Abstract
Genetically encoded Ca2+ indicators are important tools that enable the measurement of Ca2+ dynamics in a physiologically relevant context. GCaMP2, one of the most robust indicators, is a circularly permutated EGFP (cpEGFP)/M13/Calmodulin (CaM) fusion protein, that has been successfully used for studying Ca2+ fluxes in vivo in the heart and vasculature of transgenic mice. Here we describe crystal structures of bright and dim states of GCaMP2 that reveal a sophisticated molecular mechanism for Ca2+ sensing. In the bright state, CaM stabilizes the fluorophore in an ionized state similar to that observed in EGFP. Mutational analysis confirmed critical interactions between the fluorophore and elements of the fused peptides. Solution scattering studies indicate that the Ca2+-free form of GCaMP2 is a compact, pre-docked state, suggesting a molecular basis for the relatively rapid signaling kinetics reported for this indicator. These studies provide a structural basis for the rational design of improved Ca2+-sensitive probes.
Introduction
Genetically encoded Ca2+ indicators (GECIs) enable the monitoring of intracellular signaling events in defined cell lineages within complex multicellular tissues, without disrupting cell-cell contacts or permeabilizing cell membranes (Ledoux et al., 2008; Mao et al., 2008; Miyawaki et al., 1999; Miyawaki et al., 1997; Nausch et al., 2008; Roell et al., 2007; Tallini et al., 2007; Tallini et al., 2006). Several distinct strategies have been employed to achieve the goals of a bright, fast, high signal-to-noise indicator that functions under in vivo conditions, including the modification of the green fluorescent protein (GFP) family from the Aequorea victoria (Kotlikoff, 2007; Mank and Griesbeck, 2008; Mao et al., 2008; Palmer and Tsien, 2006; Pologruto et al., 2004; Reiff et al., 2005).
Intact GFP has a cage-like structure with 11 β-sheets forming a barrel around a helix containing the fluorescent moiety (Brejc et al., 1997; Ormo et al., 1996; Yang et al., 1996). Protected from solvent access and coordinated by a number of water molecules and polar side chains contributed by the cage, three consecutive residues in the central helix (Ser-Tyr-Gly) undergo post-translational cyclization to form the fluorophore (Tsien, 1998). Spectroscopic and structural studies have illuminated the molecular mechanism underlying GFP fluorescence (Tsien, 1998). Wild-type GFP (class 1 GFP) has a major excitation peak at a wavelength of 395 nm and a minor peak at 475 nm, resulting from a protonated and a deprotonated fluorophore, respectively, with distinct emission properties (503 nm vs 508 nm). Neutralizing effects of close-by residues maintain the fluorophore in its protonated state. The fluorophore is subject to reversible ionization facilitated by a surrounding hydrogen bond network involving interactions with the cage wall. Mutation of Ser-65 of the chromophore to threonine yielded an enhanced GFP (EGFP) with a deprotonated fluorophore and a single excitation peak at 489 nm (GFP-S65T; class 2 GFP) (Ormo et al., 1996).
GFP is amenable to large structural rearrangements without disrupting its basic fluorescent properties, including circular permutation (Baird et al., 1999). Progressive improvements in sensor performance have been made using a central circularly permutated EGFP (cpEGFP) moiety flanked by the M13 helix of myosin light chain and Calmodulin (CaM) at the N- and C-terminus, respectively (GCaMP2 or pericam; Figure 1A) (Nagai et al., 2001; Nagai et al., 2004; Nakai et al., 2001; Souslova et al., 2007; Tallini et al., 2007). These molecules exploit Ca2+-dependent intramolecular conformational changes to control the efficiency of fluorophore function. However, the structural basis for Ca2+-dependent changes in fluorescence is not understood constraining the rational design and optimization of these proteins.
Figure 1. Crystal structures of GCaMP2•Ca2+ and cpEGFP.
(A) Domain organization of GCaMP2 and truncated derivatives. A schematic presentation of the GCaMP2 fusion protein is shown. The color scheme introduced here is maintained throughout the manuscript. Residue numbering for circularly permutated EGFP (cpEGFP) and GCaMP2ΔRSET follows the sequence of GCaMP2. (B) Crystal structure of the isolated cpEGFP moiety. The C-terminal fragment of C-EGFP is colored in light green, the N-terminal fragment is colored in dark green. Two orthogonal views are shown. (C) Crystal structure of monomeric GCaMP2ΔRSET in its Ca2+-bound state. Crystals were grown in the presence of 1 mM Ca2+. Two orthogonal views are shown. The M13 helix is shown in blue, and the calmodulin (CaM) domain is shown in red. The cpEFGP is colored as described in (B). (D) Comparison of crystal structures of GCaMP2, cpEGFP and GFP-S65T. Distance difference matrices based on Cα positions were used to compare the conformation of cpEGFP in isolation (bottom-right triangle) and as part of GCaMP2 (top-left triangle) with the structure of GFP-S65T (PDB code: 1EMA; see Supplemental Data for details). Difference matrices were regularized using a Z-score analysis and color-coded accordingly. Each entry in the matrix depicts the difference in distance between corresponding Cα atoms in the two structures. Distances that show little change are blue. Red entries represent distances that are significantly different in the two structures.
