Abstract
The radiation chemical yields of unaltered base release have been measured in three crystalline double-stranded DNA oligomers after X irradiation at 4 K. The yields of released bases are between 10 and 20% of the total free radical yields measured at 4 K. Using these numbers, we estimate that the yield of DNA strand breaks due to the direct effect is about 0.1 μmol J−1. The damage responsible for base release is independent of the base type (C, G, A or T) and is not scavenged by anthracycline drugs intercalated in the DNA. For these reasons, reactions initiated by the hydroxyl radical have been ruled out as the source of base release. Since the intercalated anthracycline scavenges electrons and holes completely but does not inhibit base release, the possibility for damage transfer from the bases to the sugars can also be ruled out. The results are consistent with a model in which primary radical cations formed directly on the sugar-phosphate backbone react by two competing pathways: deprotonation, which localizes the damage on the sugar, and hole tunneling, which transfers the damage to the base stack. Quantitative estimates indicate that these two processes are approximately equally efficient.
INTRODUCTION
The coexistence of two different pathways of radiation damage to DNA, one indirect and the other direct, has long been recognized. The indirect pathway primarily involves the reaction of DNA with water radiolysis products (1), while the direct pathway originates primarily from the direct ionization of DNA. In dilute aqueous solutions of DNA, the contribution from the direct effect is small, while in cells both effects play a comparable role due to condensation of the DNA and the high hydroxyl radical scavenging capacity of the medium surrounding DNA (1, 2). The distinction between direct and indirect becomes more difficult to define when considering the role of the waters of hydration and the origin of the excess electrons that end up as electron-gain centers in DNA.
In general, the indirect effect is much better characterized, both quantitatively and mechanistically, than its direct counterpart. Most studies on the direct effect have been done using electron paramagnetic resonance (EPR) spectroscopy on amorphous DNA samples hydrated to various levels. A number of reviews have been published on these applications (3-5). Fewer attempts have been made to analyze the stable chemical products of the direct damage (6-8).
One of the key unresolved issues is the mechanism responsible for DNA strand breaks through the direct effect and its relative contribution to the overall damage caused by this pathway. It is well known that electron attachment to DNA in solution is not an appreciable source of DNA strand breaks (1). Recently it was shown by various techniques that one-electron oxidation of guanine is not an efficient source of immediate DNA strand breaks (9-11). However, the numerous EPR studies of the direct effect referenced above have identified the one-electron reduced pyrimidines (mostly cytosine) and one-electron oxidized guanine as the major sites of initial damage. If these are not efficient precursors for strand breaks, then what is?
We approach this problem using crystalline DNA oligomers as the target and unaltered base release as an approximate assay for the total free radical damage to the sugar-phosphate backbone. Base release has been shown to correlate closely with DNA strand breaks after an appropriate postirradiation heat treatment (12). One advantage of using crystalline rather than amorphous DNA samples is that the structure and packing of DNA in these crystals is known precisely from crystallographic studies. The second advantage is that the microscopic homogeneity of the crystals ensures a better quantitative reproducibility. This advantage has been clearly demonstrated in the previous studies of free radical yields (13, 14).
EXPERIMENTAL
The protocols used in crystallization of DNA oligomers, sample preparations, irradiation and free radical yield measurements have been described. Briefly, most samples consist of a small group of crystals, but in a few cases one single crystal provided sufficient mass. The X-ray source was a Varian/Eimac OEG-76 tube with tungsten target operating at 70 kV and 20 mA. Samples were irradiated at 4 K at a dose rate of 26 kGy/h. Some of the crystals were available from the previous studies (13, 14).
