Summary
Integrins perform the critical function of signalling cell attachment to the extracellular matrix or to other cells. This signalling is done through a structural change propagated bidirectionally across the plasma membrane. Integrin activation has been extensively studied with ectodomain constructs, but the structural change within intact, membrane-bound molecules remains a subject of live debate. Using cryoelectron tomography, we examined the simplest predication of the different integrin activation models, i.e. the change in height of the molecules. Analysis using techniques that compensate for the missing wedge during alignment and averaging and that search for patterns in the structure of the aligned molecular subvolumes extracted from the tomogram reveals that the vast majority of molecules show no dramatic height change upon Mn2+ induced activation of membrane bound integrins when compared with an inactive integrin control group. Thus, the result is inconsistent with the switchblade activation model.
Keywords: Membrane protein, cell adhesion, platelets, image processing, lipid vesicle
Introduction
Integrins are a large family of heterodimeric cell surface receptors. Each peptide has a large extracellular domain (ectodomain), which is involved in binding to either extracellular matrix ligands or other cell surface proteins, a single transmembrane α-helix and usually a small cytoplasmic tail which binds to cytoplasmic components1. Integrin binding to its partners on either side of the plasma membranes is also a signalling event that can be transmitted bidirectionally across the membrane1; 2. During outside-in signalling, stimuli from integrin-ligand interactions are transduced to the integrin cytoplasmic tail through a long range structural change leading to recruitment of cytoplasmic binding partners and assembly of various machineries for further downstream signalling in processes such as apoptosis, motility, and growth3; 4. Inside-out signalling involves intracellular factors that impinge upon the integrin cytoplasmic domains, increasing ligand binding affinity of the integrin ectodomain, which is critical for wound healing, blood clotting and immune cell recruiting1; 2; 5. Activation and inside-out signalling can also be mimicked by Mn2+ binding by the integrin ectodomain, which changes the integrin to a high affinity state as judged by exposure of activation-dependant or ligand-induced antibody binding sites (LIBS) that are hidden in the inactive conformation2.
Integrin structure and conformational changes have been studied extensively using different techniques. Those methods that can resolve individual molecules have used mostly soluble ectodomain constructs. The X-ray crystal structure of the αvβ3 ectodomain with and without peptide ligand showed a compact bent-over conformation with the ligand binding “head piece” facing toward the membrane, an orientation that would appear to be unfavourable for ligand binding6; 7. The structural changes upon ligand binding were small with virtually no change at the C-termini suggesting no obvious mechanism for propagating structural change to the cytoplasmic domains.
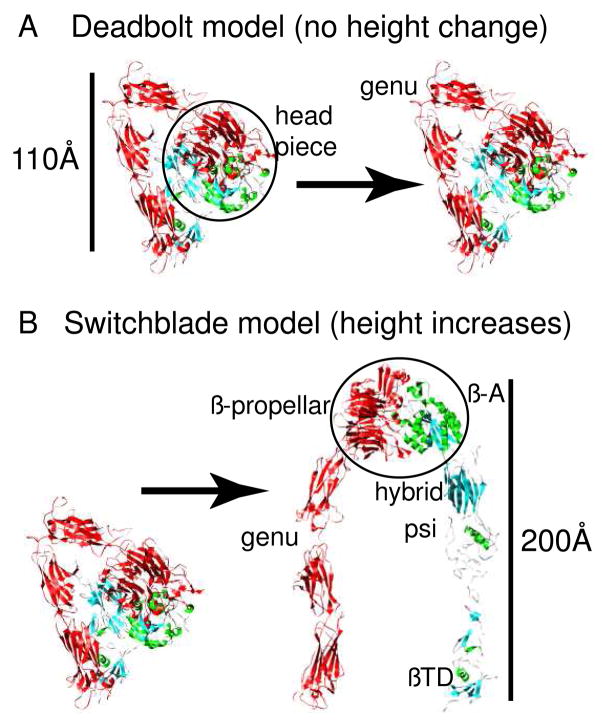
Single particle reconstructions of the negatively stained αvβ3 ectodomain, the same construct used in the crystal structure, in a complex with a fibronectin fragment showed a bent-over rather than upright conformation, suggesting that the bent-over conformation is in fact competent to bind its physiological ligand and thus is in a high affinity state8. Together with the earlier X-ray crystal structural studies, the latter study is consistent with an alternative “deadbolt” model. In this model, inside-out signalling releases an inhibitory interaction between βTD domain and βA domain and allows the bent-over model to bind to its ligand (Figure 1A). The ensuing ligand binding will then provide energy for further conformational change in outside-in signalling9.
Figure 1.
Different predications from two integrin activation models. (A) The deadbolt model predicts that the integrin remains in the bent-over conformation after activation and that no height change is coupled to the increased ligand binding affinity of activation. (B) The integrin changes to an extended conformation after activation. The height of the molecule increases dramatically as the bent over conformation straightens and the legs separate. Headpiece denotes the large globular domain comprising the C-terminal domains of both the αIIb β-propellar domain and the β3 β-A domains (circled in the upright conformation). The genu denotes the point where the leg of the αIIb chain bends in the crystal structure. The location of the other β3 domains mentioned in the text, i.e. –hybrid, -psi and -βTD are also indicated in the upright conformationwhere they are most easily identified
On the other hand, results from other studies can not be fully explained by the “deadbold” model. Electron microscopy (EM) of the negatively stained αvβ3 ectodomain with a cleavable clasp engineered into the C-terminus revealed an equilibrium shift from the bent-over conformation to an upright conformation on integrin activation, with the majority of molecules found in the bent-over conformation when inactive and the majority found in the upright conformation with two legs separated when activated10. Negative stain EM of a shorter ectodomain construct of α5β1 in complex with fibronectin showed the leg separation expected for outside-in activation11. An X-ray structure of an even shorter ectodomain construct containing the α-chain β-propellor domain, together with the β-chain β-A, -hybrid and -psi domains in complex with a small ligand also showed the expected swing-out of the hybrid domain12. Based on these microscopy results, the authors proposed a “switchblade” model for integrin activation, in which the leg separation caused by the cytoplasmic signals straightens the integrins at the “genu” or knee producing an upright, extended conformation corresponding to a high affinity state (Figure 1B). This switchblade model is different from the deadbolt model in that the authors proposed a correlation between the extension and the activation of the molecule, while in the deadbolt model the extension could be a post-activation event9. Another negative stain EM study13, with detergent solublized full length integrin, observed an extended conformation with flexible leg regions, which is similar to that observed by Takagi et al10 and supportive of the switchblade model.