The construction and optimization of novel Ca2+ indicators or other molecular sensors based on this strategy currently requires extensive random mutagensis and screening due to limited understanding of the working mechanism of the current molecules. Here, we determined structures of GCaMP2 in multiple states revealing a surprisingly sophisticated network of interactions between the cpEGFP and CaM moieties that are responsible for the high functionality of this Ca2+ probe. These studies will guide the structurally motivated improvement of current molecules and the design of novel indicators.
Results and Discussion
Crystal structures of cpEGFP and GCaMP2•Ca2+
Initial studies were carried out with the original GCaMP2 protein containing the N-terminal hexahistidine tag and RSET module (Nakai et al., 2001) (Figure 1). Removal of the purification tag and RSET motif yielded crystals of the bright, Ca2+-bound state of GCaMP2. The N-terminal truncations had no effect on basic properties and efficiency of GCaMP2 at room temperature (expression and purification, spectroscopic properties, and Ca2+ response; Supplemental Figure 1) suggesting that these domains mainly affect GCaMP2’s folded state under in vivo conditions (Tallini et al., 2006). If not indicated otherwise, the truncated form, GCaMPΔRSET, was used in subsequent experiments.
Crystallization trials carried out with native GCaMP2ΔSET and cpEGFP yielded crystal structures of monomeric GCaMP2ΔRSET•Ca2+ and the isolated cpEGFP module, respectively (Figure 1B and C; see Supplemental Figure 2 and Table 1 for details). GCaMP2ΔRSET•Ca2+ has a compact structure with the M13 helix forming an intramolecular interaction with the CaM moiety (Figure 1C). The cpEGFP module in GCaMP2 and the isolated domain are very similar in structure to GFP-S65T (rms deviation of 0.48 and 0.22 Å, respectively). To further assess the folded state and conformation of the cpEGFP module in GCaMP2 and in isolation in an unbiased way, distance difference analyses based on corresponding Cα positions were performed (Figure 1D; see Supplemental Data for details). The comparison of cpEGFP (lower part of the matrix) and GCaMP2ΔRSET (upper half of the matrix) with the structure of GFP-S65T (Ormo et al., 1996) produced featureless matrices indicating that in both structures, cpEGFP is properly folded, indistinguishable in ternary structure to intact GFP (blue: distances showing minimal difference between two structures). Minor conformational changes were restricted to a single loop (residues 66-72 in GCaMP2, corresponding to residues 154-160 in GFP) and the N-terminal three residues of the first β-sheet of the C-EGFP (Figure 1) that connect to the M13 fragment (residues 62-64 in GCaMP2/cpEGFP) (Figure 1). The latter region contributed to an intact β-barrel in cpEGFP, but is peeled away when the M13 helix was bound to the CaM domain in GCaMP2ΔSET•Ca2+, producing a hole in the cpEGFP cage (Figure 1B-D; see below). Based on these results, we can rule out major conformational changes in the cpEGFP module as the basis of Ca2+ sensitivity of GCaMP2.
Fluorophore coordination in GCaMP2 and cpEGFP
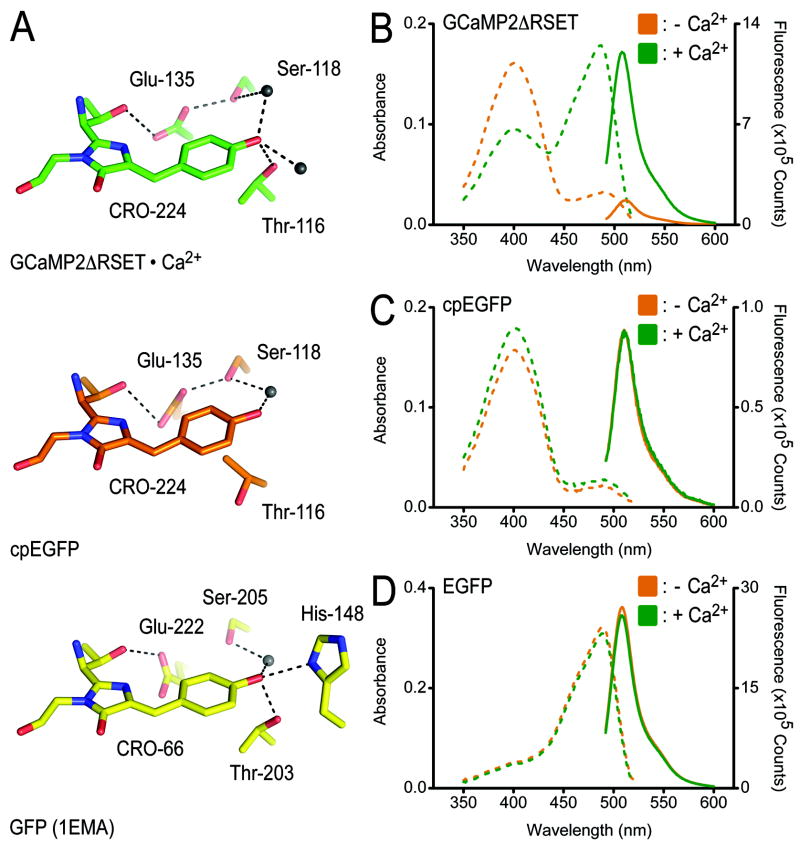
Structural and spectroscopic studies have identified hallmarks of the distinct states of GFP manifested in the coordination of the fluorophore (Brejc et al., 1997; Yang et al., 1996). In the protonated state characterized by a major absorbance peak at 395 nm, hydrogen bonds between Glu-222, Ser-205 and the fluorophore contribute to the neutralization of its phenolic oxygen. In the state with an absorbance maximum at 489 nm (e.g. GFP-S65T or EGFP), an anionic phenolate is stabilized by interactions with Thr-203 and His-148 of the β-barrel (Brejc et al., 1997; Ormo et al., 1996) (Figure 2A, bottom).
