Abstract
Fifty outbred Syrian golden hamsters were equally divided into three experimental groups and two control groups. The pouches of the experimental groups were painted bilaterally with a 0.5% 7,12-dimethylbenz[a]anthracene (DMBA) solution thrice a week for 3, 7 and 14 weeks. One of the control groups was applied with mineral oil while another control group remained untreated throughout the experiment. Neither survivin nor cIAP2 could be detected in any of the control tissues, whereas survivin and cIAP2 were found to be significantly increased in 3-, 7- and 14-week DMBA-treated pouches compared with the control pouches. Expression of XIAP, cIAP1 and NAIP were noted for both the control and 3-, 7- and 14-week DMBA-treated pouches, but levels were found to be significantly elevated in the experimental groups compared with the control pouches. p53 was not detected in any control tissues, but was significantly increased in 3-, 7- and 14-week DMBA-treated pouches. Direct sequencing revealed a point mutation (C→G) of p53 for pouch tissues treated with DMBA for 3 and 7 weeks, and there was a wide variation in the p53 sequence of the 14-week DMBA-treated pouch tissues, as compared with the control tissues. The control tissues had a survivin- and cIAP2-methylated allele, whereas the DMBA-treated tissues showed no evidence of survivin- and cIAP2-methylation. Neither the control nor DMBA-treated pouches showed evidence of XIAP-, cIAP1- or NAIP-methylation. Our results suggest that the expression of inhibitors of apoptosis family in DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis may be modulated by both genetic (mutant p53) and epigenetic mechanisms.
Keywords: cIAP1, cIAP2, DMBA carcinogenesis, hamster, IAP, NAIP, survivin, XIAP
Apoptosis, also termed programmed cell death, is a complex and highly regulated process, important for normal development and the functioning of diverse roles such as embryogenesis and tissue homeostasis, as well as tumourigenesis in multicellular organisms (Gupta 2002). Apoptosis is executed by a family of cysteine proteases known as caspases, which are produced in cells as inactive zymogens and become active proteases subsequent to proteolysis (Nicholson 1999). This caspase family of cysteine proteases performs an essential role in orchestrating the sequential breakdown of cells. The organism must tightly modulate the caspase cascade, commencing with the activation of an upstream or initiator caspase (such as caspases 8 and 9) and leading to the activation of a downstream or effector caspase (such as caspases 3, 6 and 7) (Cohen 1997).
The inhibitors of apoptosis (IAP) are a family of proteins functioning as intrinsic negative regulators of the aforementioned caspase cascade (Liston et al. 2003) and are the only known endogenous proteins that interfere with the activity of both initiator and effector caspases (Liston et al. 2003). Eight human IAP family members have been recognized so far: neuronal apoptosis inhibitory protein (NAIP), X-linked inhibitors of apoptosis protein (XIAP), cellular inhibitors of apoptosis protein 1 (cIAP1), cellular inhibitors of apoptosis protein 2 (cIAP2), survivin, baculoviral IAP repeat-containing ubiquitin-conjugating enzyme (BRUCE) (apollon), livin (ML-IAP, KIAP) and IAP-like protein 2 (ILP-2) (Liston et al. 2003). They are characterized by the presence of one or more 70–80 amino acids N-terminal domain called the baculovirus IAP repeat; some bind and potently inhibit activated caspases, including the effector caspases 3 and 7 and the initiator caspase 9 (Deveraux & Reed 1999). In addition, several IAPs also contain a carboxyl terminal really interesting new gene (RING) zinc-finger domain, which binds ubiquitin-conjugating enzymes that promote degradation of IAP caspase complexes (Yang et al. 2000). Moreover, a caspase recruitment domain (CARD), a conserved domain, has been found in cIAP1 and cIAP2, but the function of this domain in these two members is currently unknown (Salvesen & Duckett 2002). Some characteristics, as well as the number and distribution of these three kinds of domain amongst the IAP family members, are summarized in Table 1.
Table 1.
Some characteristics of the eight human inhibitors of apoptosis (IAP) family members
| IAP family members | Size (kDa) | Chromosome positions | Number of domains | Caspase specificity |
|---|---|---|---|---|
| XIAP (ILP1) | 57 | Xq25 | BIR: 3 RING: 1 | Caspase 3, 7, 9 |
| cIAP1 | 70 | 11q22-q23 | BIR: 3 RING: 1 CARD: 1 | Caspase 3, 7, 9 |
| cIAP2 | 68 | 11q22-q23 | BIR: 3 RING: 1 CARD: 1 | Caspase 3, 7, 9 |
| LIVIN (ML-IAP) | 31 | 20q13.3 | BIR: 1 RING: 1 | Caspase 3, 7, 9 |
| ILP2# | ? | 19q 13.3-q13.4 | BIR: 1 RING: 1 | Caspase 9 |
| NAIP* | 156 | 5q13.1 | BIR: 3 RING: 0 | Caspase 3, 7 |
| Survivin | 16.5 | 17q25 | BIR: 1 RING: 0 | Caspase 3, 7 |
| BRUCE | 528 | 2p21-p22 | BIR: 1 RING: 0 | Caspase 3, 7 |
The first characterized mammalian IAP.
The most recently identified IAP (a tissue-specific homologue of XIAP).
BIR, baculovirus IAP repeat; RING, really interesting new gene; CARD, caspase recruitment domain; XIAP, X-linked inhibitors of apoptosis protein; cIAP1, cellular inhibitors of apoptosis protein 1; cIAP2, cellular inhibitors of apoptosis protein 2; ILP2, IAP-like protein 2; NAIP, neuronal apoptosis inhibitory protein;. BRUCE, BIR repeat-containing ubiquitin-conjugating enzyme.
Hamster buccal-pouch mucosa provides one of the most widely accepted experimental models for oral carcinogenesis (Gimenez-Conti & Slaga 1993). Despite anatomical and histological differences between (hamster) pouch mucosa and human buccal tissue, experimental carcinogenesis protocols for the former induce premalignant changes and carcinomas that are similar to the development of premalignancy and malignancy in human oral mucosa (Morris 1961).
The p53 tumour-suppressor gene is implicated in cell cycle checkpoint mechanisms, inhibiting cell cycle progression and inducing apoptosis in reaction to DNA damage (Gottlieb & Oren 1998). Given that an association of p53 expression and survivin, as well as XIAP, has been previously noted in hamster buccal-pouch carcinogenesis (Hsue et al. 2007), the relationship of other members of the IAP family and p53 is still an interesting area for study. On the other hand, epigenetic mechanisms such as DNA methylation have been implicated in cell proliferation, differentiation and genomic integrity. Although an association of survivin expression in hamster buccal-pouch carcinomas with epigenetic alteration has been reported in our laboratory recently (Chen et al. 2005a), the regulation of IAP family members other than survivin by an epigenetic mechanism is still a promising area in the investigation of oral carcinogenesis.