The structural mechanism of integrin activation on the membrane surface is also hotly debated between those two models. Correlation of integrin inactivation with cysteine mutations that lock integrins in the bent-over conformation on the cell surface supported the switchblade model10. Fluorescence resonance energy transfer (FRET) results also favoured the switchblade model by showing a separation between the α and β cytoplasmic domains14. Other FRET studies using a peptide donor that binds to α4 integrin and octadecyl rhodamine B acceptors incorporated into the plasma membrane observed a 50 Å increase in distance consistent with a conformational change to the upright configuration on Mn2+ activation15. Ligand binding to wild type and mutant αvβ3 and αIIbβ3 integrins expressed on the surface of CHO cells showed that mutations in the “deadbolt” did not constitutively activate integrins and that disulfide crosslinks that prevent extension to the upright conformation prevented high affinity ligand binding16 a result at odds with the deadbolt model.
However, a recently published study using an antibody that specifically recognizes the “genu” region in the extended model found no increase in antibody binding when integrins become activated on the cell surface, a result that would favour the dead-bolt model17. Another earlier study showed that uPAR, which interacts with the integrin headpiece in the bent conformation, can reverse integrin binding to a fibronectin (FN) matrix on the cell surface, implying that the integrin is in the bent conformation on the cell surface when FN bound18. Furthermore, recent results measuring FRET between fluorescently labelled Fab fragments of mAbs to the βA domain of the β3 subunit and the Calf-2 domain of the αIIb subunit observed only small changes consistent with no large conformational change when platelets are activated19. These three reports are more consistent with the deadbolt model.
Most activation studies of integrins incorporated in membranes infer the upright conformation based on ligand and antibody binding without directly observing the molecular conformation. One way to test the two models is to examine their predications by directly observing the integrins incorporated into membrane vesicles. The switchblade model predicts a near doubling in molecular height upon activation while the dead-bolt model predicts no such change (Fig 1A, B). In this report, we studied the molecular height of full length αIIbβ3 integrin in a membrane environment in both the inactive and in the active state induced by Mn2+. We have used the new and developing technique of cryoelectron tomography of ice embedded specimens20 to obtain a 3-D image of the integrin incorporated into small unilamellar liposomes. Starting with a 3-D image of the liposome, which removes the superposition effects that complicate the interpretation of projection images, molecular volumes of individual integrins were extracted, aligned, subjected to multivariate data analysis (MDA)21 to group self-similar images, and class averages computed to improve signal-to-noise ratio and general feature definition. The integrin heterodimer has a relatively small molecular weight for this kind of structural analysis on specimens this thick so we have restricted our observations to the integrin height alone in the two conditions. The results suggest no height change of integrin molecules on Mn2+ induced activation, an observation that is inconsistent with predictions of the switchblade activation model.
Results
Integrin Purification, Integrin Liposome Reconstitution and Activity
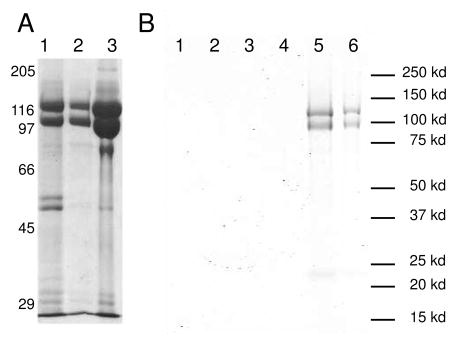
Integrins were purified from outdated human platelets. An RGD peptide conjugated column was used to separate active from inactive integrins. The final integrin preparation is are at least 95% pure based on the relative band intensities of the SDS-PAGE gel (Figure 2A). The inactive integrin fraction was then reconstituted into DMPC/DMPG unilamellar liposomes. The insertion of integrins was confirmed by SDS-PAGE gel electrophoresis of the liposome band extracted from the sucrose gradient (Figure 2B). The integrins migrated to the same position as the liposomes on the sucrose gradient, indicating that they are inserted into the liposome. The liposome band was also verified by electron microscopy of negatively stained liposomes (data not shown). The non-incorporated integrins probably formed aggregates that are less bouyant than the integrin liposomes and migrated to a lower position on the sucrose gradient (Figure 2B).
Figure 2.
(A). Gel pictures showing different steps of integrin purification. From left to right, lane 1 showing relative purity after the Con A column; lane 2 showing relative purity after the sepharose Q column; lane 3 shows relative purity after the mono Q column. (B). Gel photograph showing the reconstituted integrin liposomes used for the assays described in Figure S1 after sucrose gradient purification. Gels from other liposome preparations have the same appearance. Lanes as indicated in the figures are different layers on the sucrose gradient from top to bottom: Lane 1: Top layer; Lane 2: 2nd layer from the top; Lane 3: 3rd layer from the top; Lane 4: 4th layer from the top; Lane 5: Layer with the visible liposome band; Lane 6: Layer 1 cm beneath the visible liposome band. The gel shows that the purified integrins are reconstituted into the liposome because they migrate with the liposome. The non-incorporated integrins form irregular aggregates, are consequently less bouyant and migrate to a position below the liposome band.