Figure 2. Coordination of the fluorophore in structures of GCaMP2 and cpEGFP.
(A) Close-up view of the fluorophore-interacting residues in GCaMP2ΔRSET•Ca2+ (top), cpEGFP (middle), and GFP-S65T (bottom). Residue labeling is according to GCaMP2 numbering, except labeling of residues in the structure of GFP-S65T (PDB code: 1EMA) (Ormo et al., 1996). Hydrogen bonds are shown as dashed lines. (B) Spectroscopic properties of GCaMP2ΔRSET. Absorbance (dashed lines) and fluorescence emission (solid lines) spectra were measured at 25°C in Ca2+-free (10 mM EGTA; orange) buffer or in the presence of Ca2+ (40 μM; green). See Material and Methods for experimental details. (C) Spectroscopic properties of cpEGFP. Experimental conditions were identical to (B). (D) Spectroscopic properties of EGFP. Experimental conditions were identical to (B).
The interaction between the fluorophore and the threonine residue is preserved in the structure of Ca2+-bound GCaMP2ΔRSET (Thr-116 in GCaMP2ΔRSET; Figure 2A, top). Additional hydrogen bonds with two water molecules complete the sp3 hybridization of the phenolic oxygen, indicative of an ionic, bright fluorophore. In contrast, the structure of cpEGFP resembles features of a protonated fluorophore, as evidenced by the loss of a hydrogen bond between Thr-116 and the phenolic oxygen of the fluorophore, and the presence of a hydrogen bond between Glu-135 and Ser-118 (Glu-222 and Ser-205 in GFP-S65T) (Figure 2A, middle). The fluorophore itself and other residues involved in coordination and maturation of the fluorophore (residues Gln-252 and Arg-254 in GCaMP2; residues 94 and 96 in GFP-S65T) adopt very similar conformations in all the structures (Supplemental Figure 3A) (Ormo et al., 1996).
The structural analysis is consistent with emission and excitation properties of the proteins. GCaMP2ΔRSET behaves similarly to the parent protein GCaMP2. In the absence of Ca2+, its fluorescence intensity is low with an absorbance maximum at 399 nm, indicative of a neutral fluorophore (Figure 2B). Upon Ca2+ binding, the absorbance maximum shifts to 488 nm, approximately a 5-fold increase at this wavelength compared to the apo-state, accompanied with a 7.5-fold increase in fluorescence intensity measured at 507 nm (Figure 2B). The absolute increase in fluorescence is slightly higher than the one reported previously for GCaMP2 (Hendel et al., 2008; Tallini et al., 2006), possibly due to the usage of a homogenously monomeric sample (see below). In contrast, cpEGFP displays Ca2+-independent absorbance and emission properties and resided in a constitutively protonated state with an absorbance maximum at 399 nm and low fluorescence intensity (Tsien, 1998) (Figure 2C). Consistent with its high fluorescence and deprotonated fluorophore observed in the crystal structure, EGFP absorbed maximally at a peak wavelength of 488 nm both in the presence and absence of Ca2+ (Figure 2D). These results suggest that the Ca2+-dependent change in fluorescence derives from the fact that the destabilized fluorophore in cpEGFP is protonated at physiological pH, and that Ca2+ binding to calmodulin, and the attendant interaction with the M13 peptide, restores the deprotonated fluorophore structure, resulting in a marked increase in GFP fluorescence.
