Abstract
The bacterial transposon Tn7 maintains two distinct lifestyles, one in horizontally transferred DNA and the other in bacterial chromosomes. Access to these two DNA pools is mediated by two separate target selection pathways. The proteins involved in these pathways have evolved to specifically activate transposition into their cognate target-sites using entirely different recognition mechanisms, but the same core transposition machinery. In this review we discuss how the molecular mechanisms of Tn7-like elements contribute to their diversification and how they affect the evolution of their host genomes. The analysis of over 50 Tn7-like elements provides insight into the evolution of Tn7 and Tn7 relatives. In addition to the genes required for transposition, Tn7-like elements transport a wide variety of genes that contribute to the success of diverse organisms. We propose that by decisively moving between mobile and stationary DNA pools, Tn7-like elements accumulate a broad range of genetic material, providing a selective advantage for diverse host bacteria.
Keywords: Transposons, Tn7, target selection, genomic islands, CRISPR
1. Introduction
Transposons are mobile genetic elements that can move between locations in DNA that lack homology. Transposons play an important role in the evolution of genomes in every domain of life through activities such as horizontal gene transfer, gene disruption, gene expression modulation, and recombination (Bushman, 2002; Kazazian, 2004; Osborn and Boltner, 2002). They are common in plasmids, integrative conjugal elements (ICEs), bacteriophages, and chromosomes, and can transfer between hosts by moving from stable chromosomal sites to mobile DNA molecules. Tn7 is a bacterial transposon that possesses two separate transposition targeting systems that take advantage of both the stability of the chromosome and the mobility of plasmids and bacteriophages for propagation, persistence, and dissemination amongst bacteria (Barth et al., 1976; Craig, 2002; Finn et al., 2007; Kubo and Craig, 1990; Peters and Craig, 2001a; Peters and Craig, 2001b; Waddell and Craig, 1988; Wolkow et al., 1996). In this review we focus on the advantages of interplay between the two transposition pathways found in Tn7-like elements. We propose that the ability to move specifically between mobile DNAs and a single neutral position found in all bacterial chromosomes offer unique benefits to both host and transposon. We also analyzed over 50 Tn7-like elements, which are easily distinguished from other transposons based on the homology of at least four unique transposition gene components, distinctive arrays of transposase binding sites, and the localization of many of these elements at a specific chromosomal location. Examining the make-up of these transposons suggests ways in which Tn7-like elements have diverged from one another and how they have affected the evolution of host genomes.
From a “gene centric” perspective of evolution we can think about the specific advantages afforded a collection of genes that can move as a group in different ways (e.g. in the form of a plasmid, bacteriophage, or transposon). A collection of genes that define a plasmid gains the ability to easily “shop around” to find the best fit in a number of bacterial hosts adapted to different environments by existing as an autonomous entity. However, plasmids can be lost from the host cell more easily than genes found on the chromosome by a variety of mechanisms (i.e. amplicon exclusion or DNA restriction and modification systems). A collection of genes found on the chromosome is not as easily lost as extra chromosomally-encoded genes, but they also lose the ability to easily sample new hosts to find the best evolutionary fit in different environments. A collection of genes that possess a programmed ability to reside in the chromosome and mobilize to new hosts benefits from the advantages of both strategies. Consistent with this idea, there are many examples in nature where mechanisms have evolved for maximizing the efficient transfer of DNA between a neutral integration site in the chromosome and into a form capable of mobilizing between hosts. Bacteriophages and ICEs are both able to utilize integration sites in the chromosome, but also have the ability to move between bacteria. Bacteriophages will frequently have a single “attachment site” in the chromosome, but also the ability to form an infecting particle to transport their genes to new hosts. ICEs are collections of genes found integrated within the chromosome that can also recombine out of the chromosome as a circle and transfer by conjugation to new bacterial hosts (Osborn and Boltner, 2002). Bacteriophages and ICEs are widespread and are also credited for the formation of pathogenicity or fitness islands (also known as genomic islands) in bacterial hosts. Genomic islands are large regions of the chromosome (>30 Kb) that have originated from horizontal gene transfer and contain genes with fitness enhancing qualities (Dobrindt et al., 2004).
There are two important downsides to the life strategy of bacteriophages and ICEs. For one, these elements often need to encode 30 or more genes to carry out their elaborate form of transport to new hosts. A second downside is that each system of packaging or conjugation also comes with some limitation on host range. As we will see with the Tn7-like elements these problems have been circumvented. Tn7-like elements can access chromosomal and mobile DNAs with exceptional host-range and without the need to carry the genes for conjugation or viral infection.
Tn7 is an intricate transposon that displays many innovations distinguishing it from other transposons. It has been extensively studied as a model genetic element and many of its activities have been described in great detail. Since its discovery as a plasmid borne antibiotic resistance determinant (Barth et al., 1976; Hedges et al., 1972), Tn7 and related elements have been found in a wide variety of clinical and environmental settings (Biskri and Mazel, 2003; Oppon et al., 1998; Orman et al., 2002; Parks and Peters, 2007; Ramirez et al., 2005a; Ramirez et al., 2005b). The Tn7 element, originally called TnC, was isolated from a trimethoprim resistant Escherichia coli that had infected a calf just two years after the introduction of this synthetic antibiotic into use in veterinary settings (Hedges et al., 1972). The element was propagated on a self transmissible IncIα plasmid, R483, however it was quickly discovered that the transposon was capable of integrating into the host genome in a highly site-specific manner (Barth et al., 1976; Lichtenstein and Brenner, 1981; Lichtenstein and Brenner, 1982).
Tn7-like elements appear to carry extremely diverse arrays of genes. These elements reside in organisms in both natural environments and those modified by humans (i.e. medical, industrial, and agricultural settings) (Parks and Peters, 2007). As investigators steadily flood genome databases with new sequences, Tn7-like elements continue to appear in unique and unusual organisms and environments. The success of these elements may be in large part attributed to extremely fine-tuned regulation of transposition (Craig, 2002; Peters and Craig, 2001b).
2. Mechanism of Tn7 transposition
2.1. General overview
Of the five transposition-associated gene products produced by Tn7, three are involved in regulation of transposition, while the remaining two carry out the actual chemical steps involved in mobilizing the element (Bainton et al., 1993; Hauer and Shapiro, 1984; Waddell and Craig, 1988)(Figure 1.A.). The “core transposition machinery” of Tn7 consists of the transposase proteins TnsA and TnsB along with a regulator protein, TnsC (Bainton et al., 1993). This core machinery is directed by one of two target selecting proteins, TnsD or TnsE (Kubo and Craig, 1990; Waddell and Craig, 1988). One target recognition pathway has evolved to maximize the efficiency of vertical transmission of the element by directing transposition into the chromosome (Lichtenstein and Brenner, 1981; Lichtenstein and Brenner, 1982), and the other pathway has been optimized for horizontal gene transfer by preferentially directing transposition into mobile, or conjugal, plasmids and filamentous bacteriophages (Finn et al., 2007; Wolkow et al., 1996).
Figure 1.
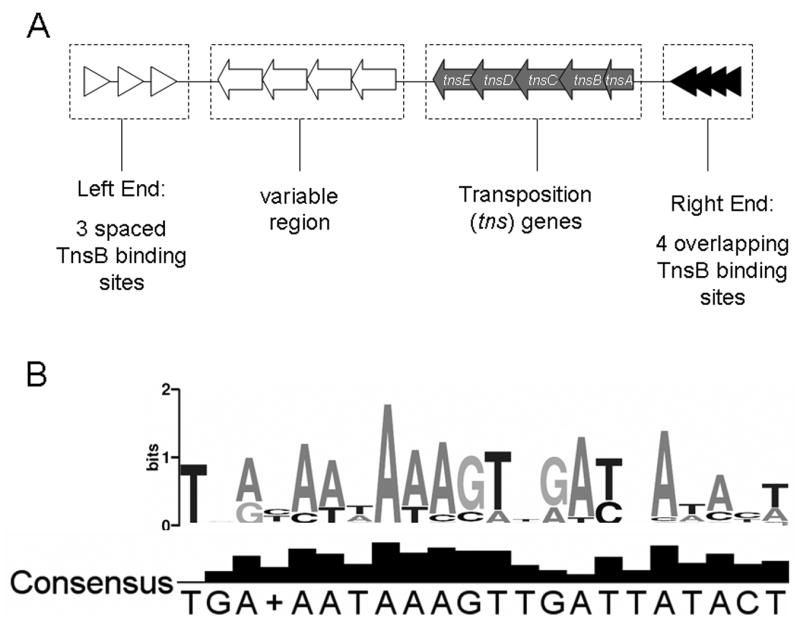
Components of Tn7-like elements. The Tn7-like elements display a high degree of diversity while maintaining conservation of components that are essential for propagation and dissemination of the element. A. A schematic representation of Tn7-like elements. The TnsB binding sites that delineate the ends of the transposon are shown as triangles. Genes involved in transposition of the Tn7 element are shown as filled arrows. The variable region in the left end of the element is represented with open arrows. B. A logo displaying the conservation of nucleotides within the TnsB binding sites within group A and B elements (see text). The logo was constructed by first aligning TnsB binding sites from the right ends of selected elements (Escherichia coli, Shigella sonnei Ss046, Pseudomonas aeruginosa, Shewanella putrefaciens CN-32, Shewanella baltica OS155, Shewanella loihica PV-4, Idiomarina loihiensis L2TR, Acidithiobacillus ferrooxidans ATCC 23270, Hahella chejuensis KCTC 2396, Pelobacter carbinolicus DSM 2380, Bacillus cereus ATCC 10987, Staphylococcus sp. 639-2, see Table S1 for accession numbers) using the ClustalW program (Chenna et al., 2003), then using the Weblogo algorithm to display homologous sites (Crooks et al., 2004).