Consequently, as the expression of the IAP family, as well as its association with p53 and epigenetic alterations, in experimental oral carcinogenesis is not completely understood, this study was designed to investigate the protein and mRNA expression of five IAP family members (survivin, XIAP, cIAP1, cIAP2 and NAIP) in 7,12-dimethylbenz[a]anthracene (DMBA)-induced hamster buccal-pouch squamous-cell carcinogenesis. The relationship between the expression of the IAP family and p53 status, as well as epigenetic alterations in DMBA-induced hamster pouch squamous-cell carcinogenesis, was also examined.
Materials and methods
Animals and treatments
Outbred, young (6-week-old), male Syrian golden hamsters (Mesocricatus auratus; 50 animals, purchased from the National Science Council Animal Breeding Center, Taipei, China), weighing approximately 100 g at the beginning of the experiment, were randomly divided into three experimental and two control groups (10 animals per group). The animals were housed under constant conditions (22 °C, 12-h light/dark cycle) and supplied with tap water and standard Purina laboratory chow ad libitum. Appropriate animal care and an approved experimental protocol ensured humane treatment, and all procedures were conducted in accordance with the NIH Guide for the Care and Use of Animals. After allowing the animals 1 week of acclimatization to their new surroundings, both pouches from all animals in the experimental groups were painted with a 0.5% DMBA solution at 9 a.m. on Monday, Wednesday and Friday of each week, using a No. 4 sable-hair brush. Bilateral pouches from each animal of one control group were similarly treated with mineral oil. Approximately 0.2 ml of the appropriate solution was applied topically to the medial walls of both pouches at each painting. The second control group of animals remained untreated throughout the experiment.
At the end of 3 weeks (3 days after the last treatment), all animals from one of the experimental groups were simultaneously killed by administration of a lethal dose of diethyl ether, at 9 a.m., to avoid any influence of diurnal variation (Lin & Chen 1997). Their pouches were exposed by dissection and examined grossly; both pouches were then excised and placed on cardboard to prevent distortion of the pouch tissues. After 7 weeks of treatment, the animals from one of the two remaining experimental groups were killed in a similar manner. Then, at 14 weeks, the animals from the last experimental group and those from the two control groups were killed, using the same procedure.
A portion of the pouch tissue was immediately frozen in liquid nitrogen for subsequent DNA/RNA extraction, RT-PCR reaction and methylation assay investigation, whilst another portion was routinely processed for light microscopy by being fixed in 10% neutral-buffered formalin solution for about 24 h, dehydrated in a series of ascending-concentration alcohol solutions, cleared in xylene, and embedded in paraffin for immunohistochemical staining.
Immunohistochemistry
Following tissue sectioning, staining was performed using a standard avidin-biotin peroxidase complex (ABC) method (Hsu et al. 1981). Antibodies for IAP proteins were obtained from Abchem Corporation, Cambridge, UK. Rabbit polyclonal antibodies against human, rat and mouse survivin (Cat. No. ab469), XIAP (Cat. No. ab21278), cIAP1 (Cat. No. ab2399), cIAP2 (Cat. No. ab23423) and NAIP (Cat. No. ab25968) were used and their specificity has been established in previous studies (Liston et al. 2001; Barnes et al. 2006; Hsue et al. 2008). Monoclonal antibody NCLp53-D07 (mAb DO7; Novocastra, Newcastle, UK) was used for the identification of p53 protein. The mAb DO7 antibody detects both wild-type and mutant forms of p53 (Vojtesek et al. 1992).
Tissue sections were mounted on gelatin–chrome alum-coated slides. Following deparaffinization in xylene (twice) and rehydration in a decreasing-concentration ethanol series (absolute, 95%, 70% and 30% ethanol, and then water), tissue sections were microwave-treated thrice (5 min each time) in a citrate buffer (10 mm; pH 6.0) to retrieve antigenicity. Endogenous peroxidase activity was blocked with 3% H2O2 in methanol for 60 min. Prior to immunohistochemical staining, a 10% solution of normal rabbit serum was applied for 60 min to tissue sections to inhibit non-specific staining. These sections were subsequently incubated with antibodies against survivin, XIAP, cIAP1, cIAP2 and NAIP (1:100 each) overnight at 4 °C. A blocking solution of 2% low-fat milk powder in Tris-buffered saline (TBS, with 0.02% sodium azide) was applied to those sections to be stained for p53 protein. These sections were then treated with mAb DO7 at a dilution of 1:200 for 2 h at room temperature. Following subsequent rinsing with TBS (thrice, 10 min each), tissue sections stained for survivin, XIAP, cIAP1, cIAP2 and NAIP were then incubated for 60 min at room temperature with biotin-conjugated goat anti-rabbit IgG (Vector, Burlingame, CA, USA; 1:100). In contrast, the sections stained for p53 were treated with biotinylated anti-mouse IgG antibody (Vector; 1:100) for 30 min. Following this, all sections were again washed with TBS (thrice, 10 min each) and then incubated with an avidin–biotin complex conjugated to horseradish peroxidase (Dako, Santa Barbara, CA, USA) for a further 60 min. After washing with PBS (thrice, 10 min each), peroxidase binding was visualized as brown reaction products via a benzidine reaction. The sections were then counterstained with Mayer's haematoxylin. Each set of experiments included a human buccal squamous-cell carcinoma specimen known to express survivin, XIAP, cIAP1, cIAP2, NAIP and p53, which served as a positive control and ensured the reproducibility of the staining process. The scores for positive staining were classified as: 0, <10%; 1, 10–24%; 2, 25–49%; 3, 50–74%; 4, 75–89%; 5, 90–100%. A negative control, in which the primary antibody step was omitted, was also included in each set of experiments.
DNA/RNA isolation
Genomic DNA was extracted from each pouch specimen by proteinase K digestion and the phenol–chloroform extraction procedure, as described elsewhere (Sambrook & Russell 2001). Total RNA was extracted by homogenizing the pouch tissue specimens in guanidium isothiocyanate, followed by ultracentrifugation in caesium, as described previously (Chomczynski & Sacchi 1987).
