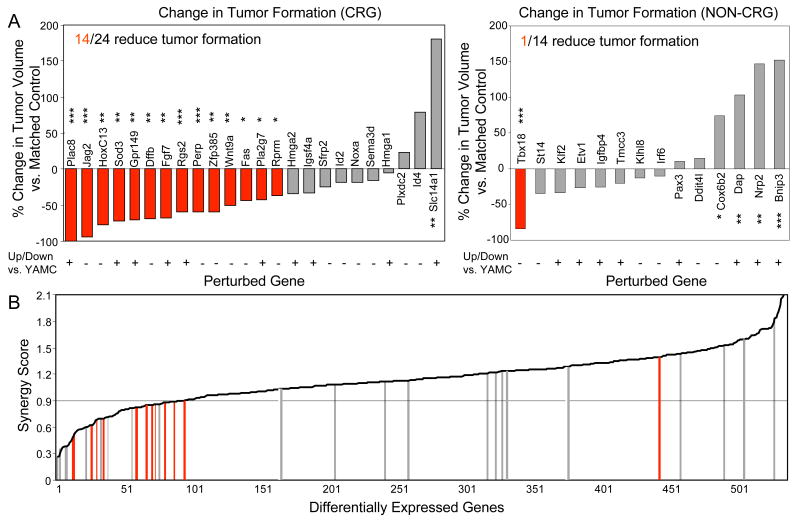
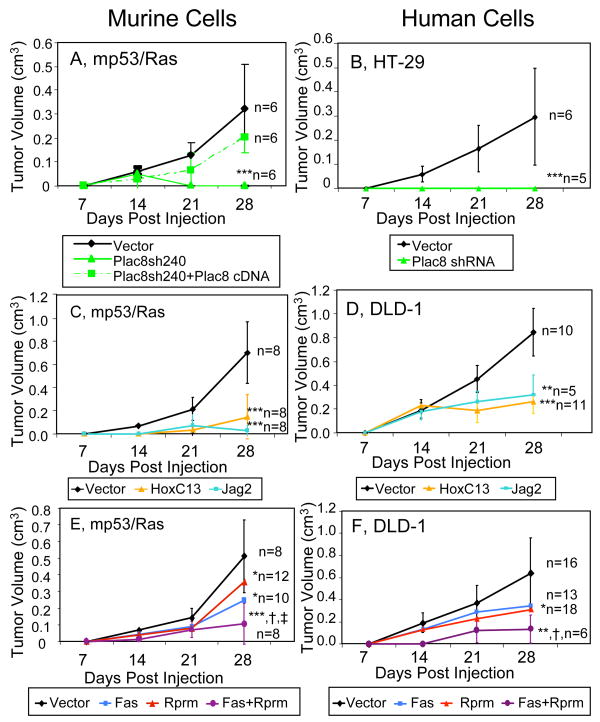
Reversal of the changes in CRG expression significantly reduced tumor formation by mp53/Ras cells in 14 out of 24 cases (Figure 3A, left panel; Figure 4A, C; Supplementary Figure 5A and Supplementary Table 3), indicating a critical role in malignant transformation for a surprisingly large fraction of these genes. Perturbation of Plac8, Jag2 and HoxC13 gene expression had the strongest effects. We also combined perturbations of two CRGs, Fas and Rprm, that alone produced significant yet milder changes in tumor formation. This yielded significantly increased efficacy in tumor inhibition as compared with the respective single perturbations (Figure 4E, Supplementary Figure 5B and Supplementary Table 4). Thus, even genetic perturbations of CRGs with relatively smaller effects when examined on their own show evidence of being essential when analyzed in combination.
Figure 3. Synergistic response of downstream genes to oncogenic mutations is a strong predictor for critical role in malignant transformation.
(A) Bar graphs indicating percent change in endpoint tumor volume following CRG and non-CRG perturbations in mp53/Ras cells (left and right panel, respectively). Perturbations significantly decreasing tumor size, as compared to matched controls are shown in red (***, p<0.001; **, p<0.01; *, p<0.05; Wilcoxn signed-rank and t-test). (B) Distribution of gene perturbations over the set of genes differentially expressed in mp53/Ras cells, rank-ordered by synergy score. Bars, color-coded as above, indicate perturbed genes. CRG cut-off synergy score (0.9) is indicated by horizontal line.
Figure 4. CRG perturbations reduce tumor formation of both mp53/Ras and human cancer cells.