2.2. The core transposition Machinery
The TnsA and TnsB proteins comprise the transposase component of the core Tn7 transposition machinery. TnsB is a DDE-type transposase that is a member of the retroviral integrase superfamily (Sarnovsky et al., 1996). This protein carries out the concerted breakage and rejoining reactions, joining the 3′-hydroxyl of the donor ends to 5′- phosphate groups at the insertion site of the target molecule (Sarnovsky et al., 1996). The TnsA protein structurally resembles a restriction endonuclease (Ronning et al., 2004) and carries out the nicking reaction on the opposite strand of the donor molecule, completely freeing the element and leaving behind a DNA double-strand break (DSB) (Bainton et al., 1991; May and Craig, 1996; Sarnovsky et al., 1996). The sites where the top and bottom strands are joined to the target molecule are offset by 5 bp, which results in gaps in the target DNA at either end of the newly inserted transposon. Repair of these gaps by host machinery after transposition results in a 5 bp duplication of the sequence at the point of the insertion; a size that is characteristic of Tn7 transposition (Bainton et al., 1991).
Normally, TnsA and TnsB will not carry out transposition alone. TnsC modulates the activity of the TnsAB transposase, and only activates transposition once complexed with target DNA and one of the target selection proteins, TnsD or TnsE (Bainton et al., 1993; Kubo and Craig, 1990; Waddell and Craig, 1988). TnsC is a double-stranded DNA binding protein that is believed to modulate activity through an ATPase domain (Stellwagen and Craig, 1997b; Stellwagen and Craig, 1998). This protein is considered the central regulator of transposition due to its involvement in a process called target-site immunity (see below) and its requirement in both pathways of target-site selection (Waddell and Craig, 1988).
The two target selecting proteins TnsD and TnsE act by identifying potential target molecules for insertion and only then activating the core transposition machinery through the formation of the nucleoprotein complex (Craig, 2002; Finn et al., 2007; Peters and Craig, 2001b). Specific protein-protein interactions between TnsC and target-selection proteins may not be necessary, as some TnsC mutants are responsive to certain alternative DNA structures (Rao et al., 2000).
The Tn7 right and left ends are distinct from one another in that they are composed of differing configurations of TnsB binding sites with varying affinities for the TnsB protein (Arciszewska et al., 1989; McKown et al., 1987) (Figure 1.A.). The right end contains four overlapping TnsB binding sites, while the left end contains three widely spaced TnsB binding sites (McKown et al., 1987; Tang et al., 1991). This elaborate configuration of binding sites imparts an asymmetry upon the transposon and somehow allows the unique behavior of Tn7 to control not only the position and timing of transposition, but also the left to right orientation of transposon end sequences with respect to the target DNA molecule (Bainton et al., 1991; Finn et al., 2007; Gringauz et al., 1988; Lichtenstein and Brenner, 1982; McKown et al., 1988; Peters and Craig, 2001a). The TnsB binding sites of the various elements are not identical, however they are all A+T rich and asymmetrical (Figure 1.B.). In the laboratory it has been shown that transposition can occur using two right ends, but not with two left ends (Arciszewska et al., 1989).
2.3. The TnsD-mediated pathway of transposition
The TnsD protein recognizes and binds to a highly conserved DNA sequence within the 3′ end of the glmS gene, directing insertions into a single position within the glmS transcriptional terminator. The GlmS protein (Glucosamine--fructose-6-phosphate aminotransferase) is responsible for catalyzing the first step in hexosamine metabolism and is essential for host cell survival (Teplyakov et al., 1999). There are some cases in which rare alternate sites can be utilized by the TnsD pathway because they have a DNA sequence that is similar to the one found in the glmS gene such as the carA gene in Proteus miabilis HI4320 (Choi and Schweizer, 2006; Kubo and Craig, 1990). However, the overwhelming preponderance of Tn7 elements that insert into bacterial chromosomes are found proximal to the glmS gene (Oppon et al., 1998; Parks and Peters, 2007). The TnsD binding site within the glmS open reading frame and the actual position of Tn7 insertion within the transcriptional terminator are collectively referred to as the Tn7 attachment site, or attTn7 (Figure 2.A.)(Gary et al., 1996; Lichtenstein and Brenner, 1982; McKown et al., 1988). Insertion into attTn7 occurs at a very high frequency (∼10-1-10-2 chromosomal insertions per donor plasmid (DeBoy and Craig, 1996) with no detectable deleterious effect and the insertion event is stable even without selection (McKenzie and Craig, 2006). When TnsD binds the attTn7 site it causes a distortion in the duplex DNA structure, which in turn recruits TnsC and activates transposition (Kuduvalli et al., 2001). Transposition into attTn7 is very specific, almost always occurring at a single set of base pair junctions within a given organism with the right end of the element proximal to the end of the glmS open reading frame (DeBoy, 1997; Gringauz et al., 1988; Lichtenstein and Brenner, 1981; Lichtenstein and Brenner, 1982). Some organisms contain more than one glmS homolog and therefore more than one attTn7 that can be utilized by the TnsD pathway (Choi et al., 2006; Kuduvalli et al., 2005). Analysis of Tn7-like elements indicates that the distance between the site of insertion and the end of the glmS open reading frame varies slightly between organisms (Craig, 1989; Parks and Peters, 2007). The TnsD protein also interacts with two host proteins, ribosomal protein L29 and acyl-carrier protein (ACP), which appear to stabilize the nucleoprotein complex and stimulate TnsABC+D transposition (Sharpe and Craig, 1998). Interaction with these proteins may provide the Tn7 element with cues regarding host cell metabolic state or growth phase, further refining regulation of transposition (Sharpe and Craig, 1998).
Figure 2.
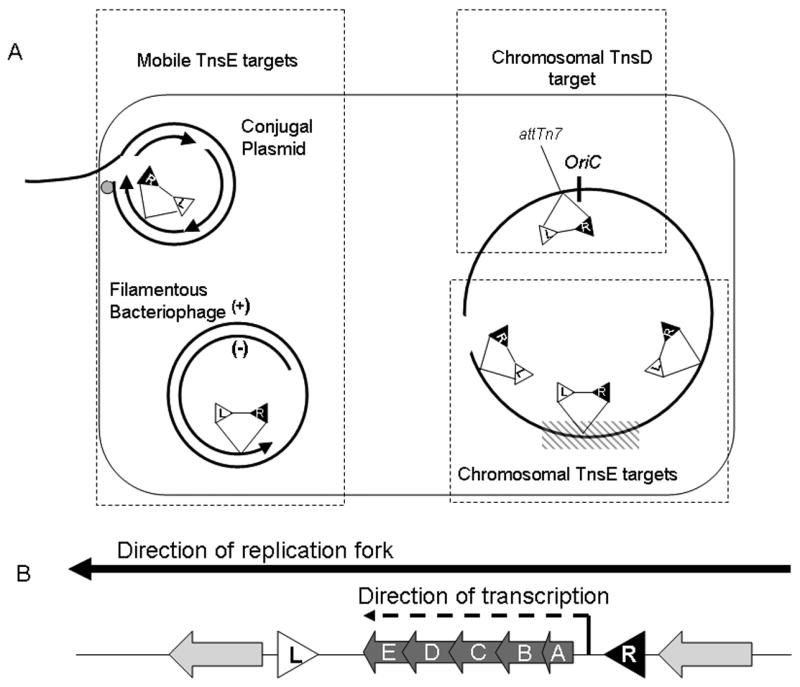
Genomic context of Tn7 insertion. A. A schematic representation of Tn7 targeting found with TnsD-mediated and TnsE-mediated transposition. TnsD insertions occur within a single site in the chromosome with a single left to right orientation (“L” represents left end, and “R” represents right end). TnsE preferentially targets mobile DNA. The small circle represents the relaxase that covalently binds the 5′ end of mobile plasmids, arrowheads represent 3′ ends, and arrow tails represent 5′ ends. The (+) and (-) distinguish the opposite strands of filamentous bacteriophages. At a low frequency TnsE can insert into the chromosome. Chromosomal targets include sites proximal to DSBs (opening in chromosome), and the region of the chromosome where DNA replication terminates (crosshatched box). The orientation bias of TnsE-mediated insertions are reversed in each replichore (right and left side of oriC in the chromosome) and is dependent on the direction of the replication fork. B. Both TnsD and TnsE mediate insertions that align the direction of the tns gene transcription with the direction of replication passage, minimizing head-to-head collisions with DNA replication machinery and transcription machinery. The tnsABCDE genes are indicated (A,B,C,D,E).