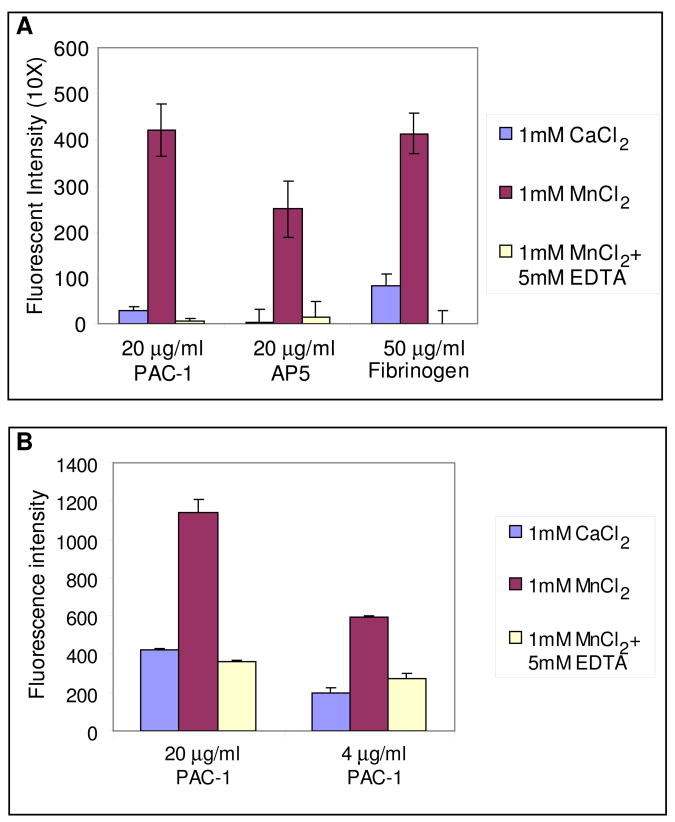
To test the activity state of the liposome-bound, inactive integrins, an ELISA style binding assay was conducted with the conformational specific antibody PAC-122, anti-LIBS antibody AP523, and the physiological ligand fibrinogen. The binding activities of integrin liposomes were tested with 1 mM CaCl2 as native state, 1 mM MnCl2 as reference for the activated state24 and 1 mM MnCl2 and 5 mM EDTA as control for the inactive state. The result clearly shows that the purified, inactive integrins have a binding affinity similar to that of the inactive control groups for PAC-1 antibody, AP5 antibody or fibrinogen, indicating that the inactive conformation remains unperturbed after reconstitution into the liposome (Figure 3A). The result also indicates that in this membrane environment MnCl2 not only activates integrins for binding to the natural ligand, fibrinogen or ligand mimicking PAC-1 antibody, but also expose the ligand-induced epitope for AP5 antibody (Figure 3A). In addition, the amount of integrin liposome binding correlates with the amount of PAC-1 coating (Figure 3B). This assay was also repeated for a different integrin preparation and using a different spectrophotometer but using the peptide integrilin 25; 26 as the negative control, and antibodies D57 and AP3 as controls for activation independent binding. These results are summarized in the supplemental material (Figure S1). The assay showed activation dependent binding for PAC-1, which could be eliminated by addition of integrilin, and activation independent binding for D57 and AP3 which was unaffected by addition of either Mn2+ or Mn2+ plus integrilin.
Figure 3.
Results of integrin activation assay using antibody PAC-1, antibody AP5 and fibrinogen. Each measurement is the average of 3 repeats and the error bars in the figure show the range of data. (A) Assay comparing binding to PAC-1, AP5 and fibrinogen. (B) Assay comparing different loadings of PAC-1 to the assay well.
Although this assay can show relative changes in integrin activation, it does not quantitate the absolute levels of activation since not all of the membrane bound integrins need bind the antibody for the vesicle to attach to the substrate and the integrins facing the inside of the vesicle are inaccessible to the antibody. Conversely, small levels of integrin activation can yield a relatively high binding of vesicles containing predominately inactive integrins. Controls show that binding is high and not activation dependent when conformationally insensitive antibodies are used (Supplemental Figure S1). Despite these disadvantages, the assay performed under several different conditions and with several sets of controls, shows that addition of MnCl2 leads to increased binding of the conformationally sensitive antibody PAC-1. Thus, in this membrane environment, MnCl2 can be used to mimic the integrin activation process.
Cryoelectron Tomograms of Integrin-Liposomes
The tomograms of frozen-hydrated, liposome suspensions confirmed the insertion of the integrins into the liposomes (Figure 4). The lipid-bound integrins were readily identified in sections from the tomogram computed parallel to the specimen plane (z-sections) that cut through the sides (Figure 4B, C) and the tops (Figure 4A) of the liposomes. In z-sections through the sides of the liposome, both lipid bilayer and the incorporated integrins are visible. Inactive integrins in these views have a compact conformation with distinctive head and leg pieces and with the head piece displaced from the bilayer (Figure 4B). The height of the molecule above the lipid bilayer is ~110 Å. The lipid bilayer is not visible on the top and bottom of the liposomes because tilt angle limitations in the electron microscope prevent collection of the edge-on views required to visualize it on these surfaces27. This missing data is commonly referred to as the “missing wedge”. The integrins are nevertheless clearly visible in the top-bottom views (Figure 4A). The utility of these views in the averages is that they give the best resolution in a direction parallel to the membrane and thus give the molecule better shape in this direction.
Figure 4.
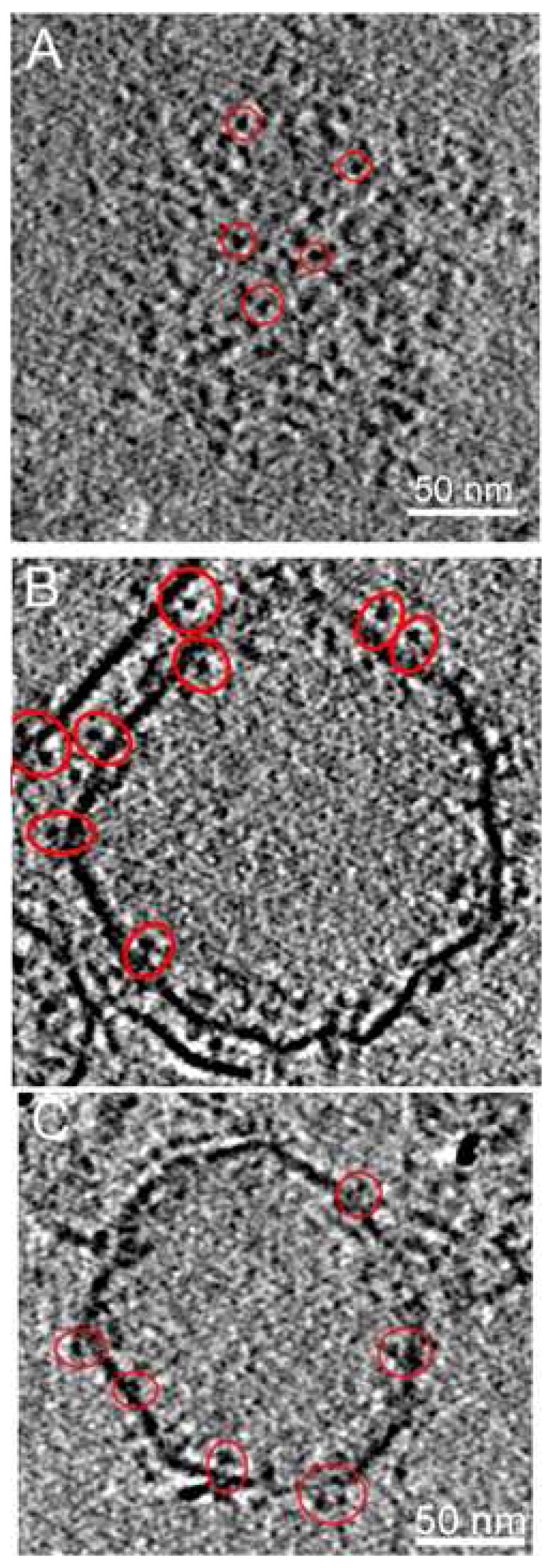
Active and inactive integrins incorporated into liposomes. (A) Inactive αIIbβ3 integrins in a z-section through the cryoelectron tomogram showing the top surface of the liposome. Representative molecules are circled in red. (B) z-Section through the middle of the liposome from the same tomogram as (A) illustrating side-views of inactive integrins. (C) z-Section through the middle of an activated αIIbβ3 integrin-liposome. Some representative molecules are circled, some of which appear to show paired leg domains.