Direct contacts between cpEGFP and CaM determine the bright state of GCaMP2
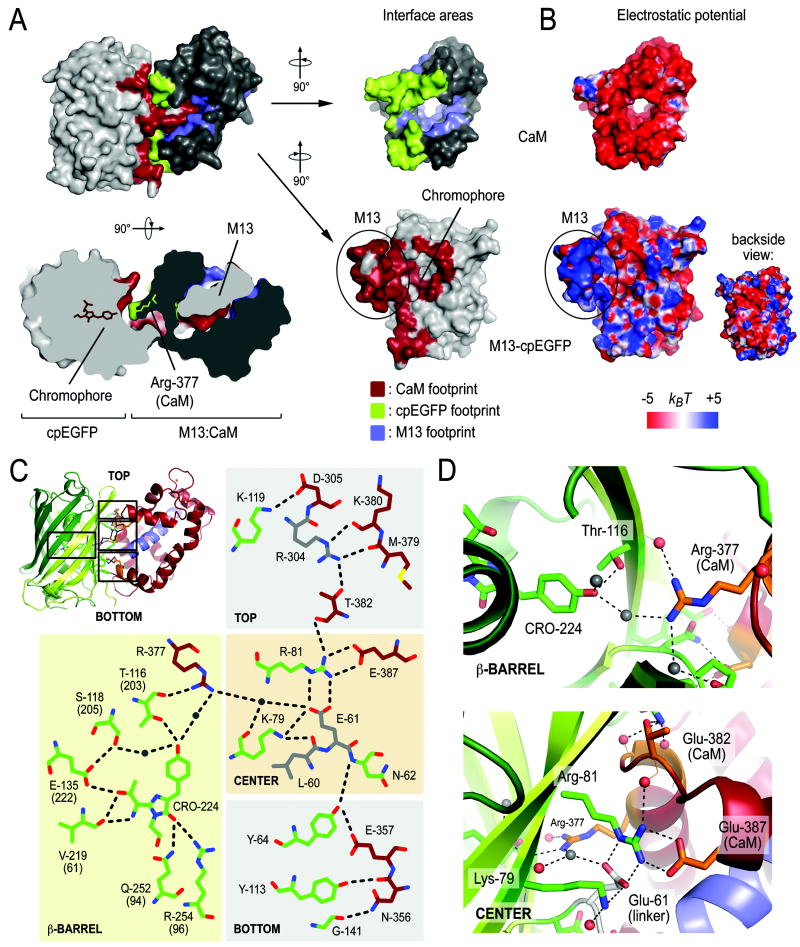
We next examined the molecular basis for the fluorophore stabilization described above. The Ca2+-bound CaM module has a tight grip on the M13 helix, bringing the CaM and cpEGFP moieties into close proximity with a buried surface area of 1581 Å2 (4287 Å2 including M13) at the interface (Figure 3A). It is interesting to note that the CaM domain forms a ring-shaped seal around a hole in the cpEGFP cage that is present due to the circular permutation and partially exposes the fluorophore to solvent (Figure 3A), a positioning that would enable protection of the exposed residues.
Figure 3. Intramolecular interfaces in monomeric GCaMP2•Ca2+.
(A) Interfaces between the cpEGFP, M13 and CaM modules in the structure of monomeric GCaMP2ΔRSET•Ca2+. Residues of the M13-cpEGFP module interacting with CaM are colored red. Interfacial residues on CaM are colored in green and blue for contacts with cpEGFP and the M13 helix, respectively. A top view, rotated 90° around the horizontal axis with respect to the view shown above, is shown as a cutaway rendition of the surface (bottom-left). The fluorophore of cpEGFP and Arg-377 of CaM are shown in stick presentation. Surface presentation of the isolated CaM domain and M13-cpEGFP unit were rotated by +90° and -90°, respectively, with respect to the view of the assembled structure (top-left). (B) Electrostatic potential of the M13-cpEGFP module and CaM mapped onto its molecular surface. Views are identical to (A). Red represents negative and blue represents positive potential (-5 to +5 kBT). (C) Schematic diagram of the fluorophore environment and the hydrogen bond network between cpEGFP and CaM. The numbering scheme for GCaMP2 was used. Corresponding residue numbers in GFP are shown in brackets. Carbon atoms of residues in cpEGFP, CaM and linker segments are shown in green, dark red, and grey, respectively. Hydrogen bonds shown in the figure are between 2.7 and 3.3Å (not drawn to scale). (D) Close-up views of the interfacial regions in GCaMP2ΔRSET•Ca2+. Water-mediated interaction between the fluorophore and Arg-377 of the CaM domain (top) and cpEGFP:CaM interfacial residues (bottom) are shown.
The interaction between CaM and cpEGFP is facilitated by a concentration of positively charged residues at the surface of cpEGFP that can form favorable contacts with an overall negatively charged CaM domain (Figure 3B). In addition, there is a considerable amount of shape complementarity in the interfacial region (Figure 3A) that allows a continuous hydrogen bond network to form along the β-barrel of cpEGFP and the point of permutation (Figure 3C). Central to the interface along the vertical axis of the cpEGFP cage are two arginine residues, Arg-81 in cpEGFP and Arg-377 in CaM. Notably, Arg-377 forms a weak hydrogen bond with the carbonyl main chain group of Thr-116 (Thr-203 in GFP-S65T), a residue involved in stabilizing the fluorophore in the ionized state (see above). His-148 in GFP-S65T, another residue involved in fluorophore coordination in the bright state, is missing in GCaMP2. Instead, Arg-377 forms a water-mediated interaction with the fluorophore, suggesting that Arg-377 is functionally equivalent to His-148 (Figure 3C and D).
Significant design efforts yielded an optimized linker sequence connecting the functional moieties of GCaMP2 (Nakai et al., 2001). Most of these regions were resolved in the crystal structure of GCaMP2•Ca2+ (except for a 15-residue fragment connecting the two parts of EGFP). Linkers were found to be peripherally involved in the packing of the domains and as such might restrict inter-domain flexibility. From the multitude of interactions at the interface, Arg-81 appears to be a central coordinator (Figure 3C and D). It is hydrogen-bonded to Glu-61 (and Glu-387), one of the two residues linking the M13 fragment to cpEGFP (Figure 1A and 3C). In addition, it is indirectly linked to Arg-304 located in the linker connecting the cpEGFP and CaM moieties (via Thr-382; Figure 3C). The linker length appears to be important as well (Nakai et al., 2001), constraining the conformational freedom of the system even further (see below).