The glmS gene is essential to the host and the primary amino acid sequence is well conserved in bacteria, making recognition of the coding sequence of this gene an elegant adaptation for Tn7. In addition, the location of the glmS gene could also be considered advantageous for Tn7. Even in diverse species of bacteria the glmS gene is nearly always located close to the chromosomal origin of replication, and is therefore replicated early in the cell cycle. Since Tn7 is a cut-and-paste transposon it leaves behind a DNA double-strand break (DSB) following excision (Bainton et al., 1991). A copy of the element is usually regenerated once the DSB is repaired by homologous recombination with the sister chromosome (Hagemann and Craig, 1993). The location of attTn7 increases the probability that the DSB created by excision of the element is able to be repaired, and the transposon regenerated, even in slowly dividing cells. This observation is supported by work in vivo which showed that an interrupted lac operon near attTn7 is restored to a functional Lac+ status by homologous recombination with an adjacent defective lac operon following the excision of Tn7 (Hagemann and Craig, 1993).
Some Tn7-like elements possess two distinct tnsD genes that range between 41% and 70% similarity (25%-54% identity) (Table S1). No other tns genes appear to have been duplicated in a similar way, so it is likely that this duplication constitutes an adaptation that in some way benefits the transposon. For example, the two TnsD proteins may be adapted to efficiently find attachment sites in phylogenetically distant organisms. Alternatively, unlike in Tn7, one of the TnsD proteins might actually allow non-specific target site recognition. This would be consistent with the observation that elements encoding two distinct TnsD proteins are common in Tn7-like elements that are not found within the attTn7 site (Table S1). The presence of two distinct tnsD genes is common and appears in ∼9/50 elements (Table S1). While it is possible that the additional TnsD proteins may identify attachment sites that are yet to be described, we find no evidence for a specific alternate attachment site in the available DNA sequences. There is no evidence of homology between TnsD and the other target-site selecting protein, TnsE, and therefore it appears unlikely that tnsE arose from tandem duplication of tnsD.
2.4. The TnsE-mediated pathway of transposition
While the TnsD pathway is exceptional at directing transposition into chromosomes and promoting vertical propagation of Tn7 to a host's progeny, the TnsE pathway maximizes horizontal transfer of the element (Finn et al., 2007; Wolkow et al., 1996). TnsE recognizes an entirely different class of target that is not at the level of nucleotide sequence (Finn et al., 2007; Peters and Craig, 2000; Peters and Craig, 2001a; Peters and Craig, 2001b; Wolkow et al., 1996). Instead TnsE-mediated transposition occurs by identifying an aspect of lagging-strand DNA synthesis (Peters and Craig, 2001a; Peters and Craig, 2001b). This pathway of transposition preferentially recognizes mobile plasmids as they enter the Tn7 containing host cell (Wolkow et al., 1996) (Figure 2.A.). By targeting a process that occurs during conjugal replication within a new host, Tn7 gains access to DNA that is actively being transported amongst bacteria, therefore increasing the likelihood that the transposon will be transported to a new host. Tn7 is able to achieve this without the evolutionary trade off that other transposons face by selecting targets at random, which will risk the disruption of essential host functions or insertion at positions that otherwise adversely effect gene expression.
It is the process of conjugal replication that activates plasmid DNAs as targets for TnsE-mediated transposition. Vegetative replication does not stimulate or preferentially attract transposition into plasmids via the TnsE pathway. However, mobilization of oriT containing plasmids by Tra functions supplied in trans is sufficient to activate transposition into the oriT-containing targets (Wolkow et al., 1996). Stimulating conjugal DNA transfer and replication also stimulates TnsE-mediated transposition. The nature of conjugal DNA replication suggests that TnsE preferentially recognizes lagging-strand DNA replication (Peters and Craig, 2001a; Peters and Craig, 2001b). As conjugal plasmids enter a new host cell they are replicated by the host DNA replication machinery using a discontinuous process akin to lagging-strand DNA synthesis (Lawley et al., 2004). The major difference between vegetative replication of a plasmid and transfer associated replication is that leading- and lagging-strand DNA synthesis occur in two physically isolated locations. Leading-strand synthesis occurs in the donor cell as the transferred strand is displaced and transported into the recipient cell, 5′ end first. Lagging-strand synthesis is carried out by the DNA replication machinery of the new recipient host cell, concurrent with transport of the DNA strand into the cell (Lawley et al., 2004). TnsE-mediated transposition events occur with the right end of the transposon in register with the 3′ end of the nascent Okazaki fragment of the conjugal plasmid within the recipient cell (Figure 2.A.)(Wolkow et al., 1996).
Analysis of TnsE-mediated transposition within the chromosome strongly supported the view that the target of the TnsE pathway is lagging-strand DNA synthesis (Peters and Craig, 2001a; Peters and Craig, 2001b). TnsE-mediated insertions occasionally occur in the chromosome, albeit at a much lower frequency than observed in conjugal plasmids. Some naturally occurring Tn7-like transposons can be found in chromosomal locations that are consistent with TnsABC+E transposition (Parks and Peters, 2007). Chromosomal TnsE-mediated insertions are enriched in the region of the chromosome where replication terminates (Peters and Craig, 2000) (Figure 2.A.), and occur with a conspicuous left to right end orientation bias that is dependent on the direction of the replication fork that has duplicated a given section of the chromosome (Peters and Craig, 2000; Peters and Craig, 2001a). All bacterial chromosomes are replicated bi-directionally by two replication forks that emanate from a single origin. In the case of circular bacterial genomes these replication forks meet at the terminus region, dividing the chromosome into two replichores (Sherratt, 2003). TnsE-mediated insertions occur with an opposite orientation of Tn7 ends in each replichore in a configuration that is consistent with the recognition of chromosomal lagging-strand DNA synthesis (Peters and Craig, 2001a). The resulting orientation of transposon ends is congruent with the right end of the transposon inserting in register with the predicted direction of the 3′ end of the nascent Okazaki fragment on the lagging-strand, just as they occur in conjugal plasmids.
In addition to Tn7 other mobile elements, including the IS200/IS605 transposon family and the RmInt1 group II intron, which utilize very different mechanisms of mobilization demonstrate lagging-strand specific insertion (Barabas et al., 2008; Nisa-Martinez et al., 2007). Interestingly, recombination with bacteriophage lambda also appears to act on the DNA strand undergoing lagging-strand DNA replication (Court et al., 2002). The evolutionary convergence of highly divergent mobile elements toward recognition of a common target suggests either a strong selective advantage for the specific recognition of these targets or a particular vulnerability of this strand.
In the laboratory it was found that half of all the TnsE-mediated insertions isolated from cells with actively conjugating plasmids were shown to insert in a single orientation near the oriT locus, within 1-2 Kb of the 5′ end that initially entered the cell (Peters and Craig, 2001a; Wolkow et al., 1996). This may benefit Tn7 by placing it on a segment of DNA that enters new host cells first, allowing early expression of transposon genes. Transposition into the attTn7 locus on the chromosome could occur as soon as possible to escape host defenses, such as DNA restriction systems, that are activated by the incoming plasmid.
There is an example of a Tn7-like element found within Escherichia coli APEC O1, pAPEC-O1-R (DQ517526), and Serratia marcescens, R478 (BX664015)(Gilmour et al., 2004; Johnson et al., 2006) that appears to lack a tnsE gene. Intriguingly, all the other components that are necessary for transposition of the element appear to be intact. In this case we cannot rule out the idea that recombination within the element could have removed the tnsE gene following insertion into the plasmid. However, there could be other explanations for Tn7-like elements inserting into conjugal plasmids without the gene encoding TnsE. For example, these insertions may have resulted from TnsE proteins produced in trans from a gene encoded by a similar, more complete, Tn7-like element. Alternatively, these elements may contain mutations in the core transposition machinery that allow transposition to occur randomly. The TnsB proteins from both of these elements contain residues that were previously shown to allow random transposition when isolated in a laboratory screen for function in the absence of target selecting proteins (see below)(Lu and Craig, 2000). It will be interesting to know how different Tn7-like elements interact and if they are able to mediate the excision and insertion of related elements.
TnsE is also capable of directing transposition into replicating filamentous bacteriophages (Finn et al., 2007). Analysis of the TnsE-mediated insertion events in bacteriophage M13 revealed that, similar to those found in mobile plasmids, they occurred almost exclusively in a single orientation (i.e. transposon ends had the same left-to-right direction in 23 out of 24 insertions with respect to the 5′-3′ direction of the (-) strand). The orientation of Tn7 ends after insertion into M13 is consistent with recognition of minus-strand DNA replication. Minus-strand synthesis produces a DNA strand complementary to the incoming single-stranded bacteriophage genome to make the double-stranded (replicative) form of the bacteriophage (Finn et al., 2007). The efficiency and rapidity of TnsE-targeted transposition is illustrated by insertions that occur into bacteriophage M13. For transposition to occur, the target DNA must be double-stranded, a condition that exists for a limited time in the lifecycle of a filamentous bacteriophage. In contrast to the F plasmid, the M13 genome is small and replication is not required to be concurrent with entry into the cell. No insertions into the M13 genome were detected using a mutant TnsABC machinery that is capable of random transposition, suggesting that a specific targeting pathway is required to achieve insertion into the short-lived duplex filamentous bacteriophage genome that represents a minority of total DNA within a cell (Finn et al., 2007).