Tumor volume was measured weekly for 4 weeks following injection into nude mice of murine (A, C, E) and human cancer cells (B, D, F) with indicated perturbations. Error bars indicate standard deviation at each time point. Number of injections (n) and significance levels as compared to matched controls are indicated (***, p<0.001; **, p<0.01; *, p<0.05). Significance of tumor reduction upon combined perturbation Fas + Rprm as compared to individual perturbations is indicated as follows: vs. Fas (†, p<0.05), vs. Rprm (‡, p<0.05).
In contrast to the multitude of CRG-related effects on tumor inhibition, out of the 14 non-CRG perturbations, only one showed a significant reduction in tumor formation of mp53/Ras cells (Figure 3A, right panel; Supplementary Figure 6, and Supplementary Table 5). Taken together, our data suggest that among the genes differentially expressed in cancer cells, malignant transformation strongly relies on the class of genes synergistically regulated by cooperating oncogenic mutations (Figure 3B, Supplementary Figure 7).
Genetic perturbation experiments were carried out utilizing retrovirus-mediated re-expression of corresponding cDNAs for down-regulated genes (Supplementary Table 6) and shRNA-dependent stable knock-down using multiple independent targets for over-expressed genes (Supplementary Table 7). In addition, Plac8 knock down was functionally rescued by expression of shRNA-resistant Plac8 (Figure 4A), confirming specificity of the Plac8 loss-of-function experiments. The extent of all gene perturbations was assessed by quantitative PCR (Supplementary Figure 8). As expected, the genetic perturbations disrupt tumor formation downstream of the initiating oncogenic mutations. Expression of both mutant p53 and activated Ras proteins remains unaffected by all genetic manipulations that alter the formation of tumors (Supplementary Figure 9). Moreover, gene perturbations distinguished tumor growth from in vitro cell proliferation, as they generally did not affect cell accumulation in tissue culture (Supplementary Fig 10).
Perturbations of CRGs in human cancer cells (Figures 4B, D, F, Supplementary Figure 11 and Supplementary Tables 8 and 9) had similarly strong tumor inhibitory effects to those in the genetically tractable murine mp53/Ras cells, as assessed by xenografts in nude mice. Perturbations of both up- and down-regulated CRGs, i.e. Dffb, Fas, HoxC13, Jag2, Perp, Plac8, Rprm, Zfp385 and Fas + Rprm were performed in human DLD-1 and/or HT-29 colon cancer cell lines using retroviruses (Supplementary Figure 12, Supplementary Tables 6 and 10) as described above. Similar to mp53/Ras cells, both human cancer cell lines have p53 mutations, whereas with K-Ras (DLD-1) and B-Raf (HT-29) mutations they express activated members of the Ras/Raf signaling pathway distinct from activated H-Ras in mp53/Ras cells. In addition, DLD-1 and HT-29 cells carry further oncogenic lesions such as APC and PIK3CA mutations, with HT-29 cells also exhibiting a mutation in Smad4 (for references, see Supplemental Methods). The genetic perturbations had no effect on mutant Ras/Raf or p53 protein expression levels in both DLD-1 and HT-29 cells (Supplementary Figure 13), indicating disruption of the cancer phenotype downstream of oncogenic mutations. Taken together, these experiments indicate the relevance of CRGs to cancer in a variety of backgrounds and genetic contexts.
The data described here indicate that the cooperative nature of malignant cell transformation, to a considerable degree, depends on a class of downstream effector genes regulated synergistically by multiple oncogenic mutations. We show that these cooperation response genes (CRGs) identified here contain a strikingly large fraction of genes (14 out of 24 tested) that are critical to the malignant phenotype, and that their perturbation, singly or in combination, can inhibit formation of tumors containing multiple oncogenic lesions, including p53 deficiency. In contrast, few of the genes differentially expressed in a non-synergistic manner (1 out of 14) significantly reduced tumor growth upon perturbation. Synergistic behavior found in gene expression data thus appears highly informative for identification of genes critically involved in malignant cell transformation (Figure 3B), and provides a rational path to discovery of both cancer cell-specific vulnerabilities and targets for intervention in cancer cells harboring multiple mutations, including p53 loss-of-function.
CRGs represent a set of 95 annotated cellular genes, many of which have been associated with human cancer by virtue of altered gene expression (Figure 2C, Supplementary Table 1). They are involved in the regulation of cell signaling, transcription, apoptosis and metabolism, and based on our data represent key control points in many facets of cancer cell behavior. We thus consider CRGs as critical nodes in gene networks underlying the malignant phenotype, providing an attractive rationale to explain why several features of cancer cells emerge simultaneously out of the interaction of a few genetic lesions17.