Semi-quantitative reverse transcription-polymerase chain reaction
Isolated total RNA (1 μg) was reverse-transcribed to cDNA in a reaction mixture (with a final volume of 20 μl) containing MgCl2 (4 μl; 5 mm), 10× reverse transcription buffer (2 μl; 10 mm Tris–HCl, pH 9.0, 50 mm KCl, 0.1% Triton® X-100), dNTP mixture (2 μl; 1 mm each), recombinant RNasin® ribnonuclease inhibitor (0.5 μl; 1 μ/μl), avian myeloblastosis virus (AMV) reverse transcriptase (15 units; High Conc.; 15 μ/μg), and oligo(dT)15 primer (0.5 μg; catalogue no. A3500; Promega, Madison, WI, USA). The reaction mixture was incubated at 42 °C for 15 min. The AMV reverse transcriptase was inactivated by heating at 99 °C for 5 min and then incubating at 0–5 °C for a further 5 min.
Oligonucleotide primers were purchased from Genset Corp. (La Jolla, CA, USA). The primer pairs were chosen from the published cDNA sequences of survivin, XIAP, cIAP1, cIAP2, NAIP, p53 and β-actin (Table 2). All these chosen primers were applicable to rodent tissues; therefore, they were justified to be used to detect hamster sequences. The 20 μl first-strand cDNA synthesis reaction product obtained from the reverse transcriptase reaction was diluted to 100 μl with nuclease-free water. The PCR amplification reaction mixture (with a final volume of 100 μl) contained diluted, first-strand cDNA reaction product (20 μl; <10 ng/μl), cDNA reaction dNTPs (2 μl; 200 μm each), MgCl2 (4 μl; 2 mm), 10× reverse transcription buffer (8 μl; 10 mm Tris–HCl, pH 9.0, 50 mm KCl, 0.1% Triton® X-100), upstream primer (50 pmol), downstream primer (50 pmol) and Taq DNA polymerase (2.5 units; Promega, Cat. No. M7660).
Table 2.
Oligoprimers 1 used for RT-PCR and oligoprimers 2 used for PCR-based methylation assay
| Primers | Sequences | Product (bp) | Accession no. |
|---|---|---|---|
| cIAP1 | Forward 1: 5-CAG CCT GAG CAG CTT GCA A-3′ | 354 | BC016174 |
| Reverse 1: 5-GCC CAT TTC CAA GGC AGA T -3′ | 674 | ||
| Forward 2: 5′ TTA TTT GTG GAT AAG AAT ATG AAG 3′ | |||
| Reverse 2: 5′ AAA AAT CCT TAT TTT AAA ACA CAA 3′ | |||
| cIAP2 | Forward 1: 5-TCC GTC AAG TTC AAG CCA GTT-3′ | 328 | BC037420 |
| Reverse 1: 5-TCT TTT TCC TCA GTT GCT CTT TCT CT-3′ | 832 | ||
| Forward 2: 5′ AGA AGA TGA AAT AAG GGA AGA GGA G 3′ | |||
| Reverse 2: 5′ TTT TAC TTC ACT TAC AAT TTC AAT AAT 3′ | |||
| NAIP | Forward 1: 5-GCT TCA CAG CGC ATC GAA-3′ | 446 | U80017 |
| Reverse 1: 5-ATG AGA GAC CCA AAA TCC GAA A -3′ | 650 | ||
| Forward 2: 5′ AAA AAG AAA AGA AAA GAA AAG AAA AAT 3′ | |||
| Reverse 2: 5′ TCT ACT AAT TCT TTT TTT TCT TTT TT 3′ | |||
| XIAP | Forward 1: 5-AGT GGT AGT CCT GTT TCA GCA TCA-3′ | 360 | U45880 |
| Reverse 1: 5′-GTT CCT CGG GTA TAT GGT GTC TGA-3′ | 884 | ||
| Forward 2: 5′ TTT AGG TGA AGG TGA TAA AGT AAA GTG 3′ | |||
| Reverse 2: 5′ TTC ACA TCA CAC ATT CAA TCA 3′ | |||
| Survivin | Forward 1: 5-AGA ACT GGC CCT TCT TG GA-3′ | 200 | BC016174 |
| Reverse 1: 5-AAG GAA AGC GCA ACC GGA CG-3′ | 584 | ||
| Forward 2: 5′ AAT AAG AAG AAA GAA TTT GAG GAA A 3′ | |||
| Reverse 2: 5′ AAA AAA ACA CAA CAA CAA AAA AAC T 3′ | |||
| β-actin | Forward 1: 5-AGA ACT GGC CCT TCT TGG A-3′ | 350 | X-00351 |
| Reverse 1: 5-AAG GAA AGC GCA ACC GGA CG-3′ |
cIAP1, cellular inhibitors of apoptosis protein 1; cIAP2, cellular inhibitors of apoptosis protein 2; NAIP, neuronal apoptosis inhibitory protein; XIAP, X-linked inhibitors of apoptosis protein.
The PCR steps were carried out on a DNA thermal cycler (TaKaRa MP, Tokyo, Japan). Thermocycling conditions included denaturing at 94 °C for 1 min (one cycle), then denaturing at 94 °C (1 min), annealing at 55 °C (1 min) for survivin and p53, at 59 °C (1 min) for XIAP, at 50 °C (1 min) for cIAP1 and cIAP2, at 56 °C (1 min) for NAIP, or at 60 °C (1 min) for β-actin, and extending at 72 °C (1 min) for 30 cycles, then a final extension at 72 °C for 7 min. The β-actin primers were utilized as positive controls. Negative controls, i.e. those conducted in the absence of RNA and reverse transcriptase, were also performed. Amplification products were analysed by electrophoresis in a 2% agarose gel along with the relevant DNA molecular-weight marker (Boehringer, Mannheim, Germany) and stained with ethidium bromide. Photographs were taken with a Polaroid DS-300 camera. The PCR products were visualized as bands with a UV transilluminator. Densitometric analysis of the PCR products was measured as the ratio of the expression of the five members of the IAP family and p53 with respect to the expression of β-actin, using the one-dscan 1-d gel analysis Software (Scanalytics, Inc., Rockville, MD, USA). The PCR products were then sequenced to confirm their identities using a T7 Sequenase kit (version 2.0; Amersham International, Little Chalfont, UK).