The resulting left-to-right alignment of the transposon ends with the direction of DNA replication may constitute a biological adaptation. The orientation bias observed both with TnsE- and TnsD-mediated insertion positions the tns genes of Tn7 in such a way that the direction of their transcription does not oppose the direction of replication (Figure 2.B.). The orientation bias may therefore limit damaging head-on collisions between DNA replication machinery and transcription machinery. This feature of transposition may be more important in organisms such as Bacillus, where coordination of the directionality of transcription and replication appeared to be more stringently enforced than in organisms such as E. coli (Wang et al., 2007). Interestingly, all of the lambda-like phage that have been examined have also evolved to insert in one orientation where transcription of the large operons occur in the same direction as DNA replication of the host (Campbell, 2002). The orientation of open reading frames other than the tns genes within Tn7-like elements is less conserved, and sometimes is found opposing the direction of tns gene transcription. Observation of the orientation of open reading frames within these elements and measurement of their transcriptional activity may serve as an indicator of an organism's tolerance to transcription and replication collisions. All open reading frames within the Tn7-like element found in the Staphylococcus plasmid pLEW6932 are oriented in the same direction (Parks and Peters, 2007), while those found in Tn7 from E. coli can be divergent from the tns genes (Craig, 2002).
TnsE-mediated transposition is stimulated by induced DNA double-strand breaks in the chromosome (Peters and Craig, 2000). In these cases insertion events occur proximal to the location of an induced DSB and tend to do so in hotspots that can be hundreds of kilobases distant from the initial break site (Shi et al., 2008). TnsE-mediated transposition appears to recognize factors associated with the repair of the DSB, such as replication mediated repair (Shi et al., 2008). Transposition into chromosomes via the TnsE pathway is possible in recA- cells, indicating that homologous recombination is not essential for TnsE-mediated transposition. It is somewhat premature to speculate about what is recognized during DSB repair in the chromosome, given that the molecular complex recognized by TnsE during replication has not been defined; however, it is possible that the mechanism that recognizes DSB repair also facilitates Tn7 transposition into bacteriophage that utilize homologous recombination as an important step in their own replication. Homologous recombination is important for replication in many bacteriophages including lambda and T4 (Mosig, 1998; Smith, 1983; Weigel and Seitz, 2006). These elements have not been thoroughly examined for their ability to serve as targets for TnsE-mediated transposition, and may be interesting topics of future research.
The mechanism of TnsE target identification has yet to be completely solved. It is known is that TnsE is a DNA binding protein that preferentially binds to DNA structures presenting a 3′ recessed end, as might be expected in incomplete Okazaki fragments (Peters and Craig, 2001a; Peters and Craig, 2001b). Where tested, randomly isolated mutations in TnsE that increase TnsABC+E transposition frequency display enhanced DNA binding ability (Peters and Craig, 2001a). Most of these mutations are located in the C-terminus of TnsE, but one increased-activity mutation (M37I) can be found in the N-terminus. The DNA binding activity of M37I is not yet known. An intriguing idea is that M37I may represent a class of mutation that alters protein-protein interactions either between TnsE and the core machinery or between TnsE and an unknown host factor.
It is likely that host factors aid in the identification of TnsE targets, just as L29 and ACP are involved in TnsD-mediated insertion (Sharpe and Craig, 1998). TnsE may interact with host proteins in such a way that provides cues based on the replication status of a given DNA molecule. Interaction with a given host factor may explain why conjugal replication stimulates TnsE-mediated transposition to a greater degree than chromosomal DNA replication. The analysis of TnsE-mediated transposition events continues to be a useful tool for understanding the replication and propagation of mobile DNA and will likely provide additional information about the modes and progression of DNA replication of diverse biological entities.
2.5. Mutant forms of the transposition machinery have been isolated that are capable of non-targeted transposition and can circumvent target-site immunity
Tn7 exhibits a property known as target-site immunity that allows the element to sense the presence of a preexisting copy of the transposon within a DNA region and prevent subsequent transposition into this region (Arciszewska et al., 1989). This principle is mediated by the ends of the transposon and by the TnsB and TnsC proteins. TnsB binds to the ends of the element and causes a redistribution of the TnsC protein away from these end sequences, presumably by stimulating the ATPase activity of TnsC (Arciszewska et al., 1989; DeBoy and Craig, 1996; Skelding et al., 2003; Stellwagen and Craig, 1997a). The ability of target site immunity to discourage transposition can even be detected between sites that are >190 Kb apart (DeBoy and Craig, 1996). The efficiency of this process is significant, but not complete. In vivo, target-site immunity prevents ∼90% of potential insertions from occurring in occupied target sites (DeBoy and Craig, 1996). Mutations in TnsB and TnsC have been isolated in the laboratory that reduce the efficiency of target-site immunity up to 20-fold (Skelding et al., 2003; Stellwagen and Craig, 1997a).
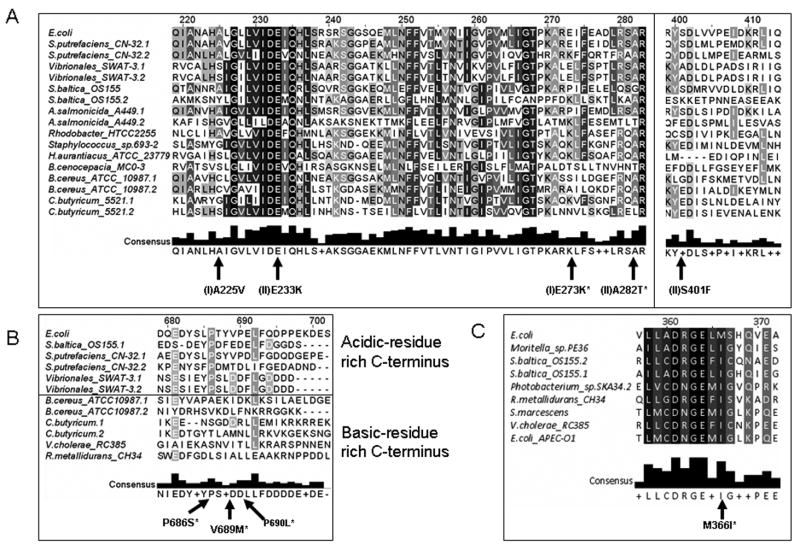
Mutations in tnsC that allow the TnsABC machinery to work in the absence of the TnsD and TnsE proteins have been generated in the laboratory and are categorized in two classes based on the ability to maintain target-site selection and target-site immunity (Stellwagen and Craig, 1997b). While these mutations were isolated in the lab, both classes of mutations appear to be represented in naturally occurring TnsC homologs (see Figure 3.A.). The class I mutations in TnsC catalyze transposition to random sites in the presence of TnsAB transposase alone, but will still avoid transposition into targets that already contain a Tn7, and can still be directed to target-sites by TnsE or TnsD when present (Figure 3.A.). Class II mutations allow transposition with TnsA and TnsB, but do not facilitate target-site immunity and are unresponsive to signals from the target selection proteins. TnsCA225V, a class I mutant, hydrolyzes ATP more slowly than wild-type, and may therefore remain in an “on” state and mediate Tn7 insertions into non-traditional targets (Stellwagen and Craig, 1997b). Insertions that occur via the TnsCA225V mutant pathways lack recognizable sequence specificity (Seringhaus et al., 2006; Stellwagen and Craig, 1997b). Altered or defective ATPase activity of TnsC could explain the existence of some Tn7-like transposons in chromosomal sites other than the attTn7 site.
Figure 3.
Alignment of selected regions of TnsB and TnsC amino acid sequences. TnsC and TnsB homologs from Tn7 and relatives were aligned using the ClustalW algorithm and edited using the Jalview software package (Chenna et al., 2003; Clamp et al., 2004). A. Alignment of selected TnsC homologs reveals possible molecular explanations for insertion into chromosomal sites other than attTn7. Sites of Class I (I) and Class II (II) mutations are indicated with arrows. Class I mutations result in random transposition, yet retain sensitivity to target selection signals. Class II mutants are insensitive to target selection signals. Sites where homologs contain the exact residue recovered in mutant screens of TnsC from E. coli are marked with an asterisk. B. Alignment of TnsB proteins reveals possible variations in interaction surfaces between TnsC and TnsB, as well as residues known to result in target-site immunity bypass. Sites resulting in target-site immunity bypass are indicated with arrows. Residues that have been found both in TnsB homologs and in mutant screens for loss of target-site immunity are indicated with asterisks. Group A elements (see text) contain TnsB proteins with C-termini dominated by acidic residues (aspartate (D) and glutatmate (E)), while Groups B and C (see text) contain elements with C-termini residues that are predominantly basic (lysine (K) and arginine (R)).