Despite the invisibility of the bilayer in the top and bottom regions, the relative position of the integrins and the membrane is easily defined because integrins extend out of both sides of the lipid bilayer. The number of z-sections, 30 spaced by 5.56 Å, separating the centers of integrin head pieces on the inside and outside in the top-bottom views gives a separation of 167 Å. This is within the range of height predicted from the side views which is about 80 to 90 Å from the center of the head piece to the center of the membrane. Integrin molecules could therefore be reliably selected from the top-bottom views.
To obtain an image of activated integrins, liposomes reconstituted with inactive integrins were activated by addition of MnCl2. Based on the activity assay (Figure 3) the integrins in the liposomes were converted in high yield to an activated form by this procedure. In addition, it has been reported that upon MnCl2 stimulation, the vast majority, about 80% to 85% of integrins undergo a dramatic conformational change to an entended form10. Therefore, if the switchblade model is correct with respect to MnCl2 activation, we should observe a height change in most integrin molecules in the presence of MnCl2. Tomograms of activated integrins showed molecules with a height very similar to the inactive molecules, ~110 Å, and with a clear head piece and a leg region (Figure 4C, 4B).
Molecular Images of Membrane Incorporated integrins
To compare the molecular images of inactive and active integrins in detail, individual integrin molecules were identified manually and subvolumes extracted. A total of 1715 molecular subvolumes from the inactive integrin liposomes, and 1255 molecular subvolumes from active integrin liposomes were selected from the inside and outside surfaces from both top-bottom and side views and aligned. Molecules from the top-bottom orientation constituted about 2/3 of all the molecules as these were easier to view as separated molecules. We estimate those selected constitute at least 50% of the available molecules. Molecule selection was biased toward examples that were clearly separated from nearest neighbours; many molecules were too close together to satisfy this requirement.
To obtain measurements of individual integrins, we used volume alignment and image classification to observe variation of the molecular height among the data. Over the course of the investigation, alignment was done by several different approaches before settling on a scheme that explicitly treats the missing wedge28. The difficulty in obtaining an average with a more defined shape is mainly due to a combination of low SNR, low resolution and a comparatively small molecule size for specimens this thick.
The picking of the molecular subvolumes selects the center of mass of the headpiece domain. The least squares fit of these centers to the equation of an ellipsoid provided the initial orientation which is given by the direction of the normal to the ellipsoid surface at that location. The large number of partially aligned subvolumes available to us could be separated into better aligned subgroups of molecules using MDA. Averages of these subgroups aligned better relative to a reference because of their higher SNR. The transformation obtained for the class average to a reference could then be applied to all members of the class. MDA then allowed for segregation into new classes using the new alignment. Our goal was simply to obtain a measure of the molecular height before and after Mn2+ activation, but the uniformity of this height was not known. To allow height variation in the data to express itself, 60 class averages were computed which resulted in an average SNR improvement due to stochastic noise of 5.4. Although missing wedge compensation was employed throughout this process, the classification tended to group those integrins selected from the side of the vesicles into a small number of classes. The classification mask (Supplemental Figure S4) was generous enough to include the membrane density thereby accounting for the tendency to segregate subvolumes containing membrane from those that did not.
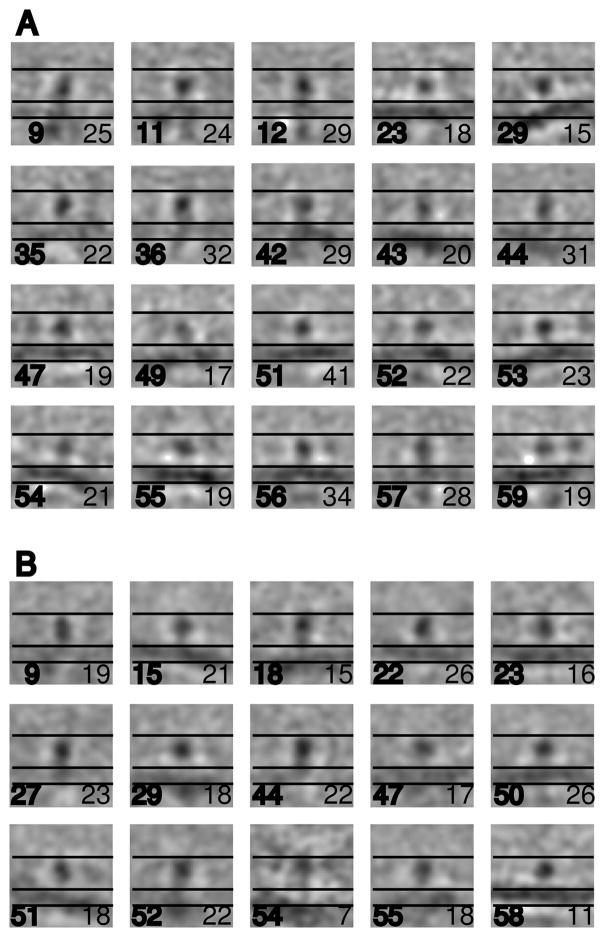
Results for the inactive integrins were quite uniform. All the class members tended to align on the head piece as a center of mass and on the leg domains to determine the direction of the membrane (Figure 5A). All classes have a pronounced headpiece and weaker leg piece. Thirteen classes show the membrane clearly (23, 29, 43, 44, 47, 49, 51–56 and 59) while another seven (9, 11, 12, 35, 36, 42, and 57) showed some weak membrane density (Figure 5A). (The complete set of 60 class averages can be seen in Supplemental Figure S2.) The main effect of the missing wedge compensation in the averaging was to maintain some membrane signature even when the relative number of class members displaying it was small. Using parallel lines set for the inner and outer surfaces of the average membrane position and a third line placed 110 Å above the membrane in displays of the class averages and aligned raw subvolumes, the heights of the vast majority of inactive integrins fell within the 110 Å boundary. Greater heights were generally reflected in the membrane density being pushed lower than its mean position in some classes. One such class, 43 containing 20 members, showed the membrane density positioned ~20 Å further from the headpiece domain than the others. The membrane did not dominate the alignment as some classes show the membrane at an angle to the integrin. Some classes (1,9,10,12, 14, 26 and 38 shown in Supplemental Figure S2) consisting predominately of top-bottom views show integrins on both sides of the supposed membrane position, with a headpiece separation of 150–160 Å corresponding to the separation estimated from the raw tomogram. Although the classes containing predominately top-bottom views are not as useful for the height measurement as the side views, they are largely consistent with the results from the side views. Integrin transmembrane and cytoplasmic domains are not visible at this resolution.