Polymerase chain reaction-based methylation assay
To investigate whether methylation of a CpG island of survivin, XIAP, cIAP1, cIAP2 and NAIP could be implicated in mRNA expression, a polymerase chain reaction-based methylation assay was used to analyse DNA samples obtained from the samples of hamster pouch mucosa. Genomic DNA was modified by bisulphite treatment, converting unmethylated cytosine to thymidine and leaving methylated cytosine unchanged (Herman et al. 1996). Approximately 10 μg of DNA with and without bisulphite treatment was digested with HindIII (1 μg/unit; Toyobo, Tokyo, Japan) at 37 °C for 12 h. The digested DNA was then precipitated with ethanol dissolved in distilled water. The DNA was further digested with mCpG-sensitive Hpa II (1 μg/unit; Toyobo) at 37 °C for 24 h. The non-digested DNA samples with and without bisulphite treatment, as well as the digested DNA samples with and without bisulphite treatment, were subsequently amplified with specific primers for survivin, XIAP, cIAP1, cIAP2 and NAIP (Table 2). The polymerase chain reactions were performed in a final volume of 25 μl, containing 1 μl of non-digested/digested DNA, 2.5 pmol of each specific primer, 50 μm of dNTPs, 10 mm Tris–HCl buffer (pH 8.3), 50 mm KCl, 1.5 mm MgCl2 and 0.5 unit of AmpliTaq Gold (Applied Biosytstems, Foster City, CA, USA). Negative controls were also performed. The amplified PCR products were separated in a 3% agarose gel and visualized by ethidium bromide.
Results
Gross observation and histopathology
The gross and histopathological changes in the untreated, mineral oil-treated and DMBA-treated pouches were similar to those described in our previous study (Chen et al. 2005b). Upon gross examination, no apparent changes for any of the mineral oil-treated or untreated pouches were observed. Thickened mucosa, with a rough surface and of whitish granular appearance, were noted in the 3 and 7 weeks DMBA-treated pouches, with 100% tumour incidence in all samples after 14 weeks of DMBA treatment. No significant histological changes were observed for any of the mineral oil-treated or untreated pouches. However, hyperkeratosis was noted in the 3 weeks DMBA-treated pouches and areas of epithelial hyperplasia were observed in the 7 weeks DMBA-treated pouches. Furthermore, infiltrative squamous-cell carcinomas were detected in the 14-week DMBA-treated mucosa.
Immunohistochemistry
The pattern of immunohistochemical staining for the five members of the IAP family in our study can be classified into two categories: (1) survivin and cIAP2; (2) XIAP, cIAP1 and NAIP.
Survivin and cIAP2
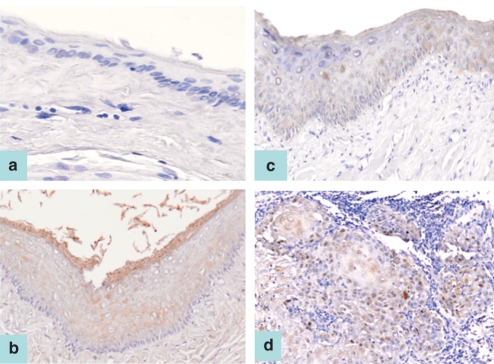
Neither survivin nor cIAP2 staining could be found in any of the untreated or mineral oil-treated hamster pouch tissue specimens (Figure 1a). Cytoplasmic staining of survivin or cIAP2 proteins was detected in the entire epithelial layer (except the basal layer) in all hamster pouch tissues treated with DMBA for 3 weeks (Figure 1b). In addition, cytoplasmic staining of survivin or cIAP2 protein was observed in all specimens of hamster buccal-pouch tissue treated with DMBA for 7 weeks (patchy staining; Figure 1c) and 14 weeks (Figure 1d). No obvious variation in the staining intensity was found for all hamster pouch tissues treated with DMBA for 3, 7 and 14 weeks respectively.
Figure 1.
cIAP2 staining was not detected in untreated hamster pouch tissue specimen (a, ABC stain ×100). Cytoplasmic staining of cIAP2 proteins was detected in the whole epithelial layer (except the basal layer) of a representative 3-week 7,12-dimethylbenz[a]anthracene (DMBA)-treated pouch tissue sample (b, ABC stain ×200). Patchy cytoplasmic staining was detected in the whole layer of a representative specimen of 7-week DMBA-treated pouch tissue (c, ABC stain ×100). Cytoplasmic cIAP2 staining was shown in a representative specimen of hamster pouch tissue treated with DMBA for 14 weeks (d, ABC stain ×100). A similar staining pattern is also observed for survivin immunostaining.
XIAP, cIAP2 and NAIP
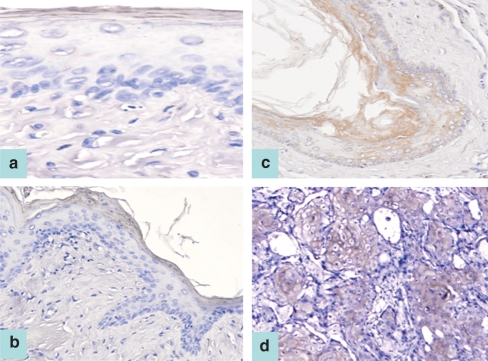
XIAP, cIAP1 or NAIP staining was located in the outermost keratinized layer of the untreated, mineral oil-treated and 3-week DMBA-treated pouch tissues (Figure 2a,b); however, the possibility of ‘edge-effect’ artefact should also be alerted. Cytoplasmic staining of XIAP, cIAP1 or NAIP proteins was found to extend downwards and was detected in the whole epithelial layer in all specimens of hamster buccal-pouch tissue treated with DMBA for 7 weeks (patchy staining; Figure 2c). Similar cytoplasmic staining of XIAP, cIAP1 or NAIP proteins was observed for all specimens of pouch tissue treated with DMBA for 14 weeks (Figure 2d). Significant deviation in the staining intensity was not being noted for all the control and experimental tissues.
Figure 2.
cIAP1 staining was located in the outermost keratinized layer of an untreated pouch tissue specimen (a, ABC stain ×200), as well as a representative 3-week 7,12-dimethylbenz[a]anthracene (DMBA)-treated pouch tissue (b, ABC stain ×100). Patchy cytoplasmic staining was detected in the whole layer of a representative specimen of 7-week DMBA-treated pouch tissue (c, ABC stain ×100). A representative specimen of pouch tissue treated with DMBA for 14 weeks demonstrated cytoplasmic cIAP1 staining (d) (ABC stain ×100). A similar immunostaining pattern is also observed for XIAP and NAIP proteins.
p53
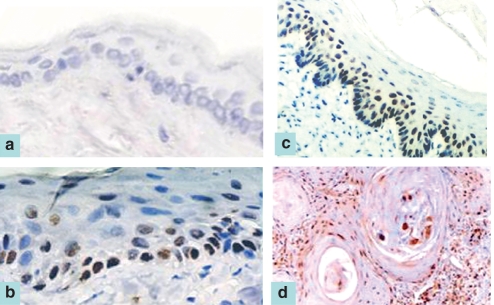
No p53 staining could be detected in any of the untreated or mineral oil-treated pouch tissue specimens (Figure 3a), whereas nuclear staining of p53 was noted for all pouch tissues treated with DMBA for 3 (Figure 3b), 7 (Figure 3c) and 14 weeks (Figure 3d). Apparent difference in the staining intensity was not found for all pouch tissues treated with DMBA for 3, 7 and 14 weeks.
