Abstract
The V3 loop of HIV-1 gp120 is considered occluded on many primary viruses. However, virus sensitivity to neutralization by different V3 mAbs often varies, indicating that access to V3 is not restricted equally for all antibodies. Here, we have sought to gain a better understanding of these restrictions by determining the neutralizing activities of 7 V3 mAbs (19b, 39F, CO11, F2A3, F530, LA21, and LE311) against 15 subtype B primary isolates and relating these activities to the fine specificity of the mAbs. Not surprisingly, we found that most mAbs neutralized the same 2–3 viruses, with only mAb F530 able to neutralize 2 additional viruses not neutralized by the other mAbs. Epitope mapping revealed that positively-charged residues in or near the V3 stem are important for the binding of all the mAbs and that most mAbs seem to require the Pro residue that forms the GPGR β hairpin turn in the V3 tip for binding. Based on the mapping, we determined that V3 sequence variation accounted for neutralization resistance of approximately half the viruses tested. Comparison of these results to those of select V3 mAbs with overall better neutralizing activities in the light of structural information suggests that an antibody’s mode of interaction with V3, driven by contact residue requirements, precludes the antibody from accessing its epitope on different viruses. Based on the data we propose an angle of interaction with V3 that is less stringent on access for antibodies with cross-neutralizing activity compared to antibodies that neutralize relatively fewer viruses.
Keywords: V3 antibodies, cross-neutralization, V3 accessibility, epitope mapping, HIV vaccine design
INTRODUCTION
An HIV vaccine will likely need to incorporate a component that elicits a humoral response capable of neutralizing a broad array of circulating virus isolates (Gallo, 2005). The target for neutralizing antibodies (NAbs) is the viral envelope spike (Burton and Montefiori, 1997) and many strategies are being pursued to elicit antibodies with the desired level of cross-neutralizing activity (reviewed in (Haynes and Montefiori, 2006; Hu and Stamatatos, 2007; Phogat and Wyatt, 2007)). However, as a result of viral evolution, driven perhaps in most part by host NAb pressure (Beaumont et al., 2001; Frost et al., 2005; Wei et al., 2003), the envelope spike harbors a remarkable plethora of features to shield conserved regions from antibody recognition (reviewed in (Karlsson Hedestam et al., 2008; Wyatt et al., 1998)). Collectively, these features represent a substantial scientific hurdle towards formulating a NAb-inducing HIV vaccine component, as evident from the relatively low cross-neutralizing activity of NAbs elicited by the large majority of immunogens developed to date.
To aid the design of an effective B-cell immunogen, and given HIV’s defenses, the antigenicity of the envelope spike on different HIV isolates from different subtypes needs to be elucidated. This process has been aided greatly by the existence of a handful of rare mAbs that can neutralize a broad range of primary viruses and protect against viral challenge in animal models, which have allowed also for the identification of potential target sites for vaccine design (Burton et al., 2004; Kramer, Siddappa, and Ruprecht, 2007; Zolla-Pazner, 2004). Unfortunately, the inability to elicit NAbs with similar broad neutralizing activity or even with modest cross-neutralizing activity suggests that there are still gaps in our understanding of features that affect efficient antibody recognition of epitopes on the envelope spike and of how to design immunogens to effectively elicit antibodies to target sites (Hu and Stamatatos, 2007).
One of the potential, albeit often debated, target sites for vaccine design is the V3 loop on the gp120 subunit of the envelope spike (Hartley et al., 2005; Haynes and Montefiori, 2006; Zolla-Pazner, 2005). The V3 loop has been studied extensively. Lower enthusiasm for seeking to target V3 is based largely on the general observation that V3 antibodies often exhibit poor cross-neutralizing activity (Hartley et al., 2005) and several factors, for example steric hindrance by glycans, V3 sequence variability, and masking of the V3 loop by V1/V2 (Gram et al., 1994; Krachmarov et al., 2005; Krachmarov et al., 2006; Pinter et al., 2004; Schonning et al., 1996), help explain why many V3 mAbs may exhibit little neutralizing activity. A recent study has suggested also that the presence (or absence) of certain V3 residues may greatly affect the neutralizing activity of V3 mAbs and the presence of subtype-specific epitopes within V3 has been proposed (Patel, Hoffman, and Swanstrom, 2008). There exists, however, a small selection of V3 mAbs that exhibit relatively better cross-neutralizing activity than other V3 mAbs (Eda et al., 2006; Gorny et al., 2006; Pantophlet et al., 2007). The neutralizing activity of these select mAbs suggests that they may be hampered less by at least some of the access-restrictive factors outlined above. The notion that such restrictions do not apply equally to all V3 antibodies is supported also by the often noted spectrum of virus sensitivities to different V3 mAbs (Gorny et al., 2004; Gorny et al., 2002).
Here, we wanted to investigate the extent to which the fine specificity of V3 mAbs affects their neutralizing activity, as part of efforts to better comprehend the often observed resistance of HIV isolates to V3-directed antibodies and broad antibody recognition in general. Efforts were made in a recent study to map the epitopes of different V3 mAbs using a phage-display peptide library expressing random dodecamer sequences (Patel, Hoffman, and Swanstrom, 2008). However, for most mAbs that were investigated, no consensus sequence could be identified that resembled V3. In this study, we applied a commonly applied scanning mutagenesis approach, utilizing a panel of V3 mutants that previously allowed mapping, with high accuracy, of residues important for the binding of the modestly cross-neutralizing V3 mAb B4e8 (Bell et al., 2008; Pantophlet et al., 2007).
RESULTS
Neutralizing activity of V3 mAbs against subtype B primary viruses
To assess the cross-neutralization potential of the V3 mAbs, we tested them against a panel of 15 viruses in a robust and highly sensitive pseudovirus-based single-round infectivity assay with luciferase readout (Table 1). The viruses were selected from a previously published 30-member panel of subtype B primary viruses (Binley et al., 2004) because they exhibit diverse sensitivities to V3 mAbs (Binley et al., 2004; Li et al., 2005). The test conditions were the same as described in the Binley et al. study. IC50 values are reported because they allow for a more reliable comparison between the potency and cross-reactivity of different antibodies, as argued previously (Binley et al., 2004).
Table 1.
Neutralizing activity of V3 mAbs against a panel of subtype B viruses
| Virusa | IC50(μg/ml)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 19b | 39F | C011 | F2A3 | F530 | LA21 | LE311 | B4e8 | 447D | 58.2 | |
| QH0515 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| QH0692 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 4.1 | 40.3 | 39.3 |
| BG1168c | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| SS1196c | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 34.6 | 1.9* | 0.5* |
| 5768-p27 | 3.2 | >50 | >50 | >50 | 5.9 | 32.7 | 41.2 | >50 | >50 | >50 |
| 6101-p15 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 91US056 | >50 | >50 | >50 | >50 | 14.8 | >50 | >50 | >50 | >50 | >50 |
| 92BR020c | 21.5 | 1.8 | 1.4 | 2.6 | 2.4 | 1.2 | 2.6 | 0.4 | >50* | >50* |
| 92BR021 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 92HT594c | >50 | >50 | >50 | >50 | 43.2 | >50 | >50 | 4.3 | 7.9 | >50 |
| 92US712c | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 0.6* | 1.4* |
| 93TH305 | >50 | 20.3 | 19.3 | 36.2 | 13.3 | >50 | 13.4 | 37.4 | 8.5 | 25.4 |
| JR-CSFc | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 7.2 | >50 | 32.8 |
| JR-FLc | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 2.4 | 32.6 | >50 |
| ME1 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
|
| ||||||||||
| aMLV | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
All viruses are quasispecies pools except where noted. Virus designations are generally denoted as year (of isolation), country (of isolation), and name/number. aMLV, amphotrophic murine leukemia virus (negative control virus). c, molecular clone.