Figure 5.
Projections of inactive integrin molecules obtained from multivariate data analysis. Each panel is 360 Å × 360 Å. The projections are computed from 11 sections (61 Å) through the center of the subvolume. (A) 20 classes from the inactive integrins that show at least some density from the lipid bilayer. All of the classes and the class members of some classes can be seen in Supplemental Figure S2. Most of the 60 classes do not display the membrane density or show it weakly due to the effect of the missing wedge27 and the fact that the majority of molecules were selected from top-bottom views. Class number is indicated in the lower left hand corner of the individual panels in bold font and the number of class members in the lower right hand corner in normal font. The three horizontal lines, which are positioned identically in all panels, indicate the top of the integrin as illustrated in Figure 1, the top of the membrane and the bottom of the membrane and are separated by 110 Å and 60 Å respectively. (B) Fifteen classes of active integrins obtained from MDA that show some density from the lipid bilayer. All of the classes and the class members of some classes can be seen in Supplemental Figure S3
To verify that the class averages were accurately reflecting the positioning of the headpiece relative to the membrane, the aligned subvolumes were displayed on the same scale as the averages and compared to the same ruler (Supplemental Figure S2, B-D). The raw integrin molecular images have very low SNR and so appear more variable than the class averages would suggest. Broadly speaking, they have a similar shape to those in the side-views of the raw tomogram and in the class averages: all have a compact conformation with the large integrin head piece sitting on top of a narrower leg piece, oriented approximately perpendicular to the bilayer surface (Figure S2, B-D). The height of the top of the headpiece relative to the membrane surface is consistent with predictions from the ecto domain X-ray structure (Figure 1A)6.
Class averages of the Mn2+activated integrins were largely indistinguishable from the inactive integrins. All 60 classes showed clearly defined head piece and weak leg piece. (The entire set of 60 classes can be seen in Supplemental Figure S3.) Nine classes (15, 18, 22, 23, 29, 50, 51, 54, and 58) showed clear membrane density while another six (9, 27, 44, 47, 52, 55) showed weak membrane density (Figure 5B). The average SNR improvement in the classes is 4.5. Most of the class averages show a height for the Mn2+ activated integrin of ~110 Å, but some classes, in particular 18, 29, 51 and 54 show a greater height above the membrane which is detectible by the membrane being displaced further from the aligned head piece. These classes account for 40 subvolumes. The class members largely reflect the class average in appearance (Supplemental Figure S3, B-F). One class, 54 containing 7 members, has the most suggestive appearance of the upright activated integrin structure. The curvature of the membrane indicates that these molecules are facing the inside of the vesicles. However, the class members appear more heterogeneous than the class average would suggest (Supplemental Figure S3,F). Two classes, 5 and 24 (Supplemental Figure S3), comprised predominately of top-bottom views show integrins on both sides of the presumed bilayer position and with a separation of 150–160 Å. Thus, there is no dramatic change in the height of Mn2+ activated integrin molecules, but there is a small tendency for more molecules to assume a more upright conformation than in the absence of Mn2+.
Discussion
This is the first study to report molecular resolution images of integrins in a membrane environment in the unstained, frozen-hydrated state. The 3-D images observed here clearly show that in the membrane environment, both inactive and Mn2+ activated integrin molecules have a height that is similar to that of the bent-over ectodomain structure determined by X-ray crystallography6; 7. One of the strongest predictions of the “switchblade” activation mechanism is that the integrin height above the membrane increases dramatically on activation concomitant with the leg separation10 and such a change has been observed with negatively stained detergent solubilized integrins13 but has not been observed directly for integrins incorporated into lipid bilayers. The increase in height predicted for integrins incorporated into a bilayer we do not see; the integrin height in our reconstructions of both the inactive and MnCl2 activated states is essentially constant. Our data is thus more consistent with the deadbolt model, where inactive and activated integrins adopt conformations of similar height9.
The molecular height we measured here is also similar to that of a detergent solubilized full length integrin αIIbβ3 structure determined by cryoEM single particle reconstruction29. This detergent solubilized molecule adopted an upright conformation different from the bent-over conformation, but with a similar height above the supposed membrane surface. At the moment our 3-D images lack the resolution to discriminate between inactive structures of similar height.
Manganese induced activation of integrin was first systematically studied in α5β124. Since then it has been shown to activate a wide variety of integrins, including α3β130, αvβ331; 32, αLβ233, αIIbβ334, α6β135, α1β1 and α2β136; 37, α4β138, and α4β739, to name a few examples. Subsequently, manganese induced activation has been used as the positive control for the level of full activation40; 41. Our result here confirmed the manganese induced activation of integrin αIIbβ3 in a reconsitituted liposome system although the type of assay used could not quantify the degree of activation.
Despite the elevated integrin activation in the liposome in the presence of manganese, we did not observe the large change in the integrin height above the bilayer predicted by the switchblade model in almost all of the class averages. Only one lightly populated class suggested an upright configuration. This observation is more consistent with previous reports with integrin αvβ3 extracellular structure7. In that study, the crystal structure of αvβ3 extracellular domain with Mn2+ showed that Mn2+ did not induce significant structural changes, despite the fact that Mn2+ replaced all six divalent cations in the structure. Subsequent ligand binding altered the incorporation of Mn2+ in the structure, most notably by additional Mn2+ binding at the MIDAS site. Some local structure changes were seen but no global conformational change occurred7. Our result is not consistent with another negative stain EM study where the engineered αvβ3 ectodomain was shown to undergo a drastic global conformational change from a bent-over to an extended shape upon stimulation of Mn2+ regardless of whether the C-terminal was clasped or not10.