Figure 3.
Negative p53 staining was observed for an untreated pouch tissue specimen (a, ABC stain ×100). Nuclear staining of p53 was noted in representative samples from 3-week (b, ABC stain ×200), 7-week (c, ABC stain ×100) and 14-week (d, ABC stain ×100) 7,12-dimethylbenz[a]anthracene-treated pouch tissue.
Statistical analyses
The scores based on percentage of positive stained cells (not staining intensity) of the five IAP family members and p53 for DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis are summarized in Table 3. Upon statistical analysis, the expression of these five IAP family members and p53 for 3-, 7- and 14-week DMBA-treated hamster pouch tissues was significantly increased compared with the expression of untreated and mineral oil-treated pouch tissues (P < 0.0001, chi-square test, Table 3).
Table 3.
Scores for immunohistochemical expression of the five members of the inhibitors of apoptosis family and p53 proteins for 7,12-dimethylbenz[a]anthracene (DMBA)-induced hamster buccal-pouch squamous-cell carcinogenesis
| cIAP1 | cIAP2 | NAIP | XIAP | Survivin | p53 | |
|---|---|---|---|---|---|---|
| Untreated | 1 | 0 | 1 | 1 | 0 | 0 |
| Mineral oil-treated | 1 | 0 | 1 | 1 | 0 | 0 |
| 3-week DMBA-treated | *2 | *4 | *3 | *4 | *3 | *3 |
| 7-week DMBA-treated | *4 | *4 | *3 | *4 | *4 | *3 |
| 14-week DMBA-treated | *4 | *4 | *3 | *5 | *4 | *4 |
Staining scores: 0, <10%; 1, 10–25%; 2, 25–50%; 3, 50–75%; 4, 75–90%; 5, 90–100%.
Statistical significance as compared with untreated or mineral oil-treated tissues (P < 0.0001, chi-square test).
cIAP1, cellular inhibitors of apoptosis protein 1; cIAP2, cellular inhibitors of apoptosis protein 2; NAIP, neuronal apoptosis inhibitory protein; XIAP, X-linked inhibitors of apoptosis protein.
Semi-quantitative reverse transcription-polymerase chain reaction
Survivin and cIAP2
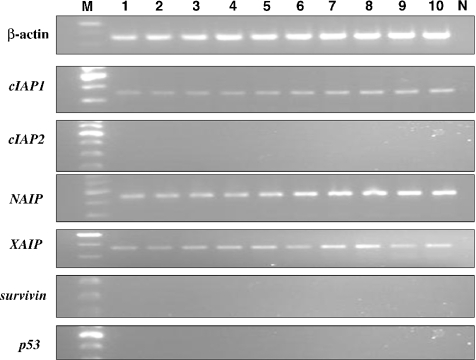
Upon RT-PCR, survivin mRNA and cIAP2 mRNA were evident as a band corresponding to a 200 and 328 bp PCR product, respectively, in all of the hamster buccal-pouch tissue specimens treated with DMBA for 3, 7 and 14 weeks (Figure 4), whereas no survivin mRNA and cIAP2 mRNA was detected in the samples of the untreated, the mineral oil-treated or the negative control animal tissue (Figure 5). All samples, except the negative control samples, revealed bands of β-actin (350 bp) (Figures 4 and 5).
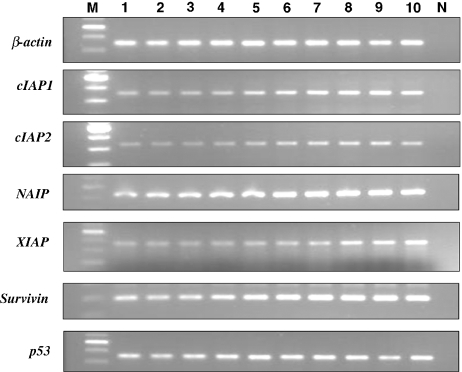
Figure 4.
Upon RT-PCR, bands corresponding to mRNA of cIAP1 (354 bp), cIAP2 (328 bp), NAIP (446 bp), XIAP (360 bp), survivin (200 bp) and p53 (370 bp) were evident in all of the hamster buccal-pouch tissue specimens treated with 7,12-dimethylbenz[a]anthracene (DMBA) for 14 weeks (lanes 1–10). All samples (lanes 1–10), except the negative control samples (lane N), revealed bands of β-actin (350 bp). Similar results were observed for hamster pouch tissue treated with DMBA for 3 and 7 weeks. Lane M: molecular weight markers.
Figure 5.
For all of the untreated samples (lanes 1–10), upon RT-PCR, bands corresponding to mRNA of cIAP1 (354 bp), NAIP (446 bp) and XIAP (360 bp) were detectable, whereas no bands for cIAP2, survivin and p53 were detected. All samples (lanes 1–10), except the negative control samples (lane N), revealed bands of β-actin (350 bp). Similar results were observed for mineral oil-treated pouch tissues. Lane M: molecular weight markers.
XIAP, cIAP1 and NAIP
Upon RT-PCR, mRNA for XIAP, cIAP1 and NAIP was detectable as a band corresponding to a 360, 354 and 446 bp PCR product, respectively, in all of the 3-, 7- and 14-week DMBA-treated hamster buccal-pouch tissue specimens (Figure 4). Messenger RNA for XIAP, cIAP1 and NAIP was also detected in the tissue derived from the untreated, mineral oil-treated tissues and negative control animals (Figure 5). All samples, except the negative control samples, revealed bands of β-actin (350 bp) (Figures 4 and 5).
p53
Messenger RNA for p53 corresponding to a band of 370 bp was observed for all the hamster buccal-pouch tissue specimens treated with DMBA for 3, 7 and 14 weeks (Figure 4). No such bands were noticed for the tissue samples of untreated, mineral oil-treated tissues and the negative-control animals (Figure 5). All samples, except the negative control samples, revealed bands of β-actin (350 bp) (Figures 4 and 5).