IC50s are color-coded as follows: white, IC50 >50 μg/ml; blue, 50 μg/ml> IC50 >10 μg/ml; yellow, 10 μg/ml> IC50 >1 μg/ml; red, IC50 <1 μg/ml. The IC50s for mAbs B4e8, 447-52D (447D), and 58.2 are taken from a previous studies (Binley et al., 2004; Pantophlet et al., 2007).
, IC50 obtained with quasispecies pool (mAbs 447D and 58.2 only); for these viruses the IC50 against the molecular clone listed has not been determined.
Overall, the mAbs were only able to neutralize a very limited number of viruses; most mAbs neutralized 2–3 viruses, whereas only mAb F530 neutralized 5 viruses (Table 1). The 92BR020 molecular clone was neutralized by all 7 mAbs and was shown previously as neutralization-sensitive to V3 mAb B4e8 (Pantophlet et al., 2007). Strikingly, the quasispecies pool of this virus was not neutralized by V3 mAbs 58.2 or 447-52D (Table 1) (Binley et al., 2004), even though the amino acid sequences of the quasispecies pool and the molecular clone share ~95% homology; the clone is also similarly sensitive to neutralization by mAbs b12, 2G12, 2F5, and 4E10 and a broadly neutralizing HIV+ serum as the quasispecies pool (T. Wrin et al., unpublished results). Although we cannot readily explain the discrepancy between the quasispecies pool and the clone for mAbs 58.2 and 447-52D, the results suggest that virus 92BR020 is likely sensitive to neutralization by several V3 antibodies.
Viruses 93TH305 and 5768-p27 were neutralized by 5 and 4 of the mAbs tested here, respectively (Table 1). For virus 93TH305 the IC50 values ranged from 13–37 μg/ml, in the same range as observed previously for V3 mAbs 447–52D, 58.2 and B4e8 (Table 1). For virus 5768-p27, IC50s ranged from 3–41 μg/ml. Virus 5768-p27 is not sensitive to neutralization by mAbs 447-52D, 58.2, or B4e8 (IC50s>50 μg/ml; Table 1), most likely due to the rather uncommon Lys at position 315; mAbs 447-52D, 58.2, and B4e8 prefer an Arg at this position for efficient binding (Keller et al., 1993; Pantophlet et al., 2007; Seligman et al., 1996; White-Scharf et al., 1993; Zolla-Pazner et al., 2004).
Aside from viruses 92BR020, 93TH305, and 5768-p27, only mAb F530 also neutralized viruses 91US056 and 92HT594 (Table 1). Virus 91US056 is not sensitive to neutralization by mAbs 447-52D, 58.2, and B4e8 (Binley et al., 2004; Pantophlet et al., 2007), because of the Ala at position 315 instead of an Arg. Virus 92HT594 is neutralized also by mAbs 447-52D and B4e8 (IC50s: 8 and 4 μg/ml, respectively; Table 1), but is resistant to neutralization by mAb 58.2. None of the 7 V3 mAbs tested here neutralized (IC50>50 μg/ml) any of the other viruses that are neutralization-sensitive to mAbs 58.2, 447-52D, and/or B4e8 (Table 1).
MAb fine specificity as determined by V3 alanine scanning
The same 18 alanine point mutants generated previously for mapping the specificity of mAb B4e8 (Pantophlet et al., 2007) were used to map the specificity of the mAbs tested here; this approach proved highly accurate for mapping B4e8’s fine specificity, as evidenced by the interactions observed between B4e8 and V3 from the subsequent structure of a B4e8:V3 peptide complex (Bell et al., 2008). As with mAb B4e8, apparent antibody binding affinities were determined and related to the apparent antibody affinity for wild-type gp120 (Pantophlet et al., 2007). We found that the binding of all mAbs could be reduced by several substitutions. None of the patterns resembled the effects of alanine substitutions on B4e8 binding (Table 2). Unexpectedly, we also observed that the binding of several mAbs –19b, CO11, F2A3, F530, and LA21—was increased upon changing select residues to Ala; the largest number of increased binding was observed for mAb F2A3 (Table 2). The following specific observations were also made:
Table 2.
Epitope mapping of V3 mAbs
| MAb | Apparent affinity (%)a relative to wild-type gp120JRCSF for indicated V3 mutants located in noted V3 segments
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| baseb |
stem
|
tip
|
stem
|
|||||||||||||||
| R298A | P299A | N301A | N302A | R304A | K305A | S306A | I307A | H308A | I309A | P313A | R315A | F317A | Y318A | T319A | T320A | E322A | D325A | |
| 19b | 71 | 146 | 113 | 66 | 5 | 23 | 167 | 0.3 | 128 | 54 | 4 | 7 | 0.3 | 0.3 | 72 | 123 | 95 | 201 |
| 39F | 66 | 156 | 148 | 73 | 66 | 0.5 | 40 | 6 | 75 | 50 | 108 | 95 | 75 | 119 | 107 | 164 | 104 | 160 |
| CO11 | 130 | 258 | 58 | 114 | 74 | 79 | 190 | 84 | 17 | 71 | 0.2 | 0.2 | 79 | 162 | 97 | 120 | 90 | 177 |
| F2A3 | 76 | 168 | 135 | 114 | 86 | 11 | 90 | 50 | 40 | 219 | 38 | 0.1 | 211 | 257 | 161 | 176 | 148 | 223 |
| F530 | 57 | 218 | 151 | 59 | 11 | 7 | 143 | 30 | 55 | 59 | 36 | 78 | 158 | 158 | 103 | 171 | 162 | 89 |
| LA21 | 77 | 118 | 124 | 97 | 130 | 135 | 128 | 216 | 41 | 197 | 0.4 | 54 | 56 | 29 | 137 | 220 | 66 | 169 |
| LE311 | 68 | 140 | 109 | 78 | 64 | 2 | 127 | 44 | 35 | 106 | 3 | 3 | 136 | 127 | 122 | 103 | 108 | 138 |
| B4e8 | 79 | 96 | 124 | 85 | 88 | 83 | 87 | 74 | 88 | 23 | 91 | 3 | 49 | 76 | 81 | 127 | 77 | 117 |
Apparent affinities were determined as the antibody concentration at half-maximal binding based on ELISA binding curves using the program Graphpad Prism (v.4.0). Apparent affinities relative to wild-type gp120JRCSF were calculated with the formula (apparent affinity for wild type/apparent affinity for mutants) ×100%. Red: <10% relative binding affinity; blue: >10% to <50% relative binding affinity; white: >50% to <200% relative affinity between; black: >200% relative affinity. The mapping results for mAb B4e8 are taken from (Pantophlet et al., 2007). Residue numbering is based on the HXB2 reference sequence (http://www.hiv.lanl.gov/). The mutants are arranged from the N- to C-terminal portion of the V3 region (left to right).