The following abbreviations are used throughout in this paper: DNA10 for a self-complementary d(GCACGCGTGC) oligomer crystallized in the presence of Co(NH3)63+, DNA6 for the d(CACGCG·GTGCGC) sequence, crystallized as a magnesium salt, and DNA6·2D and DNA6·2A for the intercalation complexes of the self-complementary d(CGATCG) hexamer with daunomycin and adriamycin, respectively. Cobalt appears to incorporate in the DNA10 lattice as an impurity (the degree is variable), and it does not significantly influence the free radical distribution at 4 K (13). Both intercalation complexes, DNA6·2D and DNA6·2A, have effectively the same crystal structures. The latter two were a gift from the laboratory of Loren Williams at the Georgia Institute of Technology. The crystal structures for all three sequences are available from the literature: DNA10 (15), DNA6 (16), DNA6·2D (17-19) and DNA6·2A (18, 19). All of the samples were warmed to room temperature after the 4 K irradiation and subsequently stored at ~−°C. It is known from EPR measurements (Debije, personal communication) that warming to room temperature reduces the free radical concentration by an order of magnitude or more. For example, the ratio of radicals lost to radicals at 4 K is ~96% and ~100% in DNA10 and DNA6·2D, respectively.
For the base release assay, the irradiated crystals were dissolved in a 20 mM phosphate buffer (pH 6.8; 50 μl per 100 μg of the crystals) containing 1 μM of 1-methylcytosine employed as an internal standard. The solutions were heated at 80°C for 15 min, chilled and analyzed by HPLC using a Phenomenex C18 4.6 mm × 250 mm × 5-μm column washed with 25 mM ammonium acetate at a flow rate of 1 ml min−1 and applying a 10% linear acetonitrile gradient over 20 min. The bases were detected and quantified by their absorbance by their absorbance at 254 nm.
RESULTS
Dose–response curves have been obtained for the release of all four DNA bases. A representative data set for the DNA10 crystals is shown in Fig. 1. The dose dependencies for the total release calculated from these and similar data for other sequences are shown in Fig. 2. The linear portion of the dose–response curves, corresponding to doses below 20 kGy, was used to calculate the radiation chemical yields of the free bases reported in Table 1. Figure 2 also includes a few data points available for the DNA6·2D and DNA6·2A crystals and the DNA10 crystals partially dehydrated over P2O5 where 10–20% of the total water was removed as measured by weight.
FIG. 1.
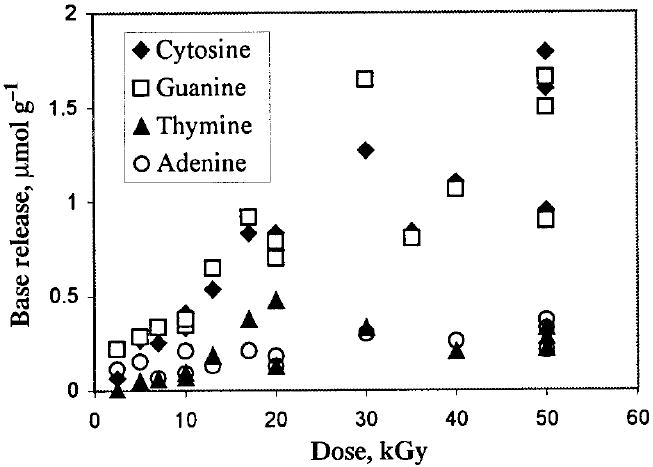
The release of individual DNA bases from irradiated DNA10 crystals as a function of radiation dose. For irradiation conditions and the postirradiation treatment, see the text.
FIG. 2.
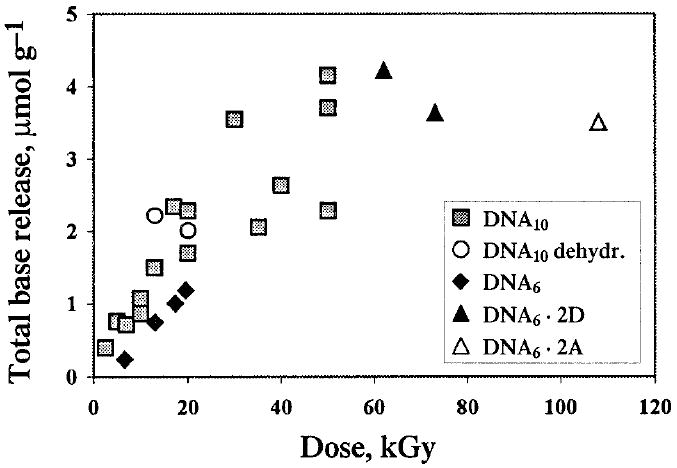
Total base release from five crystalline DNA samples as a function of radiation dose. “DNA10 dehydr.” stands for the DNA10 crystals dehydrated over P2O5; up to 10–15% of the total water is removed. Other abbreviations are explained in the text.