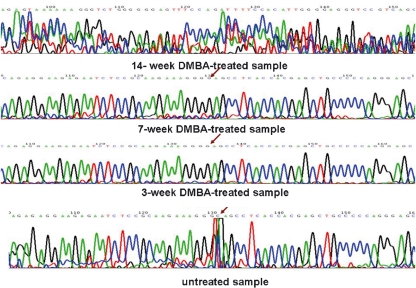
On the other hand, upon direct sequencing of this 370 bp band, a point mutation of C to G was noted for the hamster buccal-pouch tissue specimens treated with DMBA for 3 and 7 weeks, as compared with the untreated tissue specimens (Figure 6). Furthermore, there was a great variation for the sequence of the hamster buccal-pouch tissue specimens treated with DMBA for 14 weeks as compared with the untreated tissue specimens (Figure 6).
Figure 6.
Upon direct sequencing of the PCR product of p53 (370 bp), a point mutation of C to G was noted for the hamster buccal-pouch tissue specimens treated with 7,12-dimethylbenz[a]anthracene (DMBA) for 3 and 7 weeks, as compared with the untreated tissue specimens (arrow). A great variation of the sequence of p53 for hamster buccal-pouch tissue specimens treated with DMBA for 14 weeks as compared with the untreated pouch tissue specimens was found.
Statistical analyses
The mean RNA expression of these five members of the IAP family and p53 for DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis is summarized in Table 4. It was noted that the average mRNA expression in mineral oil-treated control tissues for cIAP1 is higher than that for the 3-week DMBA-treated experimental tissues. Also, the mean mRNA expression of survivin for the 14-week DMBA-treated tissues was lower than that for both the 3- and 7-week DMBA-treated tissues. It might be due to the fact that a wide range of variation was noted within these data categories (cIAP1, 3-week DMBA treatment; survivin, 14-week DMBA treatment). Upon statistical analysis, the mean RNA expression of these five members of the IAP family and p53 was significantly elevated for 3-, 7- and 14-week DMBA-treated pouch tissues compared with the untreated and mineral oil-treated tissues (P < 0.0001, one-way anova, Table 4) with the exception for comparison of the average mRNA expression in mineral oil-treated tissues for cIAP1 with that for the 3-week DMBA-treated tissues.
Table 4.
Mean mRNA expression of the five inhibitors of apoptosis family members and p53 for 7,12-dimethylbenz[a]anthracene (DMBA)-induced hamster buccal-pouch squamous-cell carcinogenesis
| Untreated | Mineral oil-treated | 3-week DMBA-treated* | 7-week DMBA-treated* | 14-week DMBA-treated* | |
|---|---|---|---|---|---|
| cIAP1 | 0.1108 ± 0.0365 | 0.2008 ± 0.0640 | 0.1518 ± 0.0602 | 0.3590 ± 0.0951 | 0.6313 ± 0.1166 |
| cIAP2 | 0 | 0 | 0.1293 ± 0.0325 | 0.2124 ± 0.0670 | 0.2759 ± 0.0887 |
| NAIP | 0.4264 ± 0.2456 | 0.4935 ± 0.0599 | 0.7286 ± 0.2263 | 0.9867 ± 0.2868 | 1.9495 ± 0.4451 |
| XIAP | 0.1294 ± 0.0376 | 0.3042 ± 0.1266 | 4.3607 ± 0.5956 | 4.6138 ± 0.5818 | 5.3556 ± 0.9270 |
| Survivin | 0 | 0 | 1.3858 ± 0.5409 | 2.3059 ± 0.3641 | 1.1561 ± 0.2815 |
| p53 | 0 | 0 | 0.1227 ± 0.0382 | 1.0079 ± 0.2870 | 1.9559 ± 0.3381 |
Statistical significance as compared with untreated or mineral oil-treated tissues with the exception for comparison of the average mRNA expression in mineral oil-treated tissues for cIAP1 with that for the 3 weeks DMBA-treated tissues (P < 0.0001, one-way anova).
cIAP1, cellular inhibitors of apoptosis protein 1; cIAP2, cellular inhibitors of apoptosis protein 2; NAIP, neuronal apoptosis inhibitory protein; XIAP, X-linked inhibitors of apoptosis protein.
On the other hand, the mean RNA expression of these five members of the IAP family for DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis was correlated to each other (linear regression analysis, Table 5); furthermore, it was significantly correlated to that of p53, with a decreasing order of correlation of NAIP, XIAP, cIAP2, cIAP1 and survivin (linear regression analysis, Table 6).
Table 5.
Correlation of the mean mRNA expression of the five inhibitors of apoptosis family members for 7,12-dimethylbenz[a]anthracene-induced hamster buccal-pouch squamous-cell carcinogenesis
| Variables | By variables | Mean ± standard deviation | Correlation coefficients (r)* |
|---|---|---|---|
| cIAP1 | 0.2871 ± 0.2004 | ||
| NAIP | 0.9168 ± 0.6198 | 0.8339 | |
| cIAP2 | 0.1235 ± 0.1227 | 0.7918 | |
| XIAP | 2.9527 ± 2.3436 | 0.6596 | |
| survivin | 0.9696 ± 0.9398 | 0.4399 |
P < 0.0001, linear regression analysis.
cIAP1, cellular inhibitors of apoptosis protein 1; cIAP2, cellular inhibitors of apoptosis protein 2; NAIP, neuronal apoptosis inhibitory protein; XIAP, X-linked inhibitors of apoptosis protein.
Table 6.
Correlation of mean mRNA expression of the five inhibitors of apoptosis family members with p53 for 7,12-dimethylbenz[a]anthracene-induced hamster buccal-pouch squamous-cell carcinogenesis (linear regression analysis)
| Variable | By variables | Mean ± standard deviation | Correlation coefficients (r)* |
|---|---|---|---|
| p53 | 0.6173 ± 0.7992 | ||
| NAIP | 0.9168 ± 0.6198 | 0.9310 | |
| cIAP1 | 0.2871 ± 0.2004 | 0.8949 | |
| cIAP2 | 0.1235 ± 0.1227 | 0.8402 | |
| XIAP | 2.9527 ± 2.3436 | 0.7183 | |
| survivin | 0.9696 ± 0.9398 | 0.4847 |
cIAP1, cellular inhibitors of apoptosis protein 1; cIAP2, cellular inhibitors of apoptosis protein 2; NAIP, neuronal apoptosis inhibitory protein; XIAP, X-linked inhibitors of apoptosis protein.