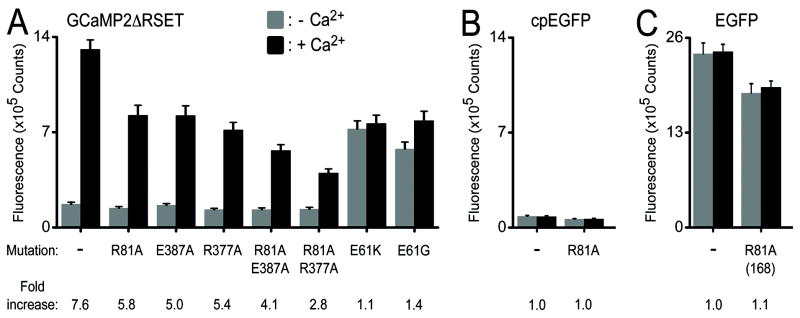
Mutating Arg-377, Arg-81 or its binding partner Glu-387 (to alanine) resulted in a dimished fluorescence signal upon Ca2+ binding without affecting the emission intensity in the absence of Ca2+ (Figure 4A), while equivalent mutations of Arg-81 in cpEGFP and EGFP affected their fluorescence intensity only slightly (Figure 4B and C). A double-mutant Arg-81-Ala/Glu-387-Ala, removing both side chains of the interacting residues, showed only moderate affects on Ca2+-dependent fluorescence increase compared to the single mutation, consistent with either modification disrupting the bright state conformation. In contrast, the double-mutant Arg-81-Ala/Arg-377-Ala exhibited markedly reduced Ca2+ sensitivity indicative of additive contributions of Arg-81 and Arg-377 to the interdomain interface and overall mechanism. Mutating the linker residue Glu-61 to either glycine or lysine generated a defective probe with intermediate fluorescence intensity but lacked Ca2+ responsiveness (Figure 4A). Therefore, the negative charge of Glu-61 appears to stabilize the neutral state of the fluorophore under Ca2+-free conditions reducing its background fluorescence. Taken together, the mutagenesis data suggests that contributions from several residues collectively account for the performance of GCaMP2.
Figure 4. Mutational analysis of GCaMP2•Ca2+.
(A) Mutations in GCaMP2ΔRSET. Interfacial residues shown in Figure 3C and D were mutated. The fluorescence emission spectra were recorded in the presence (black) and absence (grey) of Ca2+. Bar diagrams show emission at 507 nm. (B) Fluorescence emission of corresponding mutations in the isolated cpEGFP module. (C) Fluorescence emission of corresponding mutations in EGFP. The GCaMP2 numbering scheme was used. Arg-81 is equivalent to Arg-168 in GFP and EGFP.
Consistent with a shift in fluorophore protonization as the general mechanism of Ca2+-dependent fluorescence, the interactions between cpEGFP and CaM described above contributed to a decrease in the apparent acidity of the fluorophore in Ca2+-bound GCaMP2ΔREST (Supplemental Figure 3B) (Nakai et al., 2001). The apparent pKa value for the high-fluorescent state was 6.9, ~0.8 units lower than under Ca2+-free conditions, consistent with data reported for GCaMP1 (Nakai et al., 2001). In contrast, cpEGFP was less sensitive to pH, with pKa values of 7.4 in the absence of Ca2+ and 7.9 in the presence of Ca2+. These values were approximately 1.5 units higher than the one determined for EGFP (Patterson et al., 1997) (Supplemental Figure 3C), further confirming that increased solvent accessibility due to circular permutation is the major determinant for the neutral low-fluorescent state (Baird et al., 1999).
Taken together, Ca2+ sensing by GCaMP2 can be described by a switching from a class-1 towards a class-2 GFP by restricting solvent access of the fluorophore and stabilizing it in an ionized form (Tsien, 1998).
Structural characterization of the Ca2+-bound and Ca2+-free state of GCaMP2 in solution
Analysis of GCaMP2 by size exclusion chromatography suggested that Ca2+-bound GCaMP2 adopted a similar but not identical conformation as the Ca2+-free state (Figure 5A), with the Ca2+-bound state being slightly more compact based on its retention time. To further explore the conformational changes occurring upon Ca2+ binding, we carried out small angle X-ray scattering experiments, yielding molecular geometry parameters such as the radius of gyration (Rg) and maximum diameter of a molecule or assembly (Dmax) (Table 2). The scattering profiles for the Ca2+-free and Ca2+-bound state were very similar at low angles (corresponding to the range of momentum transfer of 0.019 < S < 0.13 Å-1) (Figure 5B). Radii of gyration and maximum diameters of the two states calculated from the scattering profiles were comparable (Rg[+Ca2+]= 23.1±0.5 vs Rg[-Ca2+]= 23.7±0.5; Dmax[+Ca2+]= 70.0±2.0 vs Dmax[-Ca2+]= 68.0±2.0), indicating an overall similar shape. Distance distribution functions [P(r)] corroborated this finding but also revealed a conformational change between GCaMP2 in the presence or absence of Ca2+. In the Ca2+-free state, the peak of the P(r) function shifted about 5Å towards larger dimension compared to the P(r) function for GCaMP2•Ca2+ or the corresponding profile calculated from the coordinates of the crystal structure (Figure 5C). This apparent compaction of GCaMP2 upon Ca2+ binding is consistent with the gel filtration experiments (see above).
Figure 5. SAXS-based shape reconstruction of GCaMP2 in the presence or absence of Ca2+.