V3 is divided into 3 structural segments based on the V3 structure in the context of gp120 (Huang et al., 2005): (I) the base (residues 296–300 (N-terminal portion) and 326–331 (C-terminal portion)), (II) the stem (residues 301–305 (N-terminal portion) and 321–325 (C-terminal portion)), and (III) the tip (residues 306–320).
Binding of mAb 19b was strongly reduced (>10-fold) upon substitution of Arg304, Ile307, Pro313, Arg315, Phe317, or Tyr318, and moderately reduced (~4-fold) upon substitution of Lys305 (Table 2). These results are consistent overall with those from previous mapping studies of mAb 19b utilizing Ala-substituted peptides and various gp120s (Moore et al., 1995; Scott et al., 1990). We note that Arg304 and Lys305, identified here as important for 19b binding, have not been investigated in the previous Scott et al. or Moore et al. studies. We did not find the side chain of Thr320 to be important for 19b binding, in agreement with results from the Moore et al. study showing that residues with physicochemical properties different from threonine can occupy position 320 in phage-displayed peptides selected with mAb 19b (Moore et al., 1995). A recent study by Patel et al. proposed Ile307, His308, Gly312, Pro313, and Ala316 as important for mAb 19b binding, based on a peptide sequence selected with 19b from a peptide phage display library (Patel, Hoffman, and Swanstrom, 2008). However, this sequence was only 1 of 10 different sequences selected and, thus, may not have been representative of the 19b epitope. We also found that substituting Asp325 with Ala increased the binding affinity of mAb 19b by 2-fold (Table 2). Asp325 forms a salt bridge with Arg419 located within the coreceptor binding site on the gp120 core (Huang et al., 2005). This interaction may restrict the structural flexibility of the C-terminal stem of the V3 loop; introducing the Ala may prevent formation of this salt bridge, thus, allowing for a better formation or presentation of the 19b epitope.
The apparent binding affinity of mAb 39F, unlike all the other mAbs, was diminished solely by substitutions at the juncture of the V3 stem and tip but not by substituting residues located central to the V3 tip, such as Pro313 or Arg315 (Table 2). Specifically, binding affinity was strongly diminished by replacing Lys305 or Ile307 with Ala, and moderately decreased upon substitution of Ser306 and Ile309, suggesting that this mAb interacts principally with the N-terminal flank of the V3 loop. This notion is supported further by the observation that substitution of Arg298 and Arg304 also diminished binding, although not substantially (Table 2).
The binding of mAbs CO11, F2A3, LA21, and LE311 was diminished by many of the same substitutions (Table 2). The apparent affinities of all four mAbs were diminished upon substitution of His308 and binding of mAbs CO11, LA21 and LE311 was substantially reduced upon substitution of Pro313. The extremely strong reduction in the apparent binding affinity of mAbs CO11 and LA21 for mutant P313A (500- and 250-fold, respectively; Table 2) suggests that interaction with the aliphatic side chain of the Pro and/or a precise conformation of the V3 β-hairpin turn may be critical for efficient binding of these two mAbs. The apparent binding affinities of the two other mAbs, F2A3 and LE311, for mutant K305A were strongly diminished (>10-fold) relative to wild-type binding. This observation suggests that the epitopes of these two antibodies may overlap mostly with the N-terminal flank of the V3 region. It should be remarked that the residues identified here as important for binding of mAb CO11 (His308, Pro313, and Arg315) are consistent with the sequence proposed recently in a Patel et al. study as the (core) epitope of mAb CO11 (Patel, Hoffman, and Swanstrom, 2008); contrary to the above-noted low frequency of the proposed 19b epitope sequence, the CO11 sequence was identified in 11 of 20 phage-displayed peptide sequences selected with mAb CO11.
As noted above, the binding of mAb F2A3 was substantially enhanced by several substitutions: I309A, F317A, Y318A and D325A (Table 2). We posit that the influence of Asp325 is the same as postulated above for mAb 19b. Based on the projection of the mapping results onto the gp120+V3 structure (see below), we posit that the other 3 residues affect the precise alignment of critical residues required for the interaction with F2A3; as with Asp325, their interaction with neighboring residues likely affects how well the F2A3 epitope is presented.
The effects of the Ala-substitutions on mAb F530 binding were very similar to the previous four mAbs (Table 2), which likely explains its ability to neutralize the same viruses as the other 4 mAbs (cf. Table 1). However, unlike the other 4 mAbs, the apparent binding affinity of mAb F530 was not reduced as substantially by the His308→Ala change.
V3 mAb fine specificity related to neutralizing activity
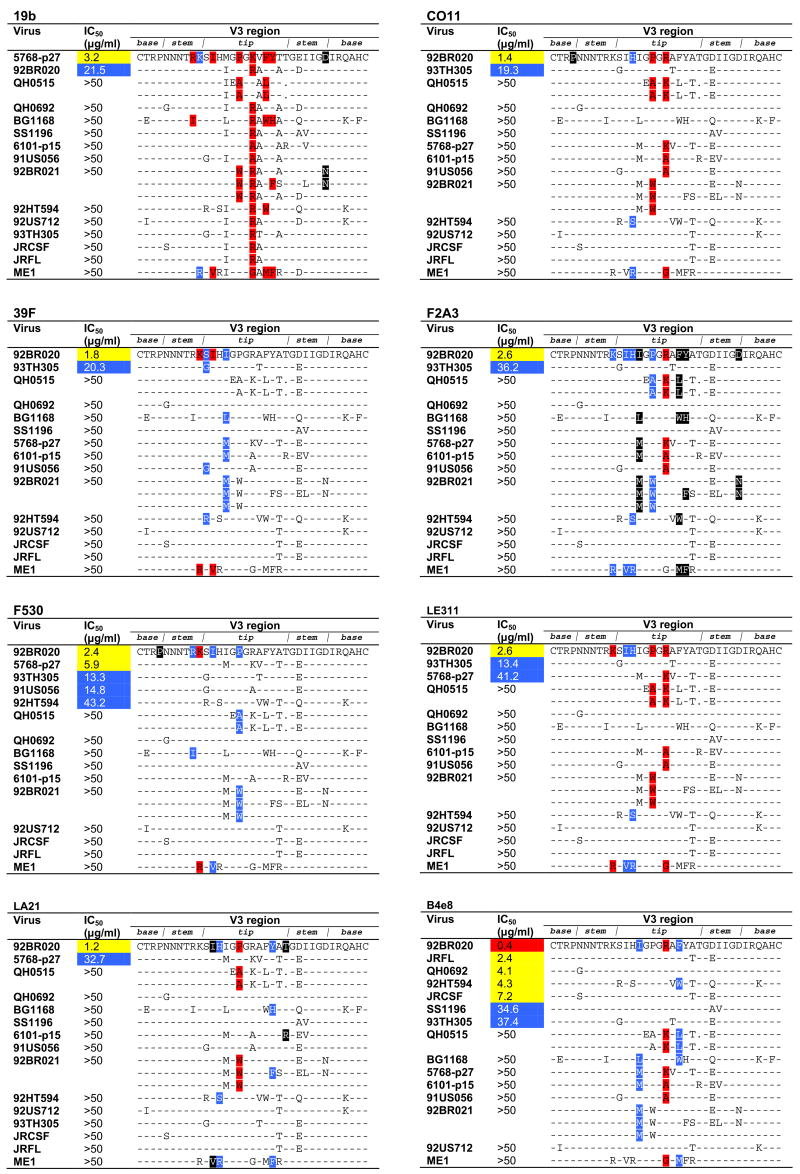
We ranked, for each mAb, the viruses that were most potently neutralized followed by those viruses that were not neutralized, to determine which residues are permissible (Fig. 2). Because some of the viruses used are from quasispecies pools, we listed all variant V3 sequences reported for a given virus; in our panel, this applied to viruses QH0515 and 92BR021.