TABLE 1.
Radiation Chemical Yields of Unaltered Base Release in Crystalline Compared to Amorphous Hydrated DNA (7)
| Yield (μmol J−1)
|
|||
|---|---|---|---|
| Base | DNA10 | DNA6 | Hydrated DNAb |
| Cytosine | 0.041 (0.002)a | 0.020 (0.001) | 0.015 (0.001) |
| Guanine | 0.043 (0.003) | 0.030 (0.002) | 0.0094 (0.0006) |
| Thymine | 0.015 (0.003) | 0.003 (0.001) | 0.020 (0.002) |
| Adenine | 0.010 (0.002) | 0.005 (0.001) | 0.017 (0.001) |
| Total | 0.109 (0.007) | 0.058 (0.003) | 0.061 (0.002) |
Standard error at the 95% confidence level.
32.7 waters per nucleotide; the values in parentheses are standard deviations for two or three independent measurements (7).
There are two problems to be addressed regarding the data presented above. The crystals employed in this study are all rich in cytosine and guanine, as are most of the oligonucleotides crystallized so far. In addition, there is a lower level of impurities eluted with the retention times close to those of C and G compared to T and A. These two factors make quantification of C and G generally more accurate.
Comparison of the yields in the crystals with similar data for hydrated DNA irradiated at room temperature (7) clearly reveals at least two major differences. First, in the crystals, the bases are released in proportions that closely match their abundance in the oligonucleotides. This is in contrast to hydrated DNA, where the relative amount of guanine released is much lower than would be predicted from the relative guanine content. The same phenomenon occurs with aqueous solutions of DNA (12, 20). Second, the base release in crystalline DNA is surprisingly insensitive to the presence of anthracycline intercalators even at the loading of one intercalator per three base pairs. This result is quite remarkable since intercalated anthracyclines completely suppress the production of both electron-gain and electron-loss centers on DNA bases even at 4 K (21). Finally, partial dehydration of the DNA10 crystals does not have any significant impact on the yields of base release. These features emphasize the distinction between the direct and indirect effects and will be discussed below.
DISCUSSION
It is widely accepted that the majority of DNA lesions induced by ionizing radiation have a free radical origin (1). In crystalline DNA irradiated at 4 K, most of the free radical chemistry occurs either during irradiation or upon warming. For example, the DNA10 crystals irradiated at 4 K lose 94% of their initial free radical population upon annealing to room temperature (Debije, personal communication). Therefore, most free radical reactions occur in the solid state, and those that occur during dissolution of the crystals are not likely to play more than a minor role in our experiments. We also note that all the key reactions leading to DNA strand breakage and base release in anoxic solutions either are unimolecular or require water as a second reactant (22). Therefore, these reactions could take place in crystals as well since the DNA in crystals is hydrated (see below).
Numerous studies have shown [for a recent review, see ref. (22)] that immediate precursors for unaltered base release from DNA are radicals localized on the sugar-phosphate backbone. The major point of the following discussion is to identify the mechanism of backbone radical formation through the direct effect, and its relative contribution to the overall mechanism of radiation damage to crystalline DNA.