(A) Size exclusion chromatographic (SEC) analysis of GCaMP2 in the presence and absence of Ca2+. GCaMP2ΔRSET was purified in its monomeric state (see Material and Methods for details). Proteins were analyzed on a S200 gel filtration column (10/300; GE Healthcare) in gel filtration buffer containing EGTA (10 mM; orange trace) or Ca2+ (1 mM; green trace). (B) Solution scattering data for GCaMP2ΔRSET in its Ca2+-bound and Ca2+-free state. Small-angle X-ray scattering curves of GCaMP2ΔRSET•Ca2+ (green) and its EGTA-treated form (orange) are shown after averaging and solvent-subtraction. Theoretical scattering profiles calculated from the ab initio models with the lowest χ values are shown (black line). The inset shows Guinier plots (including linear fits) at the low angle region (Smax*Rg<1.3). (C) Distance distribution [P(r)] functions for GCaMP2ΔRSET. P(r) curves of GCaMP2ΔRSET•Ca2+ (green) and its EGTA-treated form (orange) were calculated from SAXS data shown in (B) or from the crystal structure of GCaMP2ΔRSET•Ca2+ (dashed line). (D) SAXS-based shape reconstruction of GCaMP2ΔRSET•Ca2+. The overall volume from shape reconstructions after averaging (grey envelope) was calculated from 40 independent models. The crystal structure of monomeric GCaMP2ΔRSET•Ca2+ was docked into the envelope manually. Two orthogonal views are shown. (E) SAXS-based shape reconstruction of Ca2+-free GCaMP2ΔRSET. Details are as described in (C). The crystal structures of cpEGFP and Ca2+-free CaM (PDB code: 3CLN) (Babu et al., 1988) were docked into the envelope manually.
Table 2.
SAXS data statistics
| Rg (Å)a | Dmax (Å)b | MMseq(kDa)c | MMexp(kDa)d | Vp (nm3)e | Ipnorm/Istdnorm,f | NSDg | |
|---|---|---|---|---|---|---|---|
| 10 mM EGTA | 23.7±0.5 | 68.0±2.0 | 50.7 | 45.9±5 | 79±8 | 0.91±0.10 | 0.63±0.02 |
| 40 μM Ca2+ | 23.1±0.5 | 70.0±2.0 | 50.7 | 46.2±5 | 66±7 | 0.92±0.10 | 0.54±0.01 |
Rg determined from Grunier equation and data that satisfies QmaxxRg<1.3; similar values were obtained using the program GNOM (Svergun, 1992).
Dmax was determined using the program GNOM (Svergun, 1992).
MMseq is the molecular mass calculated from the primary sequence.
Vp is the excluded volume (Porod volume) calculated using PRIMUS (Konarev et al., 2003). Data with S>0.25 were excluded from the calculation (Petoukhov and Svergun, 2006; Porod, 1982).
After superimposing the independently modeled envelopes, pair-wise normalized spatial discrepancy (NSD) values (Volkov and Svergun, 2003) were calculated as part of the DAMAVER routine. NSD values close to unity indicate good agreement between individual models.
To investigate the origin of these differences, we calculated low-resolution ab initio shape reconstructions (see Material and Methods for details) (Figure 5D and E). Envelopes (shown in grey) represent the overall shape of GCaMP2 at a maximum resolution of ~30Å. The shape reconstruction of the Ca2+-bound state agreed very well with the crystal structure obtained for GCaMP2ΔRSET•Ca2+ (Figure 5D). In contrast, Ca2+-free GCaMP2 adopted a donut-shaped conformation with a central hole in the center of the envelopes. While one side of the envelope could accommodate the structure of cpEGFP, the other side had dimensions that fit the structure of Ca2+-free apo-CaM very well (Babu et al., 1988) (Figure 5E).
The overall similar dimensions of Ca2+-free and Ca2+-bound GCaMP2 and the shape reconstructions suggest that apo-GCaMP2 exists in a pre-docked state, likely to be stabilized by electrostatic interactions and the short nature of the linker segments connecting the domains in GCaMP2 (see above). Such a conformation might explain the rapid kinetics of Ca2+-induced changes in GCaMP2 that happen at milli-second timescale (Nakai et al., 2001; Tallini et al., 2006).