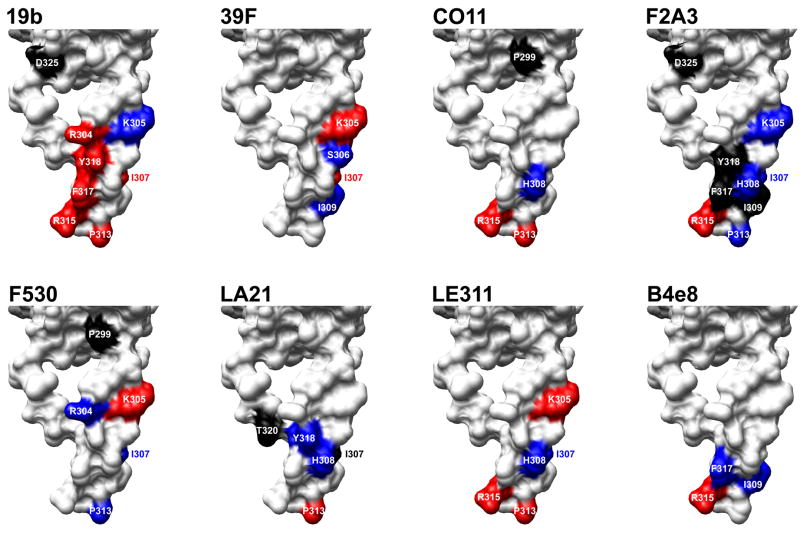
Fig. 2.
Effects of alanine substitutions on antibody binding mapped onto the structure of V3 in the context of gp120. Substitutions that significantly diminished or increased binding of the V3 mAbs tested here (Table 2) were mapped onto the structure of gp120JR-FL-core with V3 attached (Huang et al., 2005); only the V3 structure is depicted (surface rendering). The color scheme is the same as in Table 2. Molecular graphics images were produced using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera), then compiled and labeled in Adobe Photoshop.
Only mAb 19b neutralized virus 5768-p27 more potently than virus 92BR020 (Table 1; Fig. 1); although the residues identified as important for 19b binding are present in the V3 sequence of both viruses, a notable difference between them is the residue at position 309, which is a Met in virus 5768-p27 and an Ile in virus 92BR020. It has been shown previously that consensus peptide sequences selected with 19b from a phage-display library predominantly contain a Met at position 309 (Moore et al., 1995); thus, 19b seems to require an aliphatic side chain at position 309, with greater preference for Met. Based on the mapping, the inability of mAb 19b to neutralize the 6 viruses QH0515, BG1168, 6101-p15, 91US056, 92BR021, and ME1 could be explained by substitution of important contact residues (Fig. 1), whereas inability of the mAb to neutralize the other 6 viruses (QH0692, SS1196, 92US712, 93TH305, JR-CSF, and JR-FL) could not be explained by this.
Fig. 1.
Epitope specificity of V3 mAbs related to neutralizing activity. The V3 sequences of the 15 subtype B viruses investigated in this study are shown. For viruses QH0515 and 92BR021, multiple sequences are listed to represent the different V3 sequences that have been reported from quasispecies pools of these 2 viruses; for the other viruses for which quasispecies pools were used only a single sequence (shown) has been reported. To depict which residues are most permissible for each mAb, viruses are ranked according to their neutralization sensitivity; viruses not neutralized are ranked in the order as they appear in Table 1. The locations of those residues that substantially diminish or enhance mAb binding are colored as in Table 2; dashes indicate sequence homology with the virus that was neutralized most potently and, to maintain clarity, are not colored. Sequence gaps relative to the V3 sequence of the most potently neutralized virus are denoted by a dot. Sequence alignment was performed using ClustalW2 (http://www.ebi.ac.uk/clustalw/) (Larkin et al., 2007) and formatted for publication using the sequence alignment publishing tool SeqPublish (http://www.hiv.lanl.gov/content/hiv-db/SeqPublish/seqpublish.html).
For mAb 39F, the sequence of the two viruses neutralized (92BR020 and 93TH305) also correlated well with the mapping results (Fig. 1); the presence of Ser at position 306 (virus 92BR020) seems preferred over the smaller Gly (virus 93TH305). Of the 13 viruses resistant to neutralization by mAb 39F, the resistance of 6 viruses (BG1168, 5768-p27, 6101-p15, 92BR021, 92HT594, and ME1) could be explained by alterations of residues important for binding (Fig. 1), whereas neutralization of the 7 other viruses (QH0515, QH0692, SS1196, 91US056, 92US712, JR-CSF, and JR-FL) could not be accounted for by sequence incompatibility.
For the remaining 5 mAbs (CO11, F2A3, F530, LA21, and LE311) we observed also that the V3 sequence of some viruses could not account for neutralization resistance. Specifically, we observed that the V3 sequences of viruses QH0692, SS1196, 92US712, JR-CSF, and JR-FL consistently could not account for neutralization resistance (Fig. 1). It should be noted that all of these viruses are sensitive to neutralization by V3 mAbs B4e8, 447-52D, and/or 58.2 (Binley et al., 2004; Pantophlet et al., 2007), indicating that their V3 loop is not inaccessible to antibody. We observed also that the V3 sequence of other select viruses could not account for neutralization resistance of some mAbs tested here, for example virus BG1168 for mAbs CO11 and LE311, virus 91US056 for mAb LA21, and virus 6101-p15 for mAb F530.
MAb fine specificity mapped onto V3 in the structural context of gp120
To obtain a better understanding of the spatial orientation of the residues identified as important for V3 mAb binding, we projected the results of the mapping onto the structure of a soluble CD4-, Fab X5-complexed gp120 core with the V3 loop attached (Huang et al., 2005). We chose this structure because the conformation of the V3 stem is not substantially influenced by interactions with the antibody (Huang et al., 2005).
The results of the projected maps are shown in Fig. 1. For mAb 19b, we observed a distinct binding site formed by Arg304 in the N-terminal V3 stem and Arg315, Phe317, and Tyr318 in the C-terminal half of the V3 tip. Pro313, which was also important for binding, is somewhat distant from the distinctive formation of the other 4 residues, suggesting that the primarily role of this residue is to properly juxtapose the residues that form the 19b epitope, as suggested previously (Moore et al., 1995). The side chain of Arg315 is not critical for 19b binding, given that efficient binding is also observed to monomeric gp120JR-CSF with a Gln substitution at the same position (Pantophlet et al., unpublished observations). Thus, the presence of both the Pro and Arg residues is required to form the V3 tip hairpin turn and juxtapose the ‘true’ contact residues. Ile307 lies somewhat distant from the epitope map and, thus, it seems unlikely that this residue interacts with mAb 19b directly. However, based on the substantial decrease in 19b binding to I307A mutant, its presence seems crucial for proper presentation of the epitope. This notion is supported by previous results showing that mAb 19b prefers residues with aliphatic side chains at position 307 (Moore et al., 1995).