Among CRGs and other differentially expressed effector genes we also have identified examples that when perturbed produce significantly larger tumors (Figure 3 and Supplementary Tables 3 and 5). This is consistent with the notion that oncogenic mutations can induce strongly anti-proliferative cellular stress responses18-21. The existence of genes that while responding to oncogenic mutations restrict tumor formation provides direct evidence to support the idea that the state of malignant transformation arises as the result of a finely tuned balance between opposing signals generated by oncogenic mutations15-17, 20, 22, 23. It is thus reasonable to speculate that tumor suppression via perturbation of CRGs, as shown here, may involve the disruption of this delicate balance. In fact, such targeted disruption downstream of oncogenic mutations may allow selective cancer cell deconstruction yielding intervention strategies with high specificity for cancer cells.
For the 14 CRGs with tumor-inhibitory perturbations, a clear causal role in tumor formation downstream of oncogenic mutations has been shown here for the first time. Moreover, our data indicate that both gene extinctions (eight genes) and gene inductions (six genes) play important roles in this process. For example, we show that re-expression of the down-regulated CRGs Jag2, a Notch ligand, or of HoxC13, a homeobox transcription factor, as well as shRNA-dependent knock down of Plac8 gene expression are each strongly tumor inhibitory in p53 defective murine and human cancer cells. Both Notch signaling24 and HoxC1325 can play oncogenic roles in haematopoietic malignancies, but are involved in promoting differentiation of epithelial cells26, 27 consistent with the tumor-inhibitory function of Jag2 and HoxC13 in the context of the solid tumor models investigated here. Plac8 is a little investigated gene encoding a cysteine-rich highly conserved peptide expressed in placenta, haematopoietic and epithelial cells that is non-essential for mouse development28. When over-expressed, Plac8 can suppress p5329. Its essential role for tumor formation of p53-deficient cancer cells, however, is novel and unexpected. Among the eight down-regulated CRGs is Zfp385, another gene of unknown function. Moreover, there is a considerable number of pro-apoptotic/anti-proliferative genes such as Perp, Rprm, Fas, Dffb and Wnt9a, indicating that Ras activation and p53 deficiency cooperate to extinguish the expression of multiple growth inhibitory genes, each of which contributes significantly to restricting tumor growth in the YAMC model when re-expressed. Out of these genes, Perp, Rprm, and Fas previously have been identified as direct p53 targets, suggesting that their regulation by p53 is highly conditional on Ras activity (Supplementary Table 1 and references therein). Most of the up-regulated CRGs contributing to tumor growth affect signal transduction. This includes Fgf7, Rgs2, Gpr149, an uncharacterized orphan seven-trans-membrane receptor, and Sod3, which acts on signaling via modulation of metabolites30. For all of these genes, including Pla2g7, a role in promoting tumor growth is reported here for the first time.
Notably, the efficacy of CRG perturbations performed in human colon cancer cells was comparable to that in the murine colon cell transformation model, suggesting dependence of the malignant state on a similar set of genes in both backgrounds. This is remarkable in light of the fact that these human cancer cells carry oncogenic mutations in genes in addition to Ras or Raf and p53, and suggests that CRGs may play key roles in the generation and maintenance of the cancer cell phenotype in a variety of contexts. CRGs thus may provide a valuable source for identification of much sought ‘Achilles heels’ in human cancer by rational means.
Methods Summary
Cells
YAMC cells and derivation of cells with multiple oncogenic lesions17 are described in supplementary materials.
Microarray Experiments, Statistical Analysis and CRG Identification
Polysomal RNA was harvested to obtain gene expression profiles reflective of protein synthesis rates. Expression values were obtained using the RMA procedure with background correction in Bioconductor (http://www.bioconductor.org). Differentially expressed genes were identified by the step-down Westfall-Young procedure in conjunction with the permutation N-test, FWER < 0.01. Genes that respond synergistically to the combination of mutant p53 and activated Ras (CRGs) were selected by the following procedure. Let a = mean expression value for a given gene in mp53 cells, b = mean expression value for the same gene in Ras cells and d = mean expression value for this gene in mp53/Ras cells. Then, the criterion defines CRGs as for genes over-expressed in mp53/Ras cells and as for genes under-expressed in mp53/Ras cells, as compared to controls. In order to assess robustness of synergy scores, jackknife sub-sampling was used to generate estimated p values for these scores. TaqMan Low-Density Arrays (Applied Biosystems) were used to independently test gene expression differences observed by Affymetrix arrays.