C. An alignment of a region from TnsB homologs containing the M366I substitution. The M366I mutation was isolated in a Tn7 laboratory screen, allowing transposition in the absence of target-site selecting proteins (Lu and Craig, 2000). The incidence of the M366I mutant allele in Tn7 relatives may explain the presence of some elements in locations that are not consistent with TnsD- or TnsE- mediated insertion.
Mutations in the tnsA and tnsB genes have also been shown to allow TnsABC transposition without the use of target-site selection proteins. However, the molecular details of how these mutants allow untargeted transposition are not entirely clear (Lu and Craig, 2000). One example, TnsBM366I, appears to be common in many (∼8/50) TnsB homologs (Figure 3.C.). This mutation represents a class of TnsB mutation that is stimulated by target-site selection proteins TnsD or TnsE and allows random transposition to occur (Lu and Craig, 2000).
Mutations were isolated in the laboratory in TnsB that abrogate immunity. TnsB mutations that reduce immunity can impair the TnsB-TnsC interaction, but do not necessarily reduce transposition (Skelding et al., 2003). Some of the same amino acid changes that have been identified in TnsB and TnsC that attenuate target-site immunity can be identified in naturally abundant Tn7-like elements (Figure 3)(Skelding et al., 2003; Stellwagen and Craig, 1997b). However, the ability to circumvent target-site immunity in these homologs has yet to be determined experimentally. TnsB mutations that allow the loss of target-site immunity may encourage the accumulation of multiple Tn7-like elements in attTn7, contributing to the formation of genomic islands in the attTn7 locus (see below)(Parks and Peters, 2007). Since it is possible to maintain the ability to transpose and lose target-site immunity (Skelding et al., 2003), there may be a dynamic relationship between Tn7-like elements, preventing or allowing tandem insertion to occur depending on selective pressures.
3. Formation of genomic Islands within the attTn7 locus
The extreme target-site specificity of the TnsD pathway has some very interesting consequences for bacterial genomes. While target-site immunity generally prevents multiple insertions of Tn7 within attTn7, many organisms contain multiple Tn7-like elements inserted in tandem within this very specifically defined DNA locus. These insertions are striking in that while some of the elements appear to be highly divergent, they still insert in the exact same position relative to glmS. This is evidenced by the fact that they duplicate exactly the same, or very slightly overlapping, 5 bp sequence upon insertion (Parks and Peters, 2007). These elements accumulate within the attTn7 locus and lead to the formation of genomic islands. In the most extreme example, found in Clostridium butyricum 5521, there are three complete or nearly complete Tn7-like elements. The remnants of a right end from a fourth element also resides adjacent to the left end of the last complete element (Table S1).
Within the Shewanella genus alone, of which many attTn7 loci have been sequenced, genomic islands can be observed in each of various stages of development (Figure 4). Shewanella oneidensis MR-1 may be seen as a typical naïve Shewanella species with no evidence of Tn7 insertion proximal to the glmS gene (Figure 4.A.). From S. oneidensis and other closely related Shewanella species the genes flanking attTn7 can be identified, indicating that no genes whatsoever are located within attTn7 locus, making it possible to define the empty attTn7 with relative certainty (Shewanella oneidensis MR-1(AE014299), S. baltica OS195 (AATK00000000), S. sp. MR-4 (CP000446), S. sp. W3-19-1 (AALN01000053), S. sp. MR-7 (AALI01000045), S. sp. SAR-1 (AACY01051759), and S. sp. ANA-3 (AALH01000050))(Parks and Peters, 2007). Shewanella putrefaciens 200 presents an example of an individual insertion (Figure 4.B.). Shewanella putrefaciens CN-32 is found to possess two Tn7-like insertions in tandem (Figure 4.C.)(Parks and Peters, 2007). An unrelated organism, Clostridium butyricum 5521, displays an example of an intermediate step in which the right end of the oldest element remains in a degenerate state while the other functional components of this particular element are missing (Figure 4.D., Table S1). Shewanella denitrificans OS217 (Figure 4.E.) maintains horizontally transferred genes that are important for its survival, including denitrification genes, within the attTn7 locus yet contains no recognizable Tn7 remnants (Parks and Peters, 2007). It is possible that acquisition of Tn7-encoded genetic material has lead to the specialization that differentiates some Shewanellae from one another.
Figure 4.
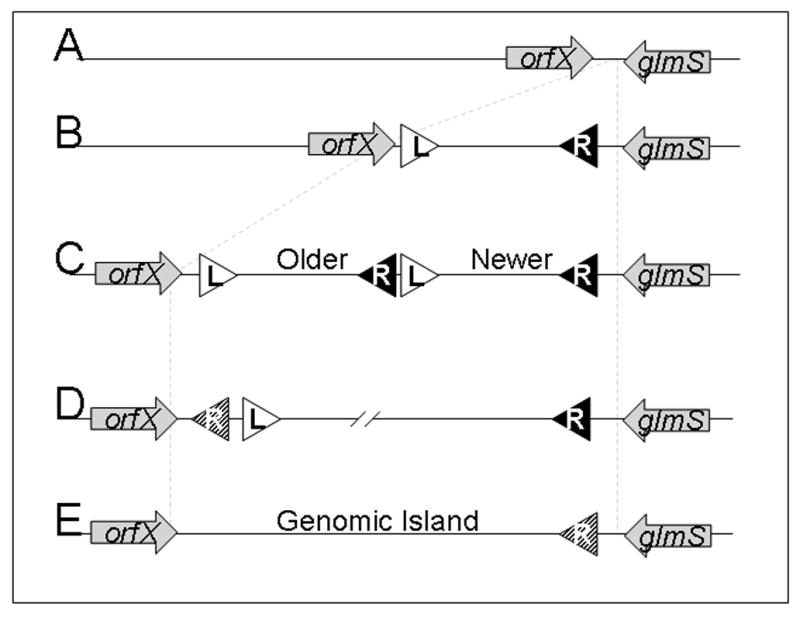
A schematic depiction of the genesis of genomic islands by Tn7-like transposons. A. A representation of a hypothetical attTn7 site flanked by glmS and a hypothetical conserved gene orfX. B. An individual insertion can be large enough alone to constitute a genomic island. Individual insertions also increase the amount of “neutral” DNA space that may allow other elements to safely insert into the chromosome. C. A tandem insertion of another Tn7-like element adjacent to glmS expands the attTn7 locus resulting in newer elements being closer to the glmS open reading frame. D. Reductive evolutionary processes, such as recombination, contracts the attTn7-associated genomic island and fixes certain genes in place by disabling critical transposon genes and transposon ends. E. Further reductive evolution fixes genes with consistent selective pressure and leaves behind only scant evidence of transposon components. Arrows represent open reading frames, while triangles represent transposon ends; (R) for right ends and (L) for left ends. Crosshatched triangles represent degenerate ends that have few TnsB binding sites and poorly conserved sequences.
The maximum distance between transposon ends that still allows transposition has not yet been experimentally defined for Tn7. However, the largest single insertion found in the environment is found within Hahella chejuensis KCTC 2396, and is 46 Kb in length (Table S1). This element is most likely able to transpose in its entirety. The entire length of the element has a significantly lower G+C content than the rest of the genome, its right and left ends appear to be intact, and all of the genes involved in transposition lack interruptions (data not shown). An interesting, but unanswered, question is whether tandem insertions can be mobilized together as a single unit using the outermost right and left ends. The presence of closely related elements within a single organism also presents the possibility of mobilization in trans. This process could allow a newly invading Tn7-like element to mobilize previously inactive Tn7 relatives, or complement missing transposon functions in multiple inactivated elements.
As explained above, target-site immunity only decreases transposition ∼90%. Repeated exposure to Tn7-like elements in the environment could overcome immunity; however, there are also several molecular explanations which could account for the tandem insertion of Tn7-like elements. First, in most cases the transposon ends and proteins are not identical to one another. Nucleotide sequence differences between TnsB binding sites may prevent heterologous TnsB proteins from recognizing binding sites of a related element. Secondly, differences in amino acid sequence of either the TnsB or TnsC proteins from different elements may lead to loss of target-site immunity in a way that maintains the productive interactions required for transposition (see above). Finally, some elements appear to contain three overlapping TnsB binding sites as opposed to the usual four, possibly reducing the local concentration of TnsB enough to have an effect on immunity, but still allowing transposition to occur.
Insertion of multiple Tn7-like elements within the attTn7 locus may lead to enhanced recombination between divergent Tn7-like elements. As described above, the DSB that is created once an element is mobilized to another location is typically fixed by homologous recombination with the sister chromosome. When multiple Tn7-like elements occupy an attachment site, recombination may occur between neighboring elements creating hybrid elements. Recombination may also be responsible for the loss of essential transposition components resulting in the immobilization of transposon encoded genes within the attachment site (Figure 4)(Parks and Peters, 2007).