Residues important for the binding of mAb 39F –Lys305, Ser306, Ile307, and Ile309—also form a distinct binding site on the N-terminal flank of V3, along the length of the V3 structure (Fig. 2), supporting the notion that this antibody interacts with the N-terminal part of V3. Projection of the Ala-scanning results for mAbs CO11, F2A3, LA21, and LE311 onto the V3 structure showed that the maps for CO11 and LA21 share a higher similarity than the maps for mAbs F2A3 and LE311, which are similar to each other. The maps for CO11 and LA21 are somewhat similar to the map for B4e8, with the notable difference that B4e8 binding is not critically dependent on Pro313 (Fig. 2). Although the residues important for CO11 and LA21 binding form a somewhat disjointed pattern, these mAbs likely also contact neighboring residues. A similar disconnected pattern is observed with residues important for the binding of mAb B4e8 (Fig. 2), yet structural analysis later revealed that B4e8 forms less-critical interactions with neighboring residues (Bell et al., 2008).
As expected, the epitope maps for mAbs F2A3 and LE311 are similar to those of mAbs CO11 and LA21, most notably His308, Pro313 and Arg315 (Fig. 2). For both mAbs F2A3 and LE311 we observed that Lys305, Ile307/His308, and Pro313, all of which are important for mAb binding, form a nearly linear arrangement on the V3 structure. In the case of mAb F2A3, the location of residues that, upon substitution to alanine, increased antibody binding are crowded around the 4-residue linear arrangement, supporting the notion that residues at the adjacent positions play a substantial role in the spatial positioning of the contact residues.
For mAb F530 we obtained a map that resembled the maps obtained with mAbs CO11, F2A3, LA21, and LE311 (Fig. 2). However, the notable ability of mAb F530 to bind V3 without requiring the presence of Arg315 suggests that this antibody, like mAb 39F, interacts mostly with the N-terminal flank of the V3 loop.
DISCUSSION
Our results illustrate how the epitope fine specificity of antibodies to V3 may affect their level of cross-neutralizing activity. All 7 mAbs tested here neutralized very few viruses; common among all mAbs was the requirement for one or more positively-charged residues at the juncture of the N-terminal flank of the V3 stem with the V3 tip for efficient binding. The mAbs also depended, to varying extents, on the presence of the Pro at the V3 tip for efficient binding. We posit that the requirement for interaction with certain residues near the V3 stem and in the V3 tip angles the mAbs such that they are restricted from interacting with their cognate epitope on many viruses, exemplified here by viruses QH0692, SS1196, 92US712, JR-CSF, and JR-FL. These 5 viruses do not have alterations in residues identified by our scanning mutagenesis approach as being important for binding but nevertheless were not neutralized by the 7 mAbs that were studied here. However, we know that the V3 loops on these viruses are not completely inaccessible to antibody, given that these viruses are sensitive to neutralizing by V3 antibodies such as B4e8 and 447-52D.
The factor(s) responsible for restricting V3 access of the 7 mAbs tested here remain to be identified. Recent studies using chimeric viruses have shown that the V1/V2 loop on some primary viruses, e.g. JR-FL an YU2, may greatly limit antibody recognition of V3 (Krachmarov et al., 2005; Krachmarov et al., 2006; Pinter et al., 2004). It is therefore tempting to speculate that V1/V2 may also be limiting V3 access on the other viruses tested here but this will need to be determined experimentally; for example, it is conceivable that specific residues outside of V3 (and V1/V2) affect the presentation of V3 antibody epitopes. It should be noted that we did not investigate further the relative importance of residues identified as critical for antibody binding in the context of different V3 sequences. It is possible that the contribution of each residue to binding affinity may vary in the context of V3 sequences from different viruses. More detailed studies, for example, by investigating changes in the sensitivity of neutralization-resistant viruses used here in which important antibody binding residues in V3 are introduced may help to further elucidate the relationship between sequence variability and neutralization sensitivity.
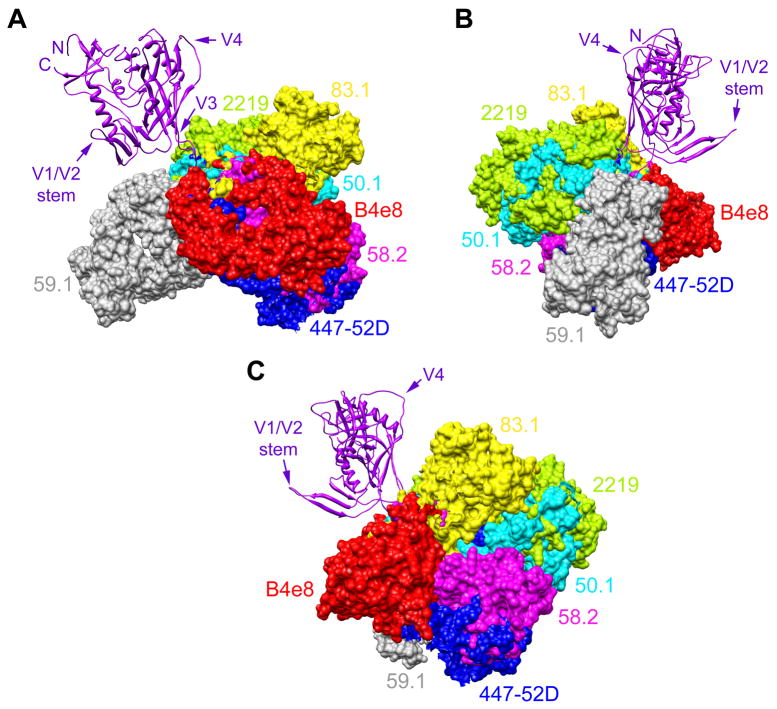
Only structural information can provide accurate insight into how the 7 mAbs tested here interact with V3. Nevertheless, even in the absence of such information, we believe structural information from other V3 mAbs for which important binding residues have been determined supports this hypothesis. For example, mAbs 59.1 (epitope: 306RIHIGPGRAFYTT320), 50.1 (epitope: 305KRIHIGP313), 83.1 (epitope: 306RIHIGPGR315), and 58.2 (epitope: 306RIHIGPGRAFY318) bind overlapping epitopes on V3 (Seligman et al., 1996) and require many of the same residues for efficient binding (important contact residues are underlined; critical residues are double underlined (White-Scharf et al., 1993)). Nonetheless, these 4 mAbs interact from markedly different angles with V3 (Fig. 3). MAbs 59.1, 50.1, and 83.1, which only neutralize viruses that are generally sensitive to antibody neutralization (Bou-Habib et al., 1994), seem to require a particular β hairpin turn conformation at the center of the V3 tip (GPGR) for binding, as judged by the substantial reduction in binding efficiency upon substituting one or both glycine residues. This hairpin turn requirement mirrors the strong dependence of mAbs 19b, LE311, CO11, and LA21 on the presence of the proline in the GPGR turn for binding, presumably to juxtapose important contact residues. Thus, mAbs 19b, LE311, CO11, and LA21 may need to interact with V3 from angles, perhaps similar to mAbs 59.1, 50.1, and/or 83.1, which do not permit access to V3 on many different primary viruses. MAb 58.2, which neutralizes more primary viruses than mAbs 19b, LE311, CO11, or LA21 (Table 1; (Binley et al., 2004)), seems less dependent on the precision of the turn given that its binding is only substantially diminished upon substituting Arg315 (White-Scharf et al., 1993), and interacts with V3 from an angle that is notably different from mAbs 59.1, 50.1, and 83.1 (Fig. 3).