The mechanism for unaltered base release through the indirect effect in aqueous solution is fairly well established (1, 22). The first step in this process involves chemical alteration of the deoxyribose moiety through hydrogen abstraction from the sugar by hydroxyl radicals. The glycosylic bond of the altered site is no longer stable and the base is released, typically along with a strand break. The presence of molecular oxygen changes the reaction mechanism but not the final outcome with respect to base release (22).
A rare exception to this scheme is hydrogen abstraction from the C4′ position of deoxyguanosine units which can result in a strand break without an immediate base release. The C4′ sugar radical undergoes fast β elimination of the 3′ phosphate to form an enol-ether radical cation (23). This intermediate is capable of oxidizing adjacent guanines through intramolecular electron transfer—the process that transfers the damage from the sugar to the base and prevents the release of free base (24). The oxidation potentials of the other three bases are too high to make this process competitive with other reaction pathways. This would explain why the yield of unaltered guanine is always reduced relative to the other bases in DNA solutions (12, 20) and in hydrated DNA irradiated at room temperature (7).
Base release from the crystals does not match the picture just described; the effect appears to be random, that is, without selectivity for any base. Moreover, the presence of intercalated anthracyclines in the DNA6·2(anthracycline) complexes has little impact on base release. This takes place despite the ability of the anthracyclines to act as efficient OH radical scavengers due to the presence of both hydroquinone and aminosugar moieties in their structure. Partial dehydration of the DNA10 crystals also does not affect the base release. The dehydration presumably removes the loosely bound water, which is the water expected to be the most important source of hydroxyl radicals (25, 26). Therefore, base release in DNA crystals is not consistent with hydroxyl radical attack on DNA, and alternative mechanisms need to be considered.
The alternative to hydrogen abstraction by hydroxyl radicals could be a free valence transfer from the base radicals to the sugar, leading to a sugar radical and subsequent base release. The involvement of such a process in DNA radiation chemistry has been discussed in the literature. A well-established example in model systems is represented by deoxycytidine, where deprotonation of the base-centered radical cation at the C1′ site leads to the C1′-centered deoxycytidine sugar radical (27). However, our results obtained with anthracycline-doped crystals clearly indicate that DNA base radicals are not, to a substantial degree, precursors to base release.
The dihydroxyanthraquinone moiety of both anthracyclines intercalates into the base stack and acts as an efficient scavenger for both electrons and holes in DNA. EPR studies have shown that, at the doping levels existing in these crystals, all excess electrons and holes in DNA transfer to the intercalators even at 4 K (21). The products of both electron and hole scavenging are relatively unreactive semiquinone radicals. Therefore, these dopants would suppress base release from the crystals if base radicals were involved in this process. But the data shown in Fig. 2 indicate that this is not the case.
The mechanism of base release in the crystals which best accounts for our findings is the formation of the sugar radicals through direct ionization of the deoxyribose moieties that takes place in parallel with ionization of the bases. The primary radical cation located on the sugar is then involved in two competitive processes: One is intra- and intermolecular hole transfer to the more oxidizable bases, and the other is deprotonation leading to a neutral sugar radical. Both pathways are expected to be very fast, though their relative rates are not known. However, the relative probability for initial ionization of sugar can be estimated from the electron density distribution of DNA. The deoxyribose moiety contains 93 electrons (including phosphate), while the average number of electrons on a DNA base is 66.75 (65 on thymine, 57 on cytosine, 69 on adenine, and 76 on guanine). Therefore, 58% of the primary radical cations should be localized on the sugar-phosphate backbone. In addition, ionization generates free electrons, which also contribute to the measurable free radical population by forming radical anions on the bases. Considering this and assuming that one radical anion is trapped per radical cation, the upper limit for radicals on the backbone should be reduced by a factor of 2. This means that the backbone-centered radical cations comprise about 29% of the total free radical population. In fact, this value may be higher, since the neutral sugar radicals appear to be less prone to recombine with the electrons than the charged electron-loss centers on the bases and therefore have more chance to survive the geminate electron–hole recombination (28).