P < 0.0001, linear regression analysis.
Polymerase chain reaction-based methylation assay
Survivin and cIAP2
The methylation status of the survivin and cIAP2 genes in all of the 3-, 7- and 14-week DMBA-treated, untreated and mineral oil-treated hamster buccal-pouch tissues was investigated. For the untreated and mineral oil-treated tissues, non-digested DNA samples with and without bisulphite conversion showed PCR products of 584 bp (survivin) and 832 bp (cIAP2), respectively, whereas digested DNA samples with and without bisulphite treatment did not show any PCR products.
On the other hand, for the 3-, 7- and 14-week DMBA-treated hamster buccal-pouch tissues, non-digested DNA samples without bisulphite treatment, as well as the digested DNA samples with and without bisulphite treatment, showed a PCR product of 584 bp (survivin) and 832 bp (cIAP2), respectively, while non-digested DNA samples with bisulphite treatment did not show a PCR product. The representative results of the RT-PCR-based methylation assay are shown in Figure 7.
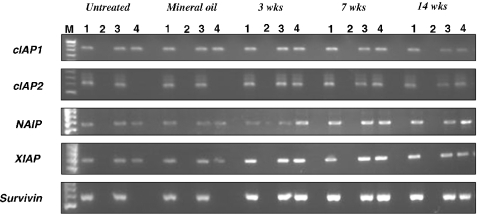
Figure 7.
Representative results of RT-PCR-based methylation assay of cIAP1, cIAP2, NAIP, XIAP and survivin genes for 3-, 7- and 14-week 7,12-dimethylbenz[a]anthracene-treated, untreated and mineral-oil treated hamster buccal-pouch tissues. Lane 1: PCR products for non-digested DNA samples without C → T conversion; lane 2: PCR products for digested DNA samples without C → T conversion; lane 3: PCR products for non-digested DNA samples with C → T conversion; lane 4: PCR products for digested DNA samples with C → T conversion; lane M: molecular weight markers.
XIAP, cIAP1 and NAIP
The methylation status of XIAP, cIAP1 and NAIP genes in all of the 3-, 7- and 14-week DMBA-treated, untreated and mineral oil-treated hamster buccal-pouch tissues was investigated. For the untreated and mineral oil-treated tissues, non-digested DNA samples with and without bisulphite treatment, as well as digested DNA samples with bisulphite treatment, showed a PCR product for XIAP (884 bp), cIAP1 (674 bp) and NAIP (650 bp), respectively, whereas non-digested DNA samples without bisulphite treatment showed no evidence of a PCR product. Similar results were obtained for all the 3-, 7- and 14-week DMBA-treated hamster buccal-pouch tissues. The representative results of the RT-PCR-based methylation assay are shown in Figure 7.
Discussion
The implication of IAP-mediated inhibition of apoptosis in some cancers has been hypothesized (LaCasse et al. 1998), and the association of IAP family members, chiefly survivin, with human oral squamous-cell carcinomas has also been studied (Tanimotoa et al. 2005; Jane et al. 2006). However, the relationship of the IAP family with chemically induced oral experimental carcinogenesis has rarely been studied. Only two IAP family members, namely survivin and XIAP proteins, have been reported to be overexpressed in DMBA-induced hamster buccal-pouch carcinogenesis recently in our laboratory (Chen et al. 2005a; Hsue et al. 2008). In the present study, besides further analysing the mRNA expression of survivin and XIAP, we also demonstrate the protein and mRNA expression of three other IAP family members (cIAP1, cIAP2 and NAIP) for DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis. This is, to the best of our knowledge, the first study to document the sequential expression (both protein and mRNA) of these five IAP family members (survivin, XIAP, cIAP1, cIAP2 and NAIP) in chemically induced oral experimental carcinogenesis. Our results indicate that expression of these five IAP family members (both protein and mRNA) is an early event in DMBA-induced pouch carcinogenesis. These findings are consistent with the results for human oral carcinogenesis (Tanimotoa et al. 2005; Jane et al. 2006). Thus, the present study further indicates that the IAP family may participate in oral carcinogenesis.
The mechanisms of IAP overexpression in cancerous tissues are largely uncertain. As all five IAP family members (both protein and mRNA) have been found to be significantly overexpressed in the present study, a synergistic effect amongst IAP family members, and therefore a probable common regulatory mechanism, might be supposed. Amplification of the survivin locus on chromosome 17 and DNA demethylation of its promoter region have been regarded as potential mechanisms of survivin overexpression in some cancers (Altieri 2001), including human oral cancer and chemically induced hamster buccal-pouch carcinoma (Tanaka et al. 2003; Chen et al. 2005a). In the present study, we have demonstrated that the control tissues (untreated and mineral oil-treated) had a survivin- and cIAP2-methylated allele, whilst the 3-, 7- and 14-week DMBA-treated tissues showed no evidence of survivin- and cIAP2-methylation. Also, neither the control nor the DMBA-treated pouches showed evidence of XIAP-, cIAP1- and NAIP-methylation. Therefore, these results indicate that overexpression of survivin and cIAP2 in DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis may be regulated by an epigenetic mechanism whereas overexpression of XIAP, cIAP1 and NAIP may also (possibly partially) modulated by the same mechanism.
On the other hand, p53 loss or mutation might result in the upregulation of survivin expression (Hoffman et al. 2002; Mirza et al. 2002), but the relevance of p53 regulation to other IAP family members has not yet been elucidated. In the present study, we demonstrate that the overexpression of these five IAP family members (both protein and mRNA) is associated with p53 mutation for DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis.
Mirza et al. (2002) and Hoffman et al. (2002) indicated that accumulation of wild-type p53 suppresses p53 promoter activity, resulting in the downregulation of survivin expression, and therefore hypothesize a negative feedback loop between survivin expression and wild-type p53. Based on the findings of the present study, overexpression of the IAP family in DMBA-induced hamster pouch squamous-cell carcinogenesis might be regulated by both genetic (mutant p53) and epigenetic mechanisms. Following gradual increasing concentration of DMBA, the induced mutant p53 in pouch keratinocytes could produce failure of the negative feedback loop originally present between IAP and wild-type p53. Consequently, the IAP family expression would become uncontrolled and hence become upregulated, a state for developing carcinoma tissues. Given that genetics and epigenetics are complementary in the field of cancer aetiology (Feinberg 2004), in the future, further study on the interaction between p53 gene expression and epigenetic alterations should be performed to evaluate the interrelationship of p53 gene mutation and epigenetic alteration in the IAP family. If an exact mechanism for the IAP family is understood, it would be a useful diagnostic marker of cancer and a potential target for the treatment of oral cancer.