Fig. 3.
Angle of interaction between V3 antibodies and V3. Structure of gp120JR-FL-core containing V3 (Huang et al., 2005) (purple; ribbon representation) shown from 3 perspectives with the Fab fragments of 7 different mAbs (surface rendering) superimposed to depict their different angles of interaction with V3. (A) Gp120 from the perspective of CD4; (B) Gp120 from the perspective of gp41 and the gp120:gp41 interface; (C) Gp120 from the approximate perspective of a coreceptor molecule (e.g. CCR5). To establish the angle for each mAb, the V3 peptide (not shown for clarity purposes) from each mAb:peptide complex structure was superimposed onto the V3 structure of gp120 using the MatchMaker feature in UCSF Chimera, which constructs a sequence alignment and then performs a least-squares fit to superimpose the aligned residue pairs; the sequence alignment is based on a combination of residue identity/similarity and secondary structure correspondence. The V3 mAbs shown are: B4e8 (red), 447-52D (blue), 58.2 (magenta), 59.1 (grey), 50.1 (cyan), 83.1 (yellow), and 2219 (green). Aside from V3, additional regions of gp120 (N- and C termini, V1/V2 stem, and V4) are denoted to orientate the reader.
The association of low cross-neutralizing activity with binding to the residues in or near the V3 stem segment is supported by other observations. For example, it has been shown that isolate-specific V3 responses in subtype B- and D-infected individuals are directed to residues located in the stem segments of the V3 loop (Lawoko et al., 2000; Schreiber et al., 1997) and weakly neutralizing serum antibody responses to V3 in individuals immunized with recombinant gp160 are preferentially directed to the N-terminal stem of V3 (Coeffier et al., 1997). Furthermore, in a recent study seeking to elicit 447-52D-like NAbs by utilizing fusion proteins containing the 447-52D contact epitope 305KSIHIGPGRAFYTT320 (Chakraborty et al., 2006), immune sera seemed to require for binding the N-terminal segment of the engrafted motif and neutralized the generally-sensitive virus MN but not the more resistant viruses BaL or JR-FL.
Fig. 3 shows that mAbs 58.2, 447-52D, and B4e8, which all neutralize several primary viruses (Binley et al., 2004; Pantophlet et al., 2007), interact with V3 within nearly the same plane of orientation; antibodies interacting with V3 from other angles tend to exhibit lower cross-neutralizing activity (Bou-Habib et al., 1994; Gorny et al., 2004; York et al., 2001). The importance of the angle of antibody interaction for effective neutralization of HIV is exemplified by insight from recent crystal structures of the broadly neutralizing mAbs 2F5, 4E10, and b12 in complex with antigen (Cardoso et al., 2005; Zhou et al., 2007), which suggest that such antibodies need to interact in with their target sites from specific angles to gain access to their epitopes on infectious virions; the difficulty in eliciting 2F5-, 4E10-, and b12-like antibodies supports the notion that these sites are not easily accessed. Our results illustrate how similar restrictive factors might apply to V3 antibodies. MAb 58.2 exhibits the lowest cross-neutralizing activity among the 3 mAbs that neutralize different primary viruses, despite interacting with V3 from nearly an identical angle as mAb 447-52D. This difference in neutralizing activity may be due to mAb 447-52D forming main-chain contacts with residues in the V3 stem region, which has been suggested to enhance its ability to neutralize a notable fraction of viral isolates (Stanfield et al., 2004). MAb B4e8, which seems to neutralize slightly better than mAb 447-52D (Keele et al., 2008; Pantophlet et al., 2007), interacts with V3 from a slightly elevated angle relative to the 2 other mAbs.
In sum, we have presented here an extensive analysis in an attempt to better understand how epitope fine specificity affects the neutralizing activity of V3 antibodies. Previous studies have shown that V3 may be shielded by the V1/V2 region (Cao et al., 1997; Krachmarov et al., 2006; Pinter et al., 2004; Wyatt et al., 1993), but such studies did not provide a clear rationale for the spectrum of sensitivities exhibited by a given primary isolate to the neutralizing activity of different V3 mAbs. Antibodies that interact with V3 from certain angles may indeed be greatly restricted from accessing their epitope on several viruses, depending on the stringency of the steric constraints. However, certain parts of V3 may still be accessible from select angles. The V3 tip in particular seems somewhat better accessible on different viruses, as judged from the appreciable neutralizing activity exhibited by mAb B4e8 (Pantophlet et al., 2007). The apparently unique binding mode of mAb B4e8, and potentially those of other V3 mAbs (Gorny et al., 2006), may provide an avenue for further investigations into V3 tip accessibility on different viruses, for example by engineering variants with specificities for V3 sequences not recognized by the parental antibody.
MATERIALS AND METHODS
Antibodies
Seven human mAbs directed to the V3 loop on HIV-1 gp120 were studied here: 19b, 39F, CO11, F2A3, F530, LA21, and LE311. MAb 19b was isolated from an asymptomatic individual infected with a clade B virus by Ebstein-Barr virus (EBV) transformation of peripheral blood lymphocytes and was shown to be specific for the V3 loop by mapping with peptides (Scott et al., 1990). MAbs 39F and F2A3 were isolated from two subtype B-infected long-term non-progressors, and mAbs CO11, LA21 and LE311 were isolated from three different patients (AC-01, AC-05, and AC-033, respectively) on anti-retroviral therapy during acute infection and who then underwent several periods of structured treatment interruptions (Montefiori et al., 2001; Rosenberg et al., 2000). These mAbs were selected as described elsewhere (Xiang et al., 2003) and have been defined as being directed to V3 based on their reactivity with select clade B V3 peptides (J. Robinson, unpublished data). MAb F530 was isolated from an HIV-1 infected patient by direct fusion of mononuclear splenocytes with HMMA human myeloma cells (Posner et al., 1989; Posner, Elboim, and Santos, 1987). Hybridomas were selected based on specific reactivity hybridoma supernatants with HIV-infected cells and ability of the antibodies to capture a relatively high degree of virions in a virus capture assay (L. Cavacini, unpublished results). The general specificity of mAb F530 for the V3 loop was determined based on its binding profile with variable-loop deleted gp120 mutants (L. Cavacini, unpublished results). Most mAbs described above have been utilized in previous antigen binding studies and were selected for study here based on their overall broad reactivity with envelope-based preparations from different clade B viruses and consensus sequences (Grundner et al., 2002; Haynes et al., 2006; Koch et al., 2003; Liao et al., 2006; Moore et al., 1994; Pantophlet, Wilson, and Burton, 2004; Patel, Hoffman, and Swanstrom, 2008; Selvarajah et al., 2005; Yang et al., 2002).