The ability of Tn7 to form genomic islands may become a useful tool in the laboratory. Tn7 has widely been used to insert genes in single copy into the chromosome of diverse organisms (Choi et al., 2006; Choi et al., 2005; McKenzie and Craig, 2006). Using elements of differing origins may enable investigators to insert various combinations of genes stably into the chromosome with minimal cloning steps.
4. Genes transported by Tn7-like elements
In most Tn7-like elements, diverse arrays of genes reside in a highly variable region in the left end of the transposon (Figure 1.A.). Maintaining the genes involved in transposition in the right end of the transposon appears to be important for the function of the transposon in some way; the tns genes are almost always localized to the extreme right end of the element and with a highly conserved synteny.
Tn7 contains a class II integron system that is composed of an integrase gene, intI2, along with antibiotic resistance gene cassettes dhfr, sat, and aadA, each flanked by class II integron recombination sequences. The integrase gene is interrupted by an ocher stop codon, which can be found at the same position in many sequenced intI2 genes. Based on its conservation, it has been proposed that the stop codon within the integrase is in fact a regulatory mechanism, not simply a random nonsense mutation, preventing wanton recombination of the integron under normal conditions (Hansson et al., 2002). Elements that appear to be otherwise identical to the original Tn7 contain additional integron cassettes that encode genes for chloramphenicol and kanamycin resistance, among others, suggesting that recombination within the integron region of Tn7 is indeed active (Biskri and Mazel, 2003; Orman et al., 2002; Ramirez et al., 2005a; Ramirez et al., 2005b). Elements that are only slightly divergent from Tn7 contain entirely different integron systems (Barlow and Gobius, 2006; Parks and Peters, 2007).
While the integron cassette system of Tn7 will allow genetic content to vary significantly through the integron-catalyzed shuffling of gene cassettes, other Tn7-like elements also encode a wide variety of genes apart from or instead of integron cassettes. The contents of the various Tn7-like elements can be determined by locating the ends of the elements and analyzing the genes encompassed by the ends and searching for homologs in various gene databases (Parks and Peters, 2007). This type of analysis reveals that elements that have nearly identical tnsABCDE genes can contain vastly different genetic cargos (Parks and Peters, 2007). Just as in Tn7, the tnsABCDE genes typically reside within the right end of the element while the genetic cargo occupies the left end (Figure 1.A.). There appears to be no unifying theme with regard to the types of genes that are carried by Tn7-like elements. Tn7-like elements contain genes involved in metal resistance and detoxification, putative DNA repair enzymes and polymerases, DNA restriction and modification systems, non-ribosomal peptide synthesis modules, siderophore production genes, efflux pumps, transcription regulators, bacteriophage associated genes, additional transposases and integrases, and many hypothetical genes of unknown function (Figure 5).
Figure 5.
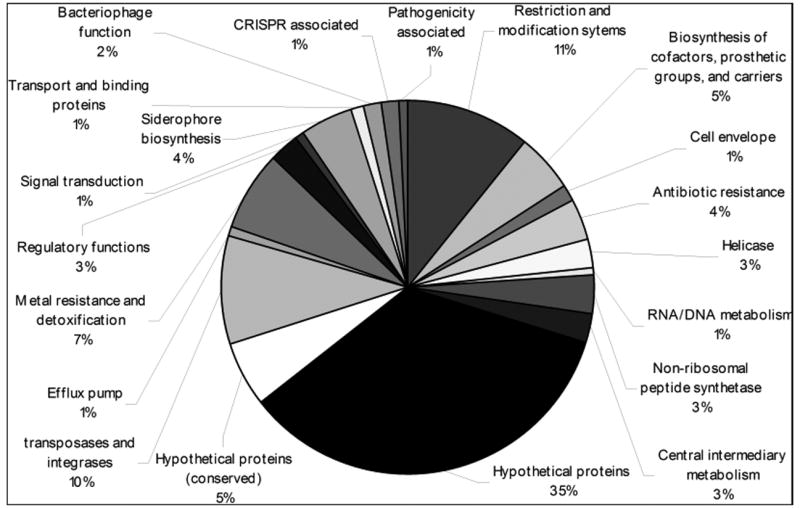
A chart describing functional categories of genes carried by Tn7-like elements. Tn7-like elements encode diverse arrays of genes with many functions. Genes were selected from elements in which the right and left ends have been determined (Table S1). Genes were then grouped by functional categories of closest matches using the BLAST algorithm to genes that reside between the right and left ends of Tn7-like elements. Functional categories of genes are loosely modeled after CMR-TIGR functional categories (Peterson et al., 2001).
DNA restriction and modification systems are an especially common feature in Tn7-like elements. These systems likely serve as host addiction systems in which the host genome is modified by a labile methyltransferase and all DNA that lacks the methylation signature is restricted by a more stable endonuclease protein, preventing loss of the transposon that encodes the methyltransferase (Kobayashi, 2004). These same systems may also help protect host cells from invasion of potentially deleterious foreign DNA by restricting intruding DNAs (i.e. bacteriophages and plasmids) before they are able to establish the methylation signature of the host genome (Kobayashi, 2001). The restriction and modification systems may also contribute to propagation of Tn7-like elements by causing DNA double-strand breaks that must be fixed by host machinery for survival. Tn7 is able to use the TnsE pathway to insert proximal to DSBs and may be able to use this facet of restriction and modification systems to move to new, potentially mobile, DNA molecules when stresses prevent sufficient methyltransferase activity. It is possible that DNAs that are easily fixed by homologous recombination once restricted, such as multicopy plasmids or lytic bacteriophages, may be activated as targets for transposition once methyltransferase activity reaches a critically low level.
Mechanisms of host protection in general appear to be a common theme in Tn7-like transposons. There is one example of a Tn7-like element within the genome of Anabaena variabilis ATCC 29413 in which a CRISPR (clustered, regularly interspaced short palindromic repeats) locus can be found. CRISPR regions have recently been shown to constitute a primitive immune system in archaea and bacteria that operates by mechanism that is not yet understood (Sorek et al., 2008). These systems are characterized by segments of short repeats that are regularly interspersed with DNA sequences that originate from and, in some way, provide protection from a broad range of invasive bacteriophages, plasmids, and transposons. The CRISPR associated genes (also found in the Tn7-like element of Anabaena variabilis ATCC 29413) are thought to affect the restriction of invading DNA and add short segments of the offending DNA sequences to the CRISPR region for future identification of foreign DNA. CRISPR loci have previously been described on plasmids (Godde and Bickerton, 2006), but to our knowledge, this is the first example in which the system has been found within a transposon.
In addition to the known medical importance of Tn7, some organisms that have emerged as pathogenic threats carry Tn7-like elements with especially intriguing genetic contents. A Tn7-like element within the virulent and multiply antibiotic resistant Acinetobacter baumanii ATCC 17978 contains a probable multidrug eflux pump, siderophore genes, and non-ribosomal peptide synthesis genes (Vallenet et al., 2008), suggesting that the transposon may contribute significantly to the pathogenicity and persistence of this organism (Iacono et al., 2008). A closely related yet non-virulent strain (Acinetobacter baumanii SDF) also contains a highly similar Tn7-like element which lacks the siderophore production and antibiotic resistance genes (Iacono et al., 2008), underscoring the variability amongst Tn7-like elements and their potential importance to host success. A Tn7-like element found in the type E toxin producing Clostridium butyricum 5521, a causative agent of botulism, contains two genes found within a lambdoid prophage of the Bacillus anthracis Ames strain (Read et al., 2003). While these genes (an acetyltransferase and a hypothetical gene) are not thought to contribute directly to pathogenicity, they may affect pathogen-host interactions and enhance the organisms ability to survive within a host (Read et al., 2003).
The integron system of Tn7 from E. coli (and others) and the CRISPR system of the Tn7-like element in Anabaena variabilis include mechanisms that add DNA sequences to specific regions. There are also unrelated mobile elements that actively target sequences associated integron cassettes (Quiroga et al., 2008). While there may be other yet-to-be-described mechanisms that Tn7-like elements exploit for the addition of genetic material, the variable region may simply provide a “neutral zone” where recombination events can occur without causing harm to the organism or disrupting genes essential for transposition, yet allowing widespread distribution of novel gene combinations.
5. Phylogeny and diversity of transposition genes
Tn7 and its related elements have been found in a wide variety of bacteria from diverse environments (Parks and Peters, 2007). They can be found in, Firmicutes, Cyanobacteria, Bacteroidetes, in nearly all classes of Proteobacteria, and in the more deeply branching Chloroflexi group (Figure 6). The ecological niches occupied by Tn7 hosts are quite diverse. These organisms can be found in deep sea hydrothermal vents (Idiomarina loihiensis, Shewanella loihica), in low pH acid mine drainage (Acidithiobacillus ferrooxidans) and high pH soda lakes (Natranaerobius thermophilus JW/NM-WN-LF), in the surface waters of the ocean (oceanic environmental metagenome libraries), in spoiled food (Bacillus cereus), in soil samples (soil environmental metagenomic libraries) and in clinical settings around the world (Escherichia coli, Helicobacter pylori, Burkholderia cenocepacia, Pseudomonas aeruginosa, Shigella sonnei, Clostridium butyricum 5521, and others) to name just a few. The G+C content of tns genes from each of the elements typically mirrors that of the host genome in which they reside and ranges from 26% (in Clostridium butyricum 5521) to 58% (in Acidithiobacillus ferrooxidans), suggesting that these elements have been maintained within similar organisms for some time. However, in some cases, such as in Hahella chejuensis, there are significant differences in G+C content. Differences in G+C content can be taken as evidence of recent arrival of the Tn7-like element (Lawrence and Ochman, 1997).