In the present study, we found that the mRNA expression of these five IAP family members is correlated to each other; however, to date, the exact pathway of interaction amongst IAP family members is not yet been completely understood. A possible interaction between IAP family members has been reported; survivin was demonstrated to bind second mitochondria-derived activator of caspase (Smac), relieving its inhibition of XIAP, and then allowing XIAP to function (Song et al. 2003). Further exploration of the interaction pathways amongst IAP family members for DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis is essential in order to elucidate the association between coexpression of IAP family members and oral experimental carcinogenesis. Moreover, the finding of overexpression of multiple IAP family members in our study raises the possibility that effective strategies will require IAP antagonists that are able to suppress multiple members of the IAP family.
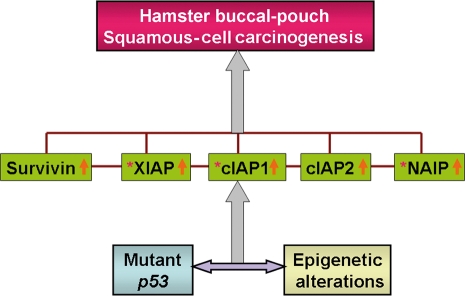
In conclusion, our findings suggest that the IAP family could play a pivotal role in DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis and what is, to the best of our knowledge, the first demonstration that their expression may be regulated by both genetic (mutant p53) and epigenetic mechanisms in this experimental model system for oral carcinogenesis, although their interrelationship and the exact interactions amongst IAP family members remain to be elucidated (Figure 8).
Figure 8.
Hypothesis of the genetic and epigenetic regulation of inhibitors of apoptosis (IAP) family members for 7,12-dimethylbenz[a]anthracene-induced hamster buccal-pouch squamous-cell carcinogenesis (brown line: interactions amongst the IAP family members remain unclear; purple double-head horizontal arrow: exact interrelationship between mutant p53 mechanism and epigenetic alteration is still uncertain; grey single-head vertical arrow: induction; orange single-head vertical arrow: overexpression of IAP members; brown asterisk: possibly partially regulated by epigenetic changes).
Acknowledgments
We wish to acknowledge the technical assistance of Ms S.M. Ji and Ms L.M. Chen. This research was supported by a grant from the National Science Council, China (NSC 95-2314-B-037-059).
References
- Altieri DC. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol. Med. 2001;7:542–547. doi: 10.1016/s1471-4914(01)02243-2. [DOI] [PubMed] [Google Scholar]
- Barnes N, Haywood P, Flint P, Knox WF, Bundred NJ. Survivin expression in in situ and invasive breast cancer relates to COX-2 expression and DCIS recurrence. Br. J. Cancer. 2006;94:253–258. doi: 10.1038/sj.bjc.6602932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YK, Hsue SS, Lin LM. Survivin expression is regulated by an epigenetic mechanism for DMBA-induced hamster buccal-pouch squamous-cell carcinomas. Arch. Oral Biol. 2005a;50:593–598. doi: 10.1016/j.archoralbio.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Chen YK, Hsue SS, Lin LM. Inhibitory effect of inducible nitric oxide synthase inhibitors on DMBA-induced hamster buccal-pouch squamous-cell carcinogenesis. Nitric Oxide. 2005b;13:232–239. doi: 10.1016/j.niox.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem. J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. IAP family proteins: suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. The epigenetics of cancer etiology. Semin. Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Gimenez-Conti IB, Slaga TJ. The hamster cheek pouch carcinogenesis model. J. Cell. Biochem. 1993;17F(Suppl.):83–90. doi: 10.1002/jcb.240531012. [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Oren M. p53 and apoptosis. Semin. Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- Gupta S. A decision between life and death during TNF-alpha-induced signaling. J. Clin. Immunol. 2002;22:185–194. doi: 10.1023/a:1016089607548. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- Hsu SM, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabelled antibody (PAP) procedures. J. Histochem. Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- Hsue SS, Chen YK, Lin LM. Expression of survivin and XIAP for DMBA-induced hamster buccal-pouch squamous cell carcinogenesis is associated with p53 accumulation. Oral Oncol. 2008;44:43–49. doi: 10.1016/j.oraloncology.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Jane C, Nerurkar AV, Shirsat NV, Deshpande RB, Amrapurkar FR, Karjodkar FR. Increased survivin expression in high-grade oral squamous cell carcinoma: a study in Indian tobacco chewers. J. Oral Pathol. Med. 2006;35:595–601. doi: 10.1111/j.1600-0714.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247–3259. doi: 10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- Lin LM, Chen YK. Diurnal variation of γ-glutamyl transpeptidase activity during DMBA-induced hamster buccal pouch carcinogenesis. Oral Dis. 1997;3:153–156. doi: 10.1111/j.1601-0825.1997.tb00027.x. [DOI] [PubMed] [Google Scholar]
- Liston P, Fong WG, Kelly NL, et al. Identification of XAF1 as an antagonist of XIAP anti-caspase activity. Nat. Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- Mirza A, McGuirk M, Hockenberry TN, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- Morris AL. Factors influencing experimental carcinogenesis in the hamster cheek pouch. J. Dent. Res. 1961;40:3–15. doi: 10.1177/00220345610400012001. [DOI] [PubMed] [Google Scholar]
- Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Preparation and analysis of eukaryotic genomic DNA. In: Sambrook J, editor. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 6.4–6.12. [Google Scholar]
- Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J. Biol. Chem. 2003;278:23130–23140. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- Tanaka C, Uzawa K, Shibahara T, Yokoe H, Noma H, Tanzawa H. Expression of an inhibitor of apoptosis, survivin, in oral carcinogenesis. J. Dent. Res. 2003;82:607–611. doi: 10.1177/154405910308200807. [DOI] [PubMed] [Google Scholar]
- Tanimotoa T, Tsudaa H, Imazekia N, et al. Nuclear expression of cIAP-1, an apoptosis protein, inhibiting in head and neck squamous cell carcinomas predicts lymph node metastasis and poor patient prognosis. Cancer Lett. 2005;224:141–151. doi: 10.1016/j.canlet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Vojtesek B, Bartek J, Midgley CA, Lane DP. An immunohistochemical analysis of the human nuclear phosphoprotein p53. New monoclonal antibodies and epitope mapping using recombinant p53. J. Immunol. Methods. 1992;151:237–244. doi: 10.1016/0022-1759(92)90122-a. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]