Several other studies suggest an explanation for the discrepancy between our cryo-ET observation and the negatively stained EM images. Integrins with mutations that lock the interaction of the α and β subunit transmembrane domain have been shown to be non-responsive to inside-out signaling, but can be easily activated to a high affinity ligand binding state from the outside by Mn2+ addition42; 43. Consistent with this, inside-out activation by talin induced the separation of the α and β transmembrane and cytoplasmic domain as measured by FRET, but Mn2+ did not14, so it is possible that Mn2+ induced activation is an intermediate step during the process of integrin conformational change. Manganese induces the highly elevated affinity to ligand as shown here and the references discussed above, but in the presence of inhibitory force from transmembrane and cytoplasmic domain association cannot induce the conformational change to the upright configuration. Full conformational change only comes as a separate and additional step after the transmembrane and cytoplasmic association is released, possibly by either inside out signaling or by deleting the entire transmembrane and cytoplasmic region as done in the negative-stain EM image study where global conformational change is observed10. The clasp engineered between the C-terminal domains might not fully mimic the effect of the transmembrane domain and cytoplasmic domain interaction10. In this proposed context, our result would be more consistent with the dead-bolt model, where the key difference with the switch-blade model is that the global conformational change to the upright orientation on the membrane is a separate step that can come after the affinity change and ligand-binding9.
In conclusion, we have observed no change in height of integrin molecules incorporated in a lipid bilayer upon MnCl2 induced activation. This observation is not as predicted by the switchblade model and is more consistent with the deadbolt model, which predicts no change in height on activation. Further work with thinner specimens, from which higher resolution could be obtained, is needed to fully resolve the density of the leg regions to get a more complete picture of integrin activation.
Materials and Methods
Purification and separation of active and inactive integrin
Integrin αIIbβ3 was purified from out-dated human platelets (Southeastern Community Blood Center) using a protocol modified from Adair et al.29. Briefly, the platelets were washed with Tyrode’s buffer and lysed by freezing and thawing. The lysed platelets were then washed with 500 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris, 0.5 mM PMSF, 0.02 mM Leupeptin, 0.2 mM DTT, pH 7.4 and then extracted with 150 mM NaCl, 2 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris, 0.5 mM PMSF, 0.02 mM Leupeptin plus 1% Triton X-100. The extracted integrins were separated from other proteins by passage successively through Con A, Q-sepharose, and if necessary, size-exclusion columns. The buffer used throughout purification and liposome formation consisted of 0.1% Triton, 50 mM NaCl, 0.5 mM CaCl2, 20 mM Tris, pH 7.4. Frozen specimens, ±Mn2+, were prepared in the same buffer but without Triton.
Activated integrin contaminants were separated from the purified integrins using a protocol from Kouns et al.44. Briefly, the purified integrins were passed twice through an RGD peptide conjugated column. In the second pass, ¼ as much integrin bound to the regenerated RGD column while the total integrin concentration remained nearly constant. The bound integrin from the two RGD purifications amounted to ~5% of the total purified integrin (data not shown). This indicates an upper limit of 5% for active integrin contamination within our inactive integrin preparation.
Reconstitution of integrins into liposomes
Purified inactive integrin was incorporated into liposomes using a protocol modified from Muller et al.45. Briefly, 0.3 mg each of DMPC and DMPG were solubilized in chloroform or chloroform/methanol, mixed thoroughly, and dried onto the wall of a glass tube under steady flow of Argon. The dried lipid mixture was then solubilized in 0.1% Triton solution and 0.1 mg of purified inactive integrin in 0.1% Triton was added to the lipid-detergent solution. Triton was removed by successive addition of bio-beads. The reconstituted liposomes were then purified by sucrose gradient centrifugation. The visible lipid band was extracted, dialyzed into liposome buffer without triton to remove sucrose, and assayed by gel electrophoresis.
Activity assay of the reconstituted integrin liposomes
The activity assay protocol was provided by Dr Mark Ginsberg (personal communication). Briefly, liposomes were made as described in the previous section but this time with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(carboxyfluorescein) (PECF) (Avanti) added to fluorescently label the liposome. The conformational specific antibody PAC-1 (Pharmingen), anti-LIBS antibody AP5 (GTI Diagnostics), or fibrinogen (Enzyme Research Laboratories) was immobilized in a 96 well plate and washed. After blocking the plate with 100 mg/ml BSA, fluorescently labelled inactive integrin liposomes were added, and incubated for 4 hours to saturate the binding. The unbound liposomes were then washed several times and fluorescent readings taken for analysis. The same experiments were repeated with 1 mM MnCl2 for positive control24, and 1 mM MnCl2 plus 5 mM EDTA for negative control.
Subsequent to the reconstruction work, we repeated the assay using additional controls, including adding Mn2+ plus integrilin, a competitive ligand inhibitor that functions like an RGD peptide 25; 26, and tested two additional antibodies, D57 and AP3, which are not conformationally specific (Supplemental Figure S1). In each case, the effect of Mn2+ plus integrilin was as predicted, it prevented binding to PAC-1 coated wells, and had no effect on wells coated with D57 or AP3.
This assay only measures activation for those integrins with the ectodomain oriented outward from the liposome. However, inspection of the tomograms indicates that there are openings on the liposome and the analysis of integrin molecule subvolumes failed to detect any difference that correlated with inside or outside orientation of the ectodomain. Probably Mn2+ leakage into the liposomes during mixing and the 2 hr incubation time facilitated integrin activation on both inside and outside of the liposome. This assay only measures relative activation of the liposome bound integrins, but does not give an absolute level of activation because it is not necessary for all molecules to be activated for the vesicles to bind.
Cryoelectron Tomography
Specimens were prepared on glow discharged, 200 mesh copper grids with reticulated carbon support film with 2 μm hole size (Quantifoil Inc., Jena, Germany). The grids were plunge frozen in liquid ethane and transferred to a Gatan Model 626 cryo-holder (Gatan, Pleasanton, NJ) and examined at a temperature of −180 °C. The tilt series were acquired at 300 keV on a Philips CM300-FEG electron microscope and images recorded on a Tem-Cam F224 2k x 2k CCD camera (Tietz Video, Gauting, Germany). The tilt angle range covered mostly −61º to 63º with Saxton scheme angle increments with an initial angle of 3°46. The EM-MENU software (Tietz Video, Gauting, Germany), was used to control the tilt series. The underfocus was ~6 μm which places the first zero of the CTF at ~35 Å resolution. However, the specimen thickness and angle increment in the tilt series are too large to realize this resolution.