Identification of a dimeric, low-fluorescent state of GCaMP2•Ca2+
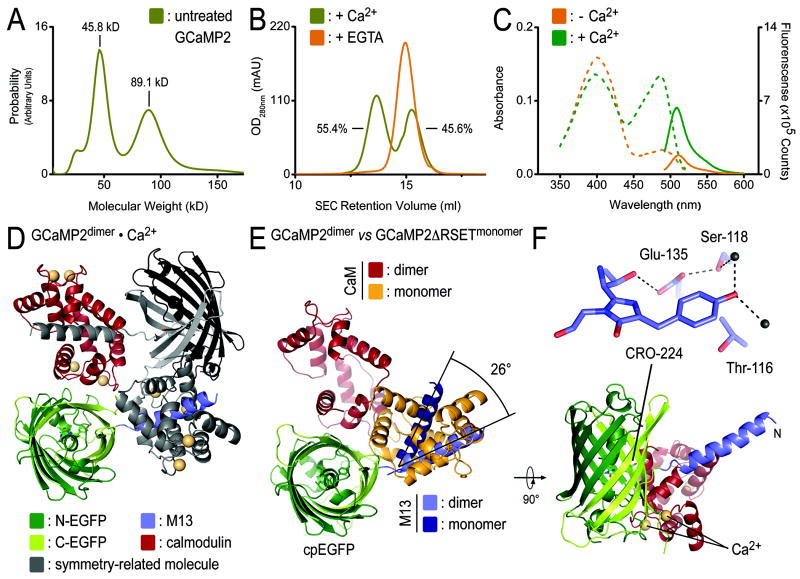
During the purification of the proteins from E. coli, we noticed a bimodal distribution of GCaMP2 and GCaMP2ΔRSET in size exclusion chromatography (Figure 6A and B). Analysis by analytical ultracentrifugation revealed that GCaMP2 coexisted in a monomeric and a dimeric state (Figure 6A), with the monomer peak eluting at 15.3 ml and the dimer peak, accounting for about 55% of the sample, eluting at 13.7 ml from a size exclusion column (Figure 6B). The dimer was sensitive to treatment with EGTA (Figure 6B). The EGTA-treated protein eluted with a slightly shorter retention time than the monomer peak of the input protein, indicating that both the dimeric and monomeric species purified from bacterial lysates were bound to Ca2+ (Figure 6B). Absorbance and emission profiles of the Ca2+-bound monomer-dimer mixture indicated a half-maximal efficiency with regard to fluorescence intensity compared to the EGTA-treated, monomeric protein (Figure 6C and 2B). The partial response could be attributed to the monomeric fraction in the sample that accounted for 45% of the sample (Figure 6A-C). The spectroscopic analysis suggested that the dimeric state of GCaMP2 is a Ca2+-bound, low-fluorescence state.
Figure 6. Characterization and crystal structure of dimeric GCaMP2•Ca2+.
(A) Identification of a dimeric GCaMP2 by analytical ultracentrifugation. Sedimentation velocity experiments using GCaMP2 purified from E. coli yielded bimodal molecular weight distributions with masses (45.8 kD and 89.1 kD), close to the theoretical mass of monomeric (50.7 kD) and dimeric GCaMP2 (101.4 kD), respectively. (B) Monomerization of GCaMP2 by EGTA-treatment. Size exclusion chromatograms for initial preparation of GCaMP2 showed bimodal distribution of the protein. Both peaks were sensitive to EGTA-treatment, converging to a single peak corresponding to Ca2+-free, monomeric protein. EGTA treatment produced protein that remained monomeric in the presence or absence of Ca2+ (see Figure 5A). (C) Spectroscopic properties of partially dimeric GCaMP2. Absorbance (dashed lines) and fluorescence emission (solid lines) spectra were measured at 25°C in Ca2+-free (10 mM EGTA; orange) buffer or in the presence of Ca2+ (40 μM; green). (D) Crystal structure of the dimeric assembly. Crystals were grown from partially dimeric Ca2+-bound GCaMP2. A crystallographic dimer of GCaMP2 is shown with protomer colored according to the scheme introduced in Figure 1A, and a crystal symmetry-related molecule shown in grey. Ca2+ is shown as yellow spheres. (C) Superposition of monomeric and dimeric GCaMP2. A single protomer of dimeric GCaMP2, colored as in (D), was superimposed on the cpEGFP domain of monomeric GCaMP2ΔRSET•Ca2+, with its M13 helix and CaM module colored in dark blue and orange, respectively. (F) Fluorophore coordination in dimeric GCaMP2•Ca2+. A close-up view of the fluorophore-interacting residues in dimeric GCaMP2•Ca2+ is shown. Residue labeling is according to GCaMP2 numbering.
The results were consistent with structural studies on the basis of crystal structures of the Ca2+-bound dimeric state (Figure 6D). Crystals were obtained in two independent trials (Supplemental Figure 4A), starting from a partially dimeric protein (Figure 6A) or from a sample that was purified in a monomeric state (Figure 4A). In solution, the latter protein remained monomeric upon Ca2+ addition suggesting that the high protein concentration and crystallization condition contributed to dimer formation. The structure showed a domain-swapped assembly, in which the M13 helix of one molecule was bound to the CaM moiety of a crystal symmetry-related molecule (Figure 6D). Comparison of the dimeric and monomeric states showed that the M13 helices and CaM domains were separated by a rotation of about 26° and 90°, respectively, relative to cpEGFP. The CaM domain of a symmetry-related molecule in the dimer occupied the approximate position of CaM in the monomeric structure (Figure 6D and E). Yet, interfacial areas in the dimer were overall less extensive and overlapped only partially with areas that were buried in the monomeric, Ca2+-bound conformation (Supplemental Figure 4B and C, Figure 3A). Most notably, interactions between cpEGFP and CaM did not occlude the hole in the β-barrel cage introduced by circular permutation, leaving the fluorophore solvent-exposed in such a conformation (Supplemental Figure 4B and C).
The fluorophore environment in the dimeric structure reflected the spectroscopic properties discussed above. The phenolic oxygen of the fluorophore formed hydrogen bonds with two water molecules. Thr-116 (Thr-203 in GFP-S65T) adopted a rotamer conformation incompetent of chromphore stabilization, characteristic of a low-fluorescent, protonated state also observed in the structure of cpEGFP (Figure 6F). The structural studies corroborated the notion based on the spectroscopic analysis that the dimeric state of GCaMP2 is Ca2+-insensitive.