Viruses
Pseudovirus preps were produced by co-transfection of HEK293 cells with an env-expressing plasmid (pCXAS) and a subgenomic plasmid (pHIVlucΔU3) as described previously (Binley et al., 2004). The test viruses (n=15; Table 1) were selected from the 30-member panel of subtype B primary viruses used in a previous study by Binley et al. (Binley et al., 2004). Eight of the test viruses were quasispecies derived from primary virus cultures and are as follows: QH0515 (Binley et al., 2004; Cleghorn et al., 2000; Hu et al., 2000; Li et al., 2005), QH0692 (Binley et al., 2004; Cleghorn et al., 2000; Li et al., 2005), 5768-p27 (Binley et al., 2004; Li et al., 2005), 6101-p15 (Binley et al., 2004; Li et al., 2005), 91US056 (Binley et al., 2004; Trkola et al., 1995), 92BR021 (Binley et al., 2004; Gao et al., 1994; Torre et al., 2000), 93TH305 (Binley et al., 2004), and ME1 (Binley et al., 2004; Chen et al., 1997); the noted references are for env sequences belonging to these viruses that have been reported in the Binley et al. study and elsewhere. The remaining 7 viruses were molecular clones. Three of these molecular clones (92HT594, JR-CSF, and JR-FL) have been described previously (Binley et al., 2004), whereas the remaining four clones (BG1168, SS1196, 92BR020, 92US712) have not been reported previously; each molecular clone was selected from a panel of 6 clones derived from the respective viral quasispecies pools reported previously (Binley et al., 2004). Each clone was chosen based on the overall similarity between its sensitivity to neutralization by the broadly neutralizing mAbs b12, 2G12, 2F5, and 4E10, and a broadly neutralizing HIV+ serum and the neutralization sensitivity profiles of the corresponding viral quasispecies to the same inhibitors (T. Wrin et al., unpublished results).
Neutralization assays
Neutralization assays were performed at Monogram Biosciences using their high-throughput neutralization assay with U87 target cells expressing CD4, CCR5, and CXCR4 and pseudotyped viruses (Richman et al., 2003; Schweighardt et al., 2007). Assay conditions were the same as described previously (Binley et al., 2004); serial dilutions of mAb, starting at 50 μg/ml, were incubated for 1 hour with virus after which the mixture was added to target cells.
Generation of V3 mutants
The V3 mutants, generated in the JR-CSF background, were the same as described recently for mapping the epitope specificity of mAb B4e8 (Pantophlet et al., 2007). All mutations were verified by DNA sequencing.
Epitope mapping by ELISA
Binding assays to determine apparent antibody binding affinities were performed using viral lysates of supernatants collected from transiently-transfected 293T cells as described (Pantophlet et al., 2007; Pantophlet et al., 2003). The mAbs were added to the ELISA plate wells in 5-fold serial dilutions and binding was detected with a peroxidase-conjugated secondary antibody and TMB substrate (Pierce). Apparent binding affinities (Kapp) were defined as the antibody concentrations at half-maximal binding; percentage changes in affinity relative to wild-type gp120 were calculated as: [Kappwild-type/Kappmutant]×100%.
Acknowledgments
We thank Rowena Aguilar-Sino for technical assistance and Susan-Zolla Pazner for critiquing early drafts of this manuscript. Molecular graphics images were produced using the Chimera package from the Resource for Biocomputing, Visualization, and Informatics at UCSF (NIH grant P41 RR-01081). This study was supported by the International AIDS Vaccine Initiative through the Neutralizing Antibody Consortium and NIH grant AI33292 (to D.R.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beaumont T, van Nuenen A, Broersen S, Blattner WA, Lukashov VV, Schuitemaker H. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J Virol. 2001;75(5):2246–52. doi: 10.1128/JVI.75.5.2246-2252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CH, Pantophlet R, Schiefner A, Cavacini LA, Stanfield RL, Burton DR, Wilson IA. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J Mol Biol. 2008;375(4):969–978. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, Norcross MA. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68(9):6006–13. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Burton DR, Montefiori DC. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl A):S87–98. [PubMed] [Google Scholar]
- Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71(12):9808–12. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22(2):163–73. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Durani V, Miranda ER, Citron M, Liang X, Schleif W, Joyce JG, Varadarajan R. Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies. Biochem J. 2006;399(3):483–91. doi: 10.1042/BJ20060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Singh MK, Balachandran R, Gupta P. Isolation and characterization of two divergent infectious molecular clones of HIV type 1 longitudinally obtained from a seropositive patient by a progressive amplification procedure. AIDS Res Hum Retroviruses. 1997;13(9):743–50. doi: 10.1089/aid.1997.13.743. [DOI] [PubMed] [Google Scholar]
- Cleghorn FR, Jack N, Carr JK, Edwards J, Mahabir B, Sill A, McDanal CB, Connolly SM, Goodman D, Bennetts RQ, O’Brien TR, Weinhold KJ, Bartholomew C, Blattner WA, Greenberg ML. A distinctive clade B HIV type 1 is heterosexually transmitted in Trinidad and Tobago. Proc Natl Acad Sci U S A. 2000;97(19):10532–7. doi: 10.1073/pnas.97.19.10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coeffier E, Excler JL, Kieny MP, Meignier B, Moste C, Tartaglia J, Pialoux G, Salmon-Ceron D, Leclerc C. Restricted specificity of anti-V3 antibodies induced in humans by HIV candidate vaccines. AIDS Res Hum Retroviruses. 1997;13(17):1471–85. doi: 10.1089/aid.1997.13.1471. [DOI] [PubMed] [Google Scholar]
- Eda Y, Murakami T, Ami Y, Nakasone T, Takizawa M, Someya K, Kaizu M, Izumi Y, Yoshino N, Matsushita S, Higuchi H, Matsui H, Shinohara K, Takeuchi H, Koyanagi Y, Yamamoto N, Honda M. Anti-V3 humanized antibody KD-247 effectively suppresses ex vivo generation of human immunodeficiency virus type 1 and affords sterile protection of monkeys against a heterologous simian/human immunodeficiency virus infection. J Virol. 2006;80(11):5563–70. doi: 10.1128/JVI.02095-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A. 2005;102(51):18514–9. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RC. The end or the beginning of the drive to an HIV-preventive vaccine: a view from over 20 years. Lancet. 2005;366(9500):1894–8. doi: 10.1016/S0140-6736(05)67395-3. [DOI] [PubMed] [Google Scholar]
- Gao F, Yue L, Craig S, Thornton CL, Robertson DL, McCutchan FE, Bradac JA, Sharp PM, Hahn BH. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retroviruses. 1994;10(11):1359–68. doi: 10.1089/aid.1994.10.1359. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J Virol. 2004;78(5):2394–404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76(18):9035–45. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006;80(14):6865–72. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram GJ, Hemming A, Bolmstedt A, Jansson B, Olofsson S, Akerblom L, Nielsen JO, Hansen JE. Identification of an N-linked glycan in the V1-loop of HIV-1 gp120 influencing neutralization by anti-V3 antibodies and soluble CD4. Arch Virol. 1994;139(3–4):253–61. doi: 10.1007/BF01310789. [DOI] [PubMed] [Google Scholar]