The raw tilt images were processed, aligned with a marker-free alignment algorithm and the 3-D reconstruction calculated by weighted back-projection47. Some specimens incorporated 5 nm gold clusters to provide high contrast image features but these were not used specifically for marker alignment. Two tomograms were obtained for inactive integrin liposomes and one for activated integrin liposomes. The back projection volumes indicated that the actual specimen thickness ranged from 151 nm to 175 nm. The automated tilt series data collection required that the specimen be made eucentric. When so done, the recorded magnification is more accurate than under normal operation.
Inactive integrin liposomes were used immediately for cryo-EM sample preparation. For the active state, the inactive integrin liposomes were activated with 1 mM MnCl2 for 2 hours at room temperature before cryo-EM sample preparation24. Some liposomes displayed a lack of spherical symmetry probably due to an osmotic effect arising from some water evaporation during freezing.
Particle selection and 3-D averaging
Integrin molecules were selected manually in the tomograms. The inactive integrin samples provided 834 molecules inside the vesicles and 881 outside the vesicles. The numbers for the active integrins were 508, and 747 respectively. An ellipsoidal surface was fitted to the coordinates of the picked molecule positions which roughly matched the shape of the vesicles. The initial orientation of the molecules was then determined by calculating the surface normals at the measured molecule positions. The orientation of the molecules inside the vesicles was obtained by inverting the normal. A global average of all subvolumes was generated and used as a starting reference for one cycle of exclusively translational alignment. This step effectively aligned all subvolumes to the center of mass of the headpiece. Translational as well as rotational alignment was carried out with similar methods that we have used previously with success48; 49; 50. However, modifications to these methods included cross-correlation alignment based on the constrained correlation function28 in order to compensate missing wedge effects on the alignment. Because the vesicles in the tomogram were rather asymmetric, the initial orientation determination by surface fitting turned out to be insufficiently accurate thereby requiring a search over a larger spherical sector than we have used previously48. To avoid the increased computational effort we took a different approach for the initial alignment. The centered molecules were first subjected to MDA and hierarchical ascendant classification to group similarly oriented molecules. The 40 class averages generated were then aligned to a single reference. The orientation search was carried out within a cone with a half-angle of 30°. The resulting alignment transformations of the averages were then applied to the individual raw subvolumes of the classes. The classification step was repeated, taking into consideration the corrected transformation of the subvolumes. The 40 new class averages were then aligned to a single reference using a cone with a 2° half angle and the resulting transformation applied to the individual raw subvolume of each class. MDA was carried out a 3rd time and 9 class averages computed. The raw subvolumes were then aligned using multireference alignment to all 9 classes. In this alignment, only the polar angle was varied. A further classification cycle provided the 60 class averages reported here. The averaging was generally carried out with the image transforms in Fourier space where each averaged Fourier coefficient was weighted according to the number of contributions, which excluded the volume falling within the missing wedge of the individual molecular transforms.
We did not measure the molecular height of the individual class members and instead produced a ruler based on the predicted height above the membrane of the integrin crystal structure, 110 Å, and for the bilayer, 60 Å, and placed over each class average and aligned subvolume (Figures 5, S2 and S3).
Multivariate Data Analysis
To avoid losing any potential integrin conformations, no molecular volumes were discarded. Instead MDA51 was used to separate conformations and selection was done by computing class averages. Experience shows that separation into similar groups leads to improved feature definition even when only a few volumes are averaged. The classification mask was a sphere centered on the bulk of the ectodomain density and including some of the membrane density (Supplemental Figure S4).
Supplementary Material
Acknowledgments
We thank Dr. Mark Ginsberg for the integrin activation assay and Drs. Brian Adair and Mark Yeager for the atomic coordinates of their integrin model. This research was supported by NIH Grant U54 GM64346 to the Cell Migration Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Miranti CK, Brugge JS. Sensing the environment: a historical perspective on integrin signal transduction. Nat Cell Biol. 2002;4:E83–90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- 4.Giancotti F, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 5.Shattil SJ, Kashiwagi H, Pampori N. Integrin signaling: the platelet paradigm. Blood. 1998;91:2645–2657. [PubMed] [Google Scholar]
- 6.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 8.Adair BD, Xiong JP, Maddock C, Goodman SL, Arnaout MA, Yeager M. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J Cell Biol. 2005;168:1109–1118. doi: 10.1083/jcb.200410068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong JP, Stehle T, Goodman SL, Arnaout MA. New insights into the structural basis of integrin activation. Blood. 2003;102:1155–1159. doi: 10.1182/blood-2003-01-0334. [DOI] [PubMed] [Google Scholar]
- 10.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–511. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 11.Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwasaki K, Mitsuoka K, Fujiyoshi Y, Fujisawa Y, Kikuchi M, Sekiguchi K, Yamada T. Electron tomography reveals diverse conformations of integrin alphaIIbbeta3 in the active state. J Struct Biol. 2005;150:259–267. doi: 10.1016/j.jsb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 15.Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular alpha4-integrin conformational activation. Biophys J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Boylan B, Luo BH, Newman PJ, Springer TA. Tests of the extension and deadbolt models of integrin activation. J Biol Chem. 2007;282:11914–11920. doi: 10.1074/jbc.M700249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta V, Gylling A, Alonso JL, Sugimori T, Ianakiev P, Xiong JP, Amin Arnaout M. The β-tail domain (βTD) regulates physiologic ligand binding to integrin CD11b/CD18. Blood. 2007;109:3513–3520. doi: 10.1182/blood-2005-11-056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Czekay RP, Robillard L, Kugler MC, Zhang F, Kim KK, Xiong JP, Humphries MJ, Chapman HA. Regulation of α5β1 integrin conformation and function by urokinase receptor binding. J Cell Biol. 2005;168:501–511. doi: 10.1083/jcb.200404112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coutinho A, Garcia C, Gonzalez-Rodriguez J, Lillo MP. Conformational changes in human integrin alphaIIbbeta3 after platelet activation, monitored by FRET. Biophys Chem. 2007;130:76–87. doi: 10.1016/j.bpc.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 21.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies - Visualization of Biological Molecules in Their Native State. 2. Oxford University Press; New York: 2006. [Google Scholar]