By comparing the upper limit of 29% with the free radical yields at 4 K and the free base yields, the relative efficiencies of hole transfer to the bases compared to deprotonation of the sugar radical cation can be estimated. The amount of total base release relative to total radical yield at 4 K [which is 0.57 μmol J−1 for DNA10 and 0.61 μmol J−1 for DNA6 (14)] is therefore 19% for the DNA10 and 10% for the DNA6 crystals. Under the assumption that each sugar radical trapped at 4 K produces a strand break and a free base on subsequent sample processing, these correspond to 66 and 34%, respectively, of the upper limit (i.e. 19/29 and 10/29). This indicates that deprotonation of the sugar radical cation and hole transfer to the bases are approximately equally efficient processes.
The direct effect is therefore viewed as producing a set of neutral sugar radicals due to the net loss of a hydrogen atom from any of the five carbons. These radicals, centered at C1′, C2′, C3′, C4′ and C5′, are structurally equivalent to those produced by the indirect effect (abstraction by the hydroxyl radical). One might expect that the effect of guanine, discussed above for the indirect effect, would also be observed for the direct effect. That is, hole transfer to guanine should compete with base elimination via radical cation intermediates that are formed on the guanine deoxyriboside and reduce the release of unaltered guanine comparatively to the other bases.
In fact, this “protection” afforded by guanine may occur for the direct effect as well. However, it would remain undetected in our experiments due to expected differences in hydrogen-abstraction distribution. For the indirect effect, solvent accessibility dictates that hydrogen abstraction by hydroxyl radicals is primarily from C4′ and C5′ (29, 30). For the direct effect, the distribution will be governed by other factors such as local electron densities (as described further below) and these factors are not likely to favor hydrogen abstraction from C4′ to the extent found for hydroxyl radical attack.
For sugar radicals produced by direct ionization and subsequent deprotonation, it is instructive to make a rough estimate of the distribution that would be expected using a simple set of assumptions. We assume that the probability of ionization corresponds to local electron densities and that deprotonation occurs at the closest C–H bond. We ignore factors such as resonance stabilization, torsion angle between the lone-electron orbital and the C–H bond, and lattice effects. As an example, the phosphate group contains 52% of the sugar-phosphate electrons. Given two hydrogens on C5′ and one on C3′, deprotonation from the phosphate radical cation is assumed to favor C5′ over C3′ by 2 to 1. Electron loss from the lactone oxygen is assumed to result in deprotonation of C1′ and C4′ with equal probability. These probabilities are added to those obtained from the number of electrons at each carbon site and adjacent noncarbon atoms. We ignore the contribution of the base completely. This gives a distribution estimate of 43% at C5′, 12% at C4′, 24% at C3′, 9% at C2′, and 12% at C1′. The point is that the fraction of C4′ radicals, produced by direct ionization, is expected to be relatively small. Therefore, any corresponding reduction in guanine release would be small and not detected by the experiments reported here.
There are a few additional points that need consideration. First, our estimate of the distribution of radical trapping sites does not include the waters of hydration and counterions, which also undergo ionizations. The hydration state of the crystals employed in this study is not known exactly because many of the water molecules and the counterions are disordered. Yet their effect can be significant since the fraction of the electron density in the water for Γ = 14 (as calculated for DNA10 using computer modeling) and sodium as a counterion is about 50% of the total (152 electrons in the water + sodium compared to 158.7 electrons per nucleotide). Water location is also important. The H2O+· radical cations generated from tightly bound water are expected to transfer holes to the DNA faster than they deprotonate to form the hydroxyl radical, while deprotonation can dominate for loosely bound waters (25, 26, 31). The H2O+· species can oxidize both the sugars and the bases; the former is favored by the shorter electron tunneling distances while the latter is favored energetically. Depending on which of the processes dominates, the waters of hydration may increase or decrease the fraction of the sugar radicals calculated for the “dry” crystals. Although the available data are too limited to draw firm conclusions, the results do raise the possibility that the hydration layer influences the damage distribution. For example, the lower fraction of the base release compared to the free radical yield found in DNA6 crystals (10% in DNA6 compared to 19% in DNA10) could be a result of its lower hydration state (Γ = 8.4 in DNA6 compared to Γ = 14 in DNA10). This observation is consistent with the water-centered holes transferring predominantly to the sugars rather than to the bases.