Figure 6.
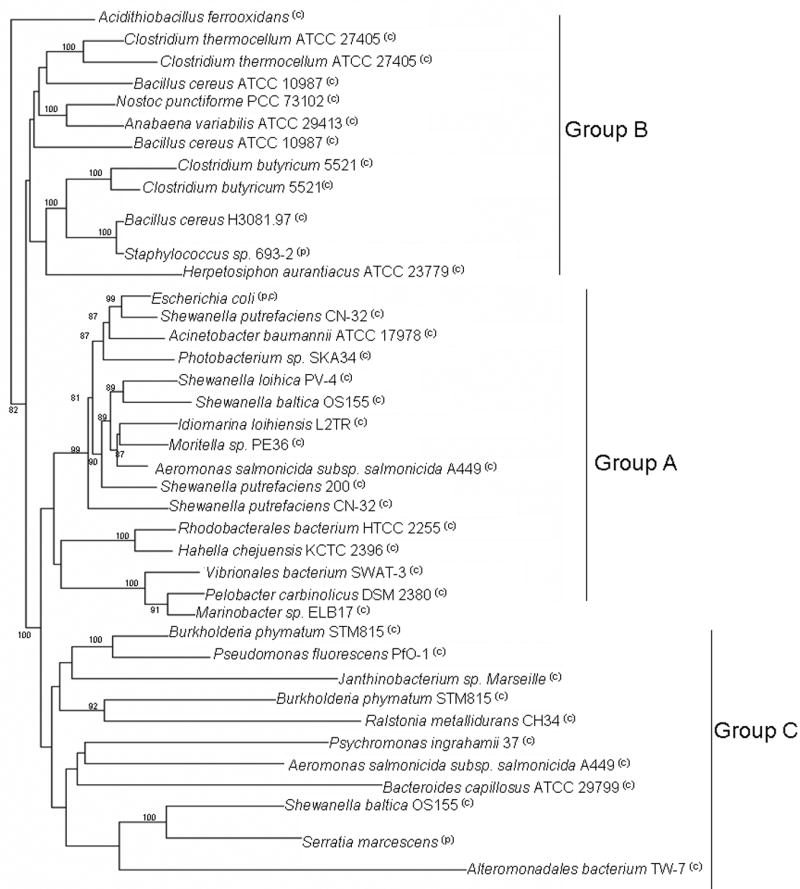
A denrogram describing the phylogeny of Tn7-like elements. Phylogenetic analysis of Tn7-like elements reveals three distinct lineages of transposons. The predicted amino acid sequences of TnsABCD from each element was concatenated and aligned using the Jalview software package and the ClustalW algorithm (see Table S1 for accession numbers) (Chenna et al., 2003; Clamp et al., 2004). Protein distances and Neighbor Joining trees were constructed using the Phylip program through the Mobyle Portal software package (Felsenstein, 1993). The tree was rooted on Acidithiobacillus ferroxidans. Bootstrap analysis was also carried out using 1000 iterations, and values greater than 80% are given at appropriate branch points. The phylogenetic tree was drawn using the MEGA4 software package (Tamura et al., 2007). “C” indicates chromosomally localized, and “P” indicates that the element is found on a plasmid.
Phylogenetic analysis of the transposition genes of Tn7-like elements reveals three distinct lineages, Groups A, B, and C (Figure 6). Group A is comprised of elements found within Proteobacteria, and includes the originally described Tn7 from E. coli. Group B elements can be found in Firmicutes, Cyanobacteria, and Chloroflexi. One representative from the Group B elements can also be found in a Gammaproteobacterium, Acidithiobacillus ferrooxidans. Group C elements are very different from those in Groups A and B in that none of them seem to contain a tnsE gene. Surprisingly, none are found within the traditional attTn7 locus even though they all contain tnsD homologs. Some of the Group C elements appear to be near the attTn7 locus, but the orientation of tns genes and their proximity to the glmS gene is not consistent with standard Group A and B Tn7-like elements. It therefore seems likely that the TnsD proteins in the Group C elements actually catalyze a more random form of transposition. These elements may represent a more primitive form of Tn7. There are at least two examples of organisms, Shewanella baltica OS155 and Aeromonas salmonicida A449, in which a Group C element inhabits the same organism as a Group A element. Further study of Group C elements and their relationship to Groups A and B may provide some interesting clues regarding the evolution of Tn7-like transposons. Interestingly, the presumed TnsC-interacting region of TnsB differs among phylogenetic groups (Figure 3.B). The C-terminus of the TnsB homologs is predominantly acidic in one group (Group A), while the same region in other TnsB proteins contain many basic residues (Figure 3.B.). In this case differences in TnsB-TnsC interface may define barriers to interaction between groups. We have yet to find any Tn7-like elements in the archaea even though gene exchange is known to occur between bacteria and archaea.
Experiments have shown that the chromosome targeting pathway of Tn7 is extremely robust (Choi et al., 2005; McKenzie and Craig, 2006). Indeed, this pathway is capable of directing transposition into a wide variety of host genomes in the laboratory, even into Human “attTn7” loci associated with gfpt-1 and gfpt-2, the glmS homologs found in our genome (Kuduvalli et al., 2005). The preponderance of Tn7 elements that have been discovered to date have been found within the presumed attTn7 locus of organisms with draft or completely sequenced genomes. This likely reflects a database bias in which there are more genomic DNA sequences present than viral or plasmid sequences. Alternatively, plasmid and bacteriophage borne Tn7s may be far more transient since excision of the transposon may destroy the vector by the resulting DNA double-strand break. There are, however, some examples, both in gram (+) and in gram (-) bacteria, of plasmids containing Tn7-like elements. No Tn7 homologs have yet been found in bacteriophages outside of the laboratory, however many chromosomal Tn7-like elements either contain genes associated with bacteriophages or are inserted in the vicinity of phage-associated genes (Parks and Peters, 2007) (Figure 4). It seems likely that bacteriophage borne elements will soon be discovered. As genome and meta-genomic sequencing projects progress, the bestiary of Tn7-like elements continues to mount in population.
6. Concluding remarks
The molecular mechanisms of Tn7 transposition contribute to the evolution of both the host genome and the transposon by maximizing the collection of genes with beneficial functions without harming the host. The ability of Tn7 to selectively direct transposition into the chromosome, mobile plasmids, and bacteriophages allows it to move in and out of stationary and mobile DNA while preventing the disruption of important host functions. This property of the transposon likely exposes it to a great diversity of environments in which various types of genes may be sampled.
While transposons are rightly considered “selfish” genetic elements that parasitize the genome of organisms to ensure their own propagation, Tn7 may tell a slightly different story. The highly evolved targeting system of Tn7 virtually ensures that deleterious insertions do not occur while allowing the host to sample genes collected through a variety of mobile DNAs. The existence of genomic islands within attTn7 where little or no Tn7 transposition functions remain underscores the ability of the host to maintain genetic information based exclusively on its value to the host.