- 22.Shattil SJ, Hoxie JA, Cunningham M, Brass LF. Changes in the platelet membrane glycoprotein IIb. IIIa complex during platelet activation. J Biol Chem. 1985;260:11107–11114. [PubMed] [Google Scholar]
- 23.Honda S, Tomiyama Y, Pelletier AJ, Annis D, Honda Y, Orchekowski R, Ruggeri Z, Kunicki TJ. Topography of ligand-induced binding sites, including a novel cation-sensitive epitope (AP5) at the amino terminus, of the human integrin beta 3 subunit. J Biol Chem. 1995;270:11947–11954. doi: 10.1074/jbc.270.20.11947. [DOI] [PubMed] [Google Scholar]
- 24.Gailit J, Ruoslahti E. Regulation of the fibronectin receptor affinity by divalent cations. J Biol Chem. 1988;263:12927–12932. [PubMed] [Google Scholar]
- 25.Tcheng JE, Harrington RA, Kottke-Marchant K, Kleiman NS, Ellis SG, Kereiakes DJ, Mick MJ, Navetta FI, Smith JE, Worley SJ, et al. Multicenter, randomized, double-blind, placebo-controlled trial of the platelet integrin glycoprotein IIb/IIIa blocker Integrelin in elective coronary intervention. IMPACT Investigators. Circulation. 1995;91:2151–2157. doi: 10.1161/01.cir.91.8.2151. [DOI] [PubMed] [Google Scholar]
- 26.Scarborough RM, Naughton MA, Teng W, Rose JW, Phillips DR, Nannizzi L, Arfsten A, Campbell AM, Charo IF. Design of potent and specific integrin antagonists. Peptide antagonists with high specificity for glycoprotein IIb-IIIa. J Biol Chem. 1993;268:1066–1073. [PubMed] [Google Scholar]
- 27.Dierksen K, Typke D, Hegerl R, Walz J, Sackmann E, Baumeister W. Three-dimensional structure of lipid vesicles embedded in vitreous ice and investigated by automated electron tomography. Biophys J. 1995;68:1416–1422. doi: 10.1016/S0006-3495(95)80314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adair BD, Yeager M. Three-dimensional model of the human platelet integrin αIIbβ3 based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci USA. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elices MJ, Urry LA, Hemler ME. Receptor functions for the integrin VLA-3: fibronectin, collagen, and laminin binding are differentially influenced by Arg-Gly-Asp peptide and by divalent cations. J Cell Biol. 1991;112:169–181. doi: 10.1083/jcb.112.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conforti G, Zanetti A, Pasquali-Ronchetti I, Quaglino D, Jr, Neyroz P, Dejana E. Modulation of vitronectin receptor binding by membrane lipid composition. J Biol Chem. 1990;265:4011–4019. [PubMed] [Google Scholar]
- 32.Yanai T, Shimo-Oka T, Ii I. Manganese ion elicits a binding activity of placenta vitronectin receptor to fibronectin cell-binding domain. Cell Struct Funct. 1991;16:149–156. doi: 10.1247/csf.16.149. [DOI] [PubMed] [Google Scholar]
- 33.Dransfield I, Cabanas C, Craig A, Hogg N. Divalent cation regulation of the function of the leukocyte integrin LFA-1. J Cell Biol. 1992;116:219–226. doi: 10.1083/jcb.116.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirchhofer D, Gailit J, Ruoslahti E, Grzesiak J, Pierschbacher MD. Cation-dependent changes in the binding specificity of the platelet receptor GPIIb/IIIa. J Biol Chem. 1990;265:18525–18530. [PubMed] [Google Scholar]
- 35.Sonnenberg A, Modderman PW, Hogervorst F. Laminin receptor on platelets is the integrin VLA-6. Nature. 1988;336:487–489. doi: 10.1038/336487a0. [DOI] [PubMed] [Google Scholar]
- 36.Kern A, Eble J, Golbik R, Kuhn K. Interaction of type IV collagen with the isolated integrins alpha 1 beta 1 and alpha 2 beta 1. Eur J Biochem. 1993;215:151–159. doi: 10.1111/j.1432-1033.1993.tb18017.x. [DOI] [PubMed] [Google Scholar]
- 37.Tuckwell D, Calderwood DA, Green LJ, Humphries MJ. Integrin alpha 2 I-domain is a binding site for collagens. J Cell Sci. 1995;108(Pt 4):1629–1637. doi: 10.1242/jcs.108.4.1629. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu Y, Mobley JL. Distinct divalent cation requirements for integrin-mediated CD4+ T lymphocyte adhesion to ICAM-1, fibronectin, VCAM-1, and invasin. J Immunol. 1993;151:4106–4115. [PubMed] [Google Scholar]
- 39.Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, Holzmann B, Weissman IL, Hamann A, Butcher EC. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 40.Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 41.Luo BH, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci USA. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu C, Takagi J, Springer TA. Association of the membrane proximal regions of the alpha and beta subunit cytoplasmic domains constrains an integrin in the inactive state. J Biol Chem. 2001;276:14642–14648. doi: 10.1074/jbc.M100600200. [DOI] [PubMed] [Google Scholar]
- 43.Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2:e153. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kouns WC, Kirchhofer D, Hadvary P, Edenhofer A, Weller T, Pfenninger G, Baumgartner HR, Jennings LK, Steiner B. Reversible conformational changes induced in glycoprotein IIb-IIIa by a potent and selective peptidomimetic inhibitor. Blood. 1992;80:2539–2547. [PubMed] [Google Scholar]
- 45.Muller B, Zerwes HG, Tangemann K, Peter J, Engel J. Two-step binding mechanism of fibrinogen to αIIbβ3 integrin reconstituted into planar lipid bilayers. J Biol Chem. 1993;268:6800–6808. [PubMed] [Google Scholar]
- 46.Saxton WO, Baumeister W, Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13:57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- 47.Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Zhu P, Liu J, Bess JJ, Chertova E, Lifson JD, Grisé H, Ofek G, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Lucaveche C, Winkler H, Baumann BA, Squire JM, Irving TC, Reedy MK, Taylor KA. Electron tomography of fast frozen, stretched rigor fibers reveals elastic distortions in the myosin crossbridges. J Struct Biol. 2004;147:268–282. doi: 10.1016/j.jsb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 51.Winkler H, Taylor KA. Multivariate statistical analysis of three-dimensional cross-bridge motifs in insect flight muscle. Ultramicroscopy. 1999;77:141–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.