The second point is that if the sugar radicals are produced in DNA crystals, why are they not seen by EPR? It has been pointed out previously that EPR studies do not in fact rule out the formation of sugar-centered radicals by direct ionization of DNA (32, 33). A detailed discussion of this issue can be found in a paper by Close (34) that addresses this problem by means of computer simulations. The answer to the question above is that if several types of sugar radicals are present in an amount less than about 20% of the total radical population, they appear as broad, low-intensity and almost structureless features, partially in the wings and partially underneath the narrower EPR spectra of the base radicals. These features are hard to identify and quantify. Nevertheless, such features are known in EPR spectra of X-irradiated amorphous DNA samples (35), and especially in those irradiated with very high doses of high-LET radiation (28, 36), where they have been attributed to sugar radicals. One characteristic feature of this signal is that it is less prone to dose saturation compared to the base radicals for the reason mentioned above (28). Because of that, the contribution of the “sugar” signal to the overall spectrum grows with dose and LET.
It is noteworthy that dose responses for base release from DNA crystals and for free radical accumulation in the same crystals remain linear up to doses of about 10 kGy. Above 10 kGy, free radical accumulation deviates from linear as the radical concentration begins to saturate (13); in contrast, base release remains almost linear up to 40 kGy. This observation indicates once again that the easy-to-saturate base radicals are unlikely to be the precursors for base release, and it is consistent with the sugar radicals being the precursors.
Based on the assumptions discussed above, the yield of free base release from crystalline DNA can be used to estimate the yield of strand breaks in DNA in vivo. The free radical yields at 4 K are 0.57 μmol J−1 for DNA10 and 0.61 μmol J−1 for DNA6 (14). Multiplying these by 19% and 10%, respectively, gives expected strand break yields of 0.12 and 0.06 μmol J−1. These values are remarkably close to the value of 0.2 μmol J−1 that was estimated by Milligan and coworkers from measurements of strand breaks in plasmids as a function of scavenger concentration (37).
CONCLUSIONS
We conclude that direct ionization of the sugar-phosphate backbone is followed by deprotonation of the primary radical cation and that deprotonation of the sugar radical cation competes (occurs in parallel) with hole transfer to the bases. Quantitative measurements indicate that 10 to 20% of the free radicals trapped in the crystals at 4 K are precursors to base release. Using these numbers, we estimate the yield of DNA strand breaks from the direct effect to be ~0.1 μmol J−1.
Acknowledgments
We thank Loren Williams and Gary Hu for providing us with the DNA6·2(anthracycline) crystals and Kermit R. Mercer for his invaluable technical assistance. The investigation was supported by PHS Grant 2- R01-CA32546, awarded by the National Cancer Institute, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; London: 1987. [Google Scholar]
- 2.Krisch RE, Flick MB, Trumbore CN. Radiation chemical mechanisms of single- and double-strand break formation in irradiated SV40 DNA. Radiat Res. 1991;126:251–259. [PubMed] [Google Scholar]
- 3.Close DM. Radical ions and their reactions in DNA constituents. ESR/ENDOR studies of radiation damage in the solid state. Radiat Res. 1993;135:1–15. [PubMed] [Google Scholar]