- Grundner C, Mirzabekov T, Sodroski J, Wyatt R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J Virol. 2002;76(7):3511–21. doi: 10.1128/JVI.76.7.3511-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS Res Hum Retroviruses. 2005;21(2):171–89. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Ma B, Montefiori DC, Wrin T, Petropoulos CJ, Sutherland LL, Scearce RM, Denton C, Xia SM, Korber BT, Liao HX. Analysis of HIV-1 subtype B third variable region peptide motifs for induction of neutralizing antibodies against HIV-1 primary isolates. Virology. 2006;345(1):44–55. doi: 10.1016/j.virol.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5(4):579–95. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- Hu QX, Barry AP, Wang ZX, Connolly SM, Peiper SC, Greenberg ML. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J Virol. 2000;74(24):11858–72. doi: 10.1128/jvi.74.24.11858-11872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SL, Stamatatos L. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr HIV Res. 2007;5(6):507–13. doi: 10.2174/157016207782418542. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Hedestam GB, Fouchier RAM, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Micro. 2008;6(2):143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PM, Arnold BA, Shaw AR, Tolman RL, Van Middlesworth F, Bondy S, Rusiecki VK, Koenig S, Zolla-Pazner S, Conard P, et al. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology. 1993;193(2):709–16. doi: 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313(2):387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79(2):780–90. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–35. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer VG, Siddappa NB, Ruprecht RM. Passive immunization as tool to identify protective HIV-1 Env epitopes. Curr HIV Res. 2007;5(6):642–55. doi: 10.2174/157016207782418506. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lawoko A, Johansson B, Ljunggren J, Fries A, Fredriksson R, Georgievska L, Malmvall B, Pipkorn R, Fenyo EM, Blomberg J. Structural prerequisites for intersubtype B and D antigenicity of the third variable envelope region (V3) of human immunodeficiency virus type 1. J Infect Dis. 2000;182(1):49–58. doi: 10.1086/315671. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, Ma BJ, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353(2):268–82. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Hill TS, Vo HT, Walker BD, Rosenberg ES. Neutralizing antibodies associated with viremia control in a subset of individuals after treatment of acute human immunodeficiency virus type 1 infection. J Virol. 2001;75(21):10200–7. doi: 10.1128/JVI.75.21.10200-10207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McCutchan FE, Poon SW, Mascola J, Liu J, Cao Y, Ho DD. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68(12):8350–64. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Trkola A, Korber B, Boots LJ, Kessler JA, 2nd, McCutchan FE, Mascola J, Ho DD, Robinson J, Conley AJ. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69(1):122–30. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Aguilar-Sino RO, Wrin T, Cavacini LA, Burton DR. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and reassessment of its epitope fine specificity by scanning mutagenesis. Virology. 2007;364(2):441–53. doi: 10.1016/j.virol.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77(1):642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Wilson IA, Burton DR. Improved design of an antigen with enhanced specificity for the broadly HIV-neutralizing antibody b12. Protein Eng Des Sel. 2004;17(10):749–58. doi: 10.1093/protein/gzh085. [DOI] [PubMed] [Google Scholar]
- Patel MB, Hoffman NG, Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J Virol. 2008;82(2):903–916. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phogat S, Wyatt R. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr Pharm Des. 2007;13(2):213–27. doi: 10.2174/138161207779313632. [DOI] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–15. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner MR, Barrach HJ, Elboim HS, Nivens K, Santos DJ, Chichester CO, Lally EV. The generation of hybridomas secreting human monoclonal antibodies reactive with type II collagen. Hybridoma. 1989;8(2):187–97. doi: 10.1089/hyb.1989.8.187. [DOI] [PubMed] [Google Scholar]
- Posner MR, Elboim H, Santos D. The construction and use of a human-mouse myeloma analogue suitable for the routine production of hybridomas secreting human monoclonal antibodies. Hybridoma. 1987;6(6):611–25. doi: 10.1089/hyb.1987.6.611. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100(7):4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg ES, Altfeld M, Poon SH, Phillips MN, Wilkes BM, Eldridge RL, Robbins GK, D’Aquila RT, Goulder PJ, Walker BD. Immune control of HIV-1 after early treatment of acute infection. Nature. 2000;407(6803):523–6. doi: 10.1038/35035103. [DOI] [PubMed] [Google Scholar]
- Schonning K, Jansson B, Olofsson S, Nielsen JO, Hansen JS. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology. 1996;218(1):134–40. doi: 10.1006/viro.1996.0173. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Wachsmuth C, Muller H, Odemuyiwa S, Schmitz H, Meyer S, Meyer B, Schneider-Mergener J. The V3-directed immune response in natural human immunodeficiency virus type 1 infection is predominantly directed against a variable, discontinuous epitope presented by the gp120 V3 domain. J Virol. 1997;71(12):9198–205. doi: 10.1128/jvi.71.12.9198-9205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighardt B, Liu Y, Huang W, Chappey C, Lie YS, Petropoulos CJ, Wrin T. Development of an HIV-1 reference panel of subtype B envelope clones isolated from the plasma of recently infected individuals. J Acquir Immune Defic Syndr. 2007;46(1):1–11. doi: 10.1097/QAI.0b013e318074eb5a. [DOI] [PubMed] [Google Scholar]
- Scott CF, Jr, Silver S, Profy AT, Putney SD, Langlois A, Weinhold K, Robinson JE. Human monoclonal antibody that recognizes the V3 region of human immunodeficiency virus gp120 and neutralizes the human T-lymphotropic virus type IIIMN strain. Proc Natl Acad Sci U S A. 1990;87(21):8597–601. doi: 10.1073/pnas.87.21.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman SJ, Binley JM, Gorny MK, Burton DR, Zolla-Pazner S, Sokolowski KA. Characterization by serial deletion competition ELISAs of HIV-1 V3 loop epitopes recognized by monoclonal antibodies. Mol Immunol. 1996;33(9):737–45. doi: 10.1016/0161-5890(96)00044-2. [DOI] [PubMed] [Google Scholar]
- Selvarajah S, Puffer B, Pantophlet R, Law M, Doms RW, Burton DR. Comparing antigenicity and immunogenicity of engineered gp120. J Virol. 2005;79(19):12148–63. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure (Camb) 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Torre VS, Marozsan AJ, Albright JL, Collins KR, Hartley O, Offord RE, Quinones-Mateu ME, Arts EJ. Variable sensitivity of CCR5-tropic human immunodeficiency virus type 1 isolates to inhibition by RANTES analogs. J Virol. 2000;74(10):4868–76. doi: 10.1128/jvi.74.10.4868-4876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, 3rd, Burton DR, Ho DD, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69(11):6609–17. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- White-Scharf ME, Potts BJ, Smith LM, Sokolowski KA, Rusche JR, Silver S. Broadly neutralizing monoclonal antibodies to the V3 region of HIV-1 can be elicited by peptide immunization. Virology. 1993;192(1):197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67(8):4557–65. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology. 2003;315(1):124–34. doi: 10.1016/s0042-6822(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York J, Follis KE, Trahey M, Nyambi PN, Zolla-Pazner S, Nunberg JH. Antibody Binding and Neutralization of Primary and T-Cell Line-Adapted Isolates of Human Immunodeficiency Virus Type 1. J Virol. 2001;75(6):2741–2752. doi: 10.1128/JVI.75.6.2741-2752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4(3):199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies. 2005;14(3–4):69–72. [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–8. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]