Supplementary Material
Acknowledgments
We are grateful to Zaoping Li and Qiaojuan Shi for comments on the manuscript and to Steve Zinder for advice on issues of phylogeny. This work was funded by the National Institutes of Health grant GM069508.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arciszewska LK, Drake D, Craig NL. Transposon Tn7 cis-acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- Bainton R, Gamas P, Craig NL. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Kubo KM, Feng JN, Craig NL. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell. 2008;132:208–20. doi: 10.1016/j.cell.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow RS, Gobius KS. Diverse class 2 integrons in bacteria from beef cattle sources. J Antimicrob Chemother. 2006;58:1133–8. doi: 10.1093/jac/dkl423. [DOI] [PubMed] [Google Scholar]
- Barth PT, Datta N, Hedges RW, Grinter NJ. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976;125:800–10. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskri L, Mazel D. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob Agents Chemother. 2003;47:3326–31. doi: 10.1128/AAC.47.10.3326-3331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. Lateral DNA transfer : mechanisms and consequences. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2002. [Google Scholar]
- Campbell AM. Preferential orientation of natural lambdoid prophages and bacterial chromosome organization. Theor Popul Biol. 2002;61:503–7. doi: 10.1006/tpbi.2002.1604. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, DeShazer D, Schweizer HP. mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nat Protoc. 2006;1:162–9. doi: 10.1038/nprot.2006.25. [DOI] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2:443–8. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with secondary, non-glmS-linked attTn7 sites: example Proteus mirabilis HI4320. Nat Protoc. 2006;1:170–8. doi: 10.1038/nprot.2006.26. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–7. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- Craig N. Transposon Tn7. Mobile DNA. 1989:211–225. [Google Scholar]
- Craig NL. Tn7. In: Lambowitz-Alan M, editor. Mobile DNA II. ASM Press; Washington, DC: 2002. pp. 423–456. [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoy R, Craig NL. Tn7 transposition as a probe of cis interactions between widely separated (190 Kilobases Apart) DNA sites in the Escherichia coli chromosome. J Bacteriol. 1996;178:6184–6191. doi: 10.1128/jb.178.21.6184-6191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoy RT. Molecular Biology and Genetics. Johns Hopkins University; Baltimore, Maryland: 1997. Transposon Tn7: target DNAs can modulate Tn7 insertion; p. 250. [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–24. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Seattle: 1993. Distributed by the author. [Google Scholar]
- Finn JA, Parks AR, Peters JE. Transposon Tn7 directs transposition into the genome of filamentous bacteriophage M13 using the element-encoded TnsE protein. J Bacteriol. 2007;189:9122–5. doi: 10.1128/JB.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary PA, Biery MC, Bainton RJ, Craig NL. Multiple DNA processing reactions underlie Tn7 transposition. J Mol Biol. 1996;257:301–316. doi: 10.1006/jmbi.1996.0164. [DOI] [PubMed] [Google Scholar]
- Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid. 2004;52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- Gringauz E, Orle K, Orle A, Waddell CS, Craig NL. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion. J Bacteriol. 1988;170:2832–2840. doi: 10.1128/jb.170.6.2832-2840.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann AT, Craig NL. Tn7 transposition creates a hotspot for homologous recombination at the transposon donor site. Genetics. 1993;133:9–16. doi: 10.1093/genetics/133.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson K, Sundstrom L, Pelletier A, Roy PH. IntI2 integron integrase in Tn7. J Bacteriol. 2002;184:1712–21. doi: 10.1128/JB.184.6.1712-1721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer B, Shapiro JA. Control of Tn7 transposition. Mol Gen Genet. 1984;194:149–158. doi: 10.1007/BF00383510. [DOI] [PubMed] [Google Scholar]
- Hedges RW, Datta N, Fleming MP. R factors conferring resistance to trimethoprim but not sulphonamides. J Gen Microbiol. 1972;73:573–5. doi: 10.1099/00221287-73-3-573. [DOI] [PubMed] [Google Scholar]
- Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. Whole genome pyrosequencing of an epidemic multidrug resistant Acinetobacter baumannii of the European clone II. Antimicrob Agents Chemother. 2008 doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TJ, Wannemeuhler YM, Scaccianoce JA, Johnson SJ, Nolan LK. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob Agents Chemother. 2006;50:3929–33. doi: 10.1128/AAC.00569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–56. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I. Genetic Addiction: a Principle of Gene Symbiosis in a Genome. Plasmid Biology. 2004:105–144. [Google Scholar]
- Kubo KM, Craig NL. Bacterial transposon Tn7 utilizes two different classes of target sites. J Bacteriol. 1990;172:2774–8. doi: 10.1128/jb.172.5.2774-2778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuduvalli P, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–32. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuduvalli PN, Mitra R, Craig NL. Site-specific Tn7 transposition into the human genome. Nucleic Acids Res. 2005;33:857–63. doi: 10.1093/nar/gki227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T, Wilkins B, Frost L. Bacterial conjugation in Gram-negative bacteria. Plasmid Biology. 2004:203–226. [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–97. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lichtenstein C, Brenner S. Site-specific properties of Tn7 transposition into the E. coli chromosome. Mol Gen Genet. 1981;183:380–387. doi: 10.1007/BF00270644. [DOI] [PubMed] [Google Scholar]
- Lichtenstein C, Brenner S. Unique insertion site of Tn7 in E. coli chromosome. Nature. 1982;297:601–603. doi: 10.1038/297601a0. [DOI] [PubMed] [Google Scholar]
- Lu F, Craig NL. Isolation and characterization of Tn7 transposase gain-of-function mutants: a model for transposase activation. EMBO J. 2000;19:3446–3457. doi: 10.1093/emboj/19.13.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May EW, Craig NL. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Craig NL. Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 2006;6:39. doi: 10.1186/1471-2180-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown RL, Orle KA, Chen T, Craig NL. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988;170:352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown RL, Waddell CS, Arciszewska LA, Craig NL. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. Proc Natl Acad Sci USA. 1987;84:7807–7811. doi: 10.1073/pnas.84.22.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- Nisa-Martinez R, Jimenez-Zurdo JI, Martinez-Abarca F, Munoz-Adelantado E, Toro N. Dispersion of the RmInt1 group II intron in the Sinorhizobium meliloti genome upon acquisition by conjugative transfer. Nucleic Acids Res. 2007;35:214–22. doi: 10.1093/nar/gkl1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppon JC, Sarnovsky RJ, Craig NL, Rawlings DE. A Tn7-like transposon is present in the glmUS region of the obligately chemoautolithotrophic bacterium Thiobacillus ferrooxidans. J Bacteriology. 1998;180:3007–3012. doi: 10.1128/jb.180.11.3007-3012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman BE, Pineiro SA, Arduino S, Galas M, Melano R, Caffer MI, Sordelli DO, Centron D. Evolution of multiresistance in nontyphoid salmonella serovars from 1984 to 1998 in Argentina. Antimicrob Agents Chemother. 2002;46:3963–70. doi: 10.1128/AAC.46.12.3963-3970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AM, Boltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48:202–12. doi: 10.1016/s0147-619x(02)00117-8. [DOI] [PubMed] [Google Scholar]
- Parks AR, Peters JE. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J Bacteriol. 2007;189:2170–3. doi: 10.1128/JB.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol Cell. 2000;6:573–582. doi: 10.1016/s1097-2765(00)00056-3. [DOI] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7 recognizes target structures associated with DNA replication using the DNA binding protein TnsE. Genes & Dev. 2001a;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7: smarter than we thought. Nature Reviews/Molecular Cell Biology. 2001b;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–5. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga C, Roy PH, Centron D. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiology. 2008;154:1341–53. doi: 10.1099/mic.0.2007/016360-0. [DOI] [PubMed] [Google Scholar]
- Ramirez MS, Quiroga C, Centron D. Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005a;49:5179–81. doi: 10.1128/AAC.49.12.5179-5181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MS, Vargas LJ, Cagnoni V, Tokumoto M, Centron D. Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob Agents Chemother. 2005b;49:4418–20. doi: 10.1128/AAC.49.10.4418-4420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JE, Miller PS, Craig NL. Recognition of triple-helical DNA structures by transposon Tn7. Proc Natl Acad Sci USA. 2000;97:3936–3941. doi: 10.1073/pnas.080061497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolonay JF, Beanan MJ, Dodson RJ, Brinkac LM, Gwinn M, DeBoy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pop M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang L, Hance IR, Weidman JF, Berry KJ, Plaut RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Koehler TM, Hanna PC, Kolsto AB, Fraser CM. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–6. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- Ronning DR, Li Y, Perez ZN, Ross PD, Hickman AB, Craig NL, Dyda F. The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA. Embo J. 2004;23:2972–81. doi: 10.1038/sj.emboj.7600311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnovsky R, May EW, Craig NL. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- Seringhaus M, Kumar A, Hartigan J, Snyder M, Gerstein M. Genomic analysis of insertion behavior and target specificity of mini-Tn7 and Tn3 transposons in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:e57. doi: 10.1093/nar/gkl184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe P, Craig NL. Host proteins can stimulate Tn7 transposition: a novel role for the ribosomal protein L29 and the acyl carrier protein. EMBO J. 1998;17:5822–31. doi: 10.1093/emboj/17.19.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–5. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- Shi Q, Parks AR, Potter BD, Safir IJ, Luo Y, Forster BM, Peters JE. DNA damage differentially activates regional chromosomal loci for Tn7 transposition in Escherichia coli. Genetics. 2008;179:1237–50. doi: 10.1534/genetics.108.088161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelding Z, Queen-Baker J, Craig NL. Alternative interactions between the Tn7 transposase and the Tn7 target DNA binding protein regulate target immunity and transposition. Embo J. 2003;22:5904–17. doi: 10.1093/emboj/cdg551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. Lambda II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1983. General recombination; pp. 175–209. [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Stellwagen A, Craig NL. Avoiding Self: Two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 1997a;16:6823–6834. doi: 10.1093/emboj/16.22.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A, Craig NL. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics. 1997b;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A, Craig NL. Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. TIBS. 1998;23:486–490. doi: 10.1016/s0968-0004(98)01325-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lichtenstein C, Cotterill S. Purification and characterization of the TnsB protein of Tn7, a transposition protein that binds to the ends of Tn7. Nucl Acids Res. 1991;19:3395–3402. doi: 10.1093/nar/19.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyakov A, Obmolova G, Badet-Denisot MA, Badet B. The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase. Protein Sci. 1999;8:596–602. doi: 10.1110/ps.8.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell CS, Craig NL. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes & Dev. 1988;2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- Wang JD, Berkmen MB, Grossman AD. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:5608–13. doi: 10.1073/pnas.0608999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol Rev. 2006;30:321–81. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes & Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.