- 4.Becker D, Sevilla MD. The chemical consequences of radiation damage to DNA. Adv Radiat Biol. 1993;17:121–180. [Google Scholar]
- 5.Sevilla MD, Becker D. Radiation damage in DNA. Electron Spin Resonance (A Specialist Periodical Report) 1994;14:130–165. [Google Scholar]
- 6.Swarts SG, Sevilla MD, Wheeler KT. Specific DNA base damage produced by irradiation of hydrated DNA. In: Chapman JD, Whitmore GF, Dewey WC, editors. Radiation Research. Vol. 1. Academic Press; San Diego: 1991. Abstract. [Google Scholar]
- 7.Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiation-induced damage as a function of hydration. I. Release of unaltered bases. Radiat Res. 1992;129:333–344. [PubMed] [Google Scholar]
- 8.Swarts SG, Becker D, Sevilla MD, Wheeler KT. Radiation-induced DNA damage as a function of hydration. II. Base damage from electron-loss centers. Radiat Res. 1996;145:304–314. [PubMed] [Google Scholar]
- 9.Wolf P, Jones GDD, O’Neill P. Induction of strand breaks in polyribonucleotides and DNA by the sulfate radical-anion-role of electron-loss centres as precursors of strand breakage. Int J Radiat Biol. 1993;64:7–18. doi: 10.1080/09553009314551061. [DOI] [PubMed] [Google Scholar]
- 10.Cullis PM, Malone ME, Merson-Davies LA. Guanine radical cations are precursors of 7,8-dihydro-8-oxo-2′-deoxyguanosine but are not precursors of immediate strand breaks in DNA. J Am Chem Soc. 1996;118:2775–2781. [Google Scholar]
- 11.Milligan JR, Aguilera JA, Nguyen TTD, Ward JF, Kow YW, He B, Cunningham RP. Yield of DNA strand breaks after base oxidation of plasmid DNA. Radiat Res. 1999;151:334–342. [PubMed] [Google Scholar]
- 12.Henle ES, Roots R, Holley WR, Chatterjee A. DNA strand breakage is correlated with unaltered base release after gamma irradiation. Radiat Res. 1995;143:144–150. [PubMed] [Google Scholar]
- 13.Debije MG, Bernhard WA. Free radical yields in crystalline DNA X-irradiated at 4 K. Radiat Res. 1999;152:575–581. [PMC free article] [PubMed] [Google Scholar]
- 14.Debije MG, Milano MT, Bernhard WA. DNA responds to ionizing irradiation as an insulator not as a “molecular wire”. Angew Chem. 1999;38:2752–2756. [PMC free article] [PubMed] [Google Scholar]
- 15.Ban C, Sundaralingam M. Crystal structure of the self-complementary 5′-purine start decamer d(GCACGCGTGC) in the A-conformation. Biophys J. 1996;71:1222–1227. doi: 10.1016/S0006-3495(96)79351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadasivan C, Gautham N. Sequence-dependent microheterogeneity of Z-DNA: The crystal and molecular structures of d(CACGCG)d(CGCGTG) and d(CGCACG)d(CGTGCG) J Mol Biol. 1995;248:918–930. doi: 10.1006/jmbi.1995.9894. [DOI] [PubMed] [Google Scholar]
- 17.Quigley GL, Wang AH-J, Ughetto G, van den Marel G, van Boom JH, Rich A. Molecular structure of an anticancer drug–DNA complex—daunomycin plus d(CpGpTpApCpG) Proc Natl Acad Sci USA. 1980;77:7204–7208. doi: 10.1073/pnas.77.12.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frederick CA, Williams LD, Ughetto G, van den Marel GA, van Boom JH, Rich A, Wang AH-J. Structural comparison of anticancer drug–DNA complexes: Adriamycin and daunomycin. Biochemistry. 1990;29:2538–2549. [PubMed] [Google Scholar]
- 19.Lipscomb LA, Peek ME, Zhou FX, Bertrand JA, Van Derveer D, Williams LD. Water ring structure at DNA interfaces: Hydration and dynamics of DNA–anthracycline complexes. Biochemistry. 1994;33:3649–3659. doi: 10.1021/bi00178a023. [DOI] [PubMed] [Google Scholar]