Genetic Perturbation of Gene Expression
cDNAs expressed via pBabe retroviral vectors or shRNA in pSuper-retro vectors were used to generate gene perturbations. These were tested by comparison of RNA expression levels in empty vector-infected cells and cells subjected to gene perturbation via SYBR Green qPCR with gene-specific primers.
Xenograft Assays
Tumor formation was assessed by sub-cutaneous injection of cells into CD-1 nude mice (Crl: CD-1-Foxn1nu, Charles River Laboratories). Tumor size was measured by caliper at 2, 3 and 4 weeks post-injection. Significance of difference in tumor size was calculated by the Wilcoxn signed-rank test and by the t-test using directly matching vector control cells for each perturbation.
Supplementary Material
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank Drs. D. Bohmann, C. Jordan and M. Noble for discussion, C. Brower, A. Cardillo, A. Petenkaya, L. Salamone and Drs. A. Brooks, Y. Xiao, S. Welle and A. Rosenberg for assistance with microarray data analysis, Drs. A. Burgess, R. Whitehead, J. Filmus, L. Milner for materials, and Dr. L. Maquat for sharing equipment. This work was supported in part by NIH grants CA90663, CA120317, GM075299 and a James P. Wilmot Cancer Center pilot grant. H.R.M. was supported in part by NIH T32 CA09363, P.S. by NIH K99 LM009477. This work is dedicated to Carmela V. Richards.
Footnotes
Author Contributions H.L. conceived and directed the project. H.R.M., E.R.S., G.C., C.K. and B.S. designed and carried out experiments. S.C and L.N. carried out experiments. P.S. consulted on and performed statistical analysis of microarray and tumor formation data. L.K. and A.Y. designed statistical methods to analyze microarray data. H.R.M. and H.L. wrote the paper. H.R.M., E.R.S., G.C. and C.K. made equal contributions to the manuscript.
Author Information Microarray data are deposited in the NCBI GEO database (Accession No. GSE9199). Reprints and permissions information is available at www.nature.com/reprints. Correspondence should be addressed to H.L. (Land@urmc.rochester.edu).
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Identification and classification of p53-regulated genes. Proc Natl Acad Sci U S A. 1999;96:14517–22. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao R, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–93. [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze A, Lehmann K, Jefferies HB, McMahon M, Downward J. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 2001;15:981–94. doi: 10.1101/gad.191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang E, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–30. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 6.Boiko AD, et al. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–52. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 8.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 9.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Viciana P, et al. Cancer targets in the Ras pathway. Cold Spring Harb Symp Quant Biol. 2005;70:461–7. doi: 10.1101/sqb.2005.70.044. [DOI] [PubMed] [Google Scholar]
- 11.Okada F, et al. Impact of oncogenes in tumor angiogenesis: mutant K-ras up-regulation of vascular endothelial growth factor/vascular permeability factor is necessary, but not sufficient for tumorigenicity of human colorectal carcinoma cells. Proc Natl Acad Sci U S A. 1998;95:3609–14. doi: 10.1073/pnas.95.7.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–97. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd AC, et al. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 1997;11:663–77. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- 17.Xia M, Land H. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat Struct Mol Biol. 2007;14:215–23. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
- 18.Ridley AJ, Paterson HF, Noble M, Land H. Ras-mediated cell cycle arrest is altered by nuclear oncogenes to induce Schwann cell transformation. Embo J. 1988;7:1635–45. doi: 10.1002/j.1460-2075.1988.tb02990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirakawa T, Ruley HE. Rescue of cells from ras oncogene-induced growth arrest by a second, complementing, oncogene. Proc Natl Acad Sci U S A. 1988;85:1519–23. doi: 10.1073/pnas.85.5.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanidi A, Harrington EA, Evan GI. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature. 1992;359:554–6. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- 21.Denoyelle C, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–63. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 22.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 23.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 24.Houde C, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104:3697–704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- 25.Panagopoulos I, et al. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36:107–12. doi: 10.1002/gcc.10139. [DOI] [PubMed] [Google Scholar]
- 26.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 27.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ledford JG, Kovarova M, Koller BH. Impaired host defense in mice lacking ONZIN. J Immunol. 2007;178:5132–43. doi: 10.4049/jimmunol.178.8.5132. [DOI] [PubMed] [Google Scholar]
- 29.Rogulski K, et al. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene. 2005;24:7524–41. doi: 10.1038/sj.onc.1208897. [DOI] [PubMed] [Google Scholar]
- 30.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med. 2003;35:236–56. doi: 10.1016/s0891-5849(03)00275-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is linked to the online version of the paper at www.nature.com/nature.