- 20.Ward JF, Kuo I. Strand breaks, base release, and postirradiation changes in DNA γ-irradiated in dilute O2-saturated aqueous solution. Radiat Res. 1976;66:485–498. [PubMed] [Google Scholar]
- 21.Milano MT, Hu GG, Williams LD, Bernhard WA. Migration of electrons and holes in crystalline d(CGATCG)–anthracycline complexes X-irradiated at 4 K. Radiat Res. 1998;150:101–114. [PubMed] [Google Scholar]
- 22.Pogozelski WK, Tullius TD. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 23.Gugger A, Batra R, Rzadek P, Rist G, Giese B. Spectroscopic evidence for a radical cation as intermediate in a model reaction of the 4′-DNA radical strand cleavage. J Am Chem Soc. 1997;119:8740–8741. [Google Scholar]
- 24.Meggers E, Kusch D, Spichty M, Wille U, Giese B. Electron transfer through DNA in the course of radical-induced strand cleavage. Angew Chem. 1998;37:460–462. doi: 10.1002/(SICI)1521-3773(19980302)37:4<460::AID-ANIE460>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.Becker D, LaVere T, Sevilla MD. ESR detection at 77 K of the hydroxyl radical in the hydration layer of gamma-irradiated DNA. Radiat Res. 1994;140:123–129. [PubMed] [Google Scholar]
- 26.LaVere T, Becker D, Sevilla MD. Yields of OH· in gamma-irradiated DNA as a function of DNA hydration: Hole transfer in competition OH· formation. Radiat Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 27.Malone ME, Cullis PM, Symons MCR, Parker AW. Biphotonic photoionization of cytosine and its derivatives with UV radiation at 248 nm: An EPR study in low-temperature perchlorate glasses. J Phys Chem. 1995;99:9299–9308. [Google Scholar]
- 28.Becker D, Razskazovskii Y, Callaghan MU, Sevilla MD. Electron spin resonance of DNA irradiated with a heavy-ion beam (16O8+): Evidence for damage to the deoxyribose phosphate backbone. Radiat Res. 1996;146:361–368. [PubMed] [Google Scholar]
- 29.Sy D, Savoye C, Begusova M, Michalik V, Charlier M, Spotheim-Maurizot M. Sequence-dependent variations of DNA structure modulate radiation-induced strand breakage. Int J Radiat Biol. 1997;72:147–155. doi: 10.1080/095530097143365. [DOI] [PubMed] [Google Scholar]
- 30.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milano MT, Bernhard WA. The effect of packing and conformation on free radical yields in films of variably hydrated DNA. Radiat Res. 1999;151:139–149. [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhard WA. Solid-state radiation of DNA: The bases. Adv Radiat Biol. 1981;9:199–280. [Google Scholar]
- 33.Kar L, Bernhard WA. Electron gain and electron loss radicals stabilized on the purine and pyrimidine of cocrystal exhibiting base–base interstacking: ESR-ENDOR of X-irradiated adenosine:5-bromouracil. Radiat Res. 1983;93:232–253. [Google Scholar]
- 34.Close DM. Where are the sugar radicals in irradiated DNA? Radiat Res. 1997;147:663–673. [PubMed] [Google Scholar]
- 35.Weiland B, Huttermann J. Free radicals from X-irradiated ‘dry’ and hydrated lyophilized DNA as studied by electron spin resonance spectroscopy: analysis of spectral components between 77K and room temperature. Int J Radiat Biol. 1998;74:341–358. doi: 10.1080/095530098141483. [DOI] [PubMed] [Google Scholar]
- 36.Weiland B, Huttermann J. Free radicals from lyophilized ‘dry’ DNA bombarded with heavy-ions as studied by electron spin resonance spectroscopy. Int J Radiat Biol. 1999;75:1169–1175. doi: 10.1080/095530099139647. [DOI] [PubMed] [Google Scholar]
- 37.Milligan JR, Aguilera JA, Ward JF. Variation of single-strand break yield with scavenger concentration for plasmid DNA irradiated in aqueous solution. Radiat Res. 1993;133:151–157. [PubMed] [Google Scholar]