Conclusions
Here, we described the molecular mechanism underlying Ca2+-dependent fluorescence of the high signal, genetically encoded Ca2+ sensor GCaMP2 based on high-resolution crystal structures and solution scattering studies. Many aspects of the particular design of GCaMP2 are likely to contribute to the success of this Ca2+ indicator, including the particular point of permutation allowing for reversible solvent exposure of the fluorophore, the complementarity between interfaces, and the functional conservation of fluorophore-coordinating residues. Our results indicate that circular permutation of EGFP destabilizes the deprotonated state of the fluorophore, and that this state is re-established in a Ca2+-dependent manner through intensive contacts between the condensed CaM:M13 structure and EGFP moieties. A pre-docked state of CaM in the absence of Ca2+ might contribute to the rapid transition between the two fluorophore states.
The complex and extensive nature of the interactions underlying this mechanism carries important implications for the design of GECIs and other molecular sensors based on the cpEGFP platform. First, achieving the reversible destabilization of a bright state with other fluorescent proteins or fusion peptides by random means will likely require extensive mutagenesis and screening, as occurred with the development of GCaMP2. Rather, forward design of structures to achieve this goal would seem more likely to yield effective signaling molecules. Second, the surface properties such as charge and shape complementarity of the fluorescent module and the fusion peptides should be considered so as to optimize the kinetics of state transition and to minimize interactions with other binding partners. Alternatively, the incorporation of binding sites (e.g. a calmodulin binding site) could also confer a pre-docked state. The results may suggest that it is unlikely, however, that marked improvement in kinetics will be achieved using current cpEGFP-based signaling proteins. While this may limit the utility of these GECIs for the most rapid cell signaling events (Hendel et al., 2008; Mao et al., 2008; Tallini et al., 2006), GCaMP2 transitions occur in less than 100 ms at physiological temperatures, sufficient for many signaling processes. Third, the discovery of a dimeric dim state indicates the importance of preventing intermolecular binding, as these proteins contribute to background fluorescence and thereby may markedly limit the dynamic range of the signaling system. In summary, our studies provide a structural basis for future improvements in the rational design of protein-based signaling molecules and identify key limitations to this strategy.
Material and Methods
Protein expression and purification
The coding regions corresponding to GCaMP2, EGFP, and cpEGFP were amplified by standard PCR and were cloned into the pRSET expression plasmid (Nakai et al., 2001). GCaMP2ΔRSET was cloned into a modified pET28a expression plasmid (Novagen) yielding N-terminally hexahistidine-tagged SUMO fusion proteins. The hexahistidine-tagged SUMO-moiety was cleavable using the protease from Ulp-1 from S. cerevisiae.
Transformed E. coli cells BL21(DE3) (Novagen) were grown in TB medium supplemented with 50 mg/l antibiotics at 37°C. At a cell density corresponding to an absorbance of 1.0 at 600 nm the temperature was reduced to 18°C, and protein production was induced with 1 mM IPTG. Proteins were expressed for 12–16 hr. Cells were collected by centrifugation, resuspended in NiNTA buffer A (25 mM Tris-Cl pH 8.2, 500 mM NaCl and 20 mM imidazole). After cell lysis by sonication, cell debris was removed by centrifugation at 40,000 × g for 1 hr at 4°C. Clear lysates were loaded onto HisTrap NiNTA columns (GE Healthcare) equilibrated in NiNTA buffer A. The resin was washed with 20 column volumes NiNTA buffer A, and proteins were eluted in a single step with NiNTA buffer A supplemented with 500 mM imidazole. GCaMP2ΔRSET was incubated with SUMO protease ULP-1 at 4°C overnight for removal of the hexahistidine-SUMO tag, and the cleaved protein was collected in the flow-through during NiNTA affinity chromatography. Proteins were further subjected to size exclusion chromatography on a Superdex200 column (16/60; GE Healthcare) equilibrated in gel filtration buffer (100 mM NaCl, 25 mM HEPES pH 7.4).
Ca2+-free proteins were produced by incubation in gel filtration buffer supplemented with 10 mM EGTA prior the size exclusion column. Fractions containing protein were pooled and concentrated on a Centricon ultrafiltration device (10 kDa cutoff; Millipore). Protein aliquots were frozen in liquid nitrogen and stored at -80°C.
Point mutations were generated using the QuikChange XL Mutagenesis Kit (Stratagene) following the manufacturer’s instructions. Expression and purification of mutant proteins was identical to the procedure described for parent proteins. Mutant proteins showed size exclusion profiles in the presence or absence of Ca2+ indistinguishable from original proteins.
Analytical size exclusion chromatography was carried out on a Superdex200 column (10/300; GE Healthcare) equilibrated in gel filtration buffer at a protein concentration of 0.2 mM (initial concentration). EGTA (“-Ca2+”; 10 mM) or CaCl2 (1 mM) was present where indicated.
Crystallization, X-ray data collection, and structure solution
Crystals were obtained by hanging drop vapor diffusion by mixing equal volumes of protein (~10-30 mg/ml) and reservoir solution at 20°C (if not indicated otherwise). For cpEGFP, the reservoir solution comprised 0.1 M Tris pH 8.5, 2.4 M di-ammonium hydrogen phosphate. Crystals for monomeric GCaMP2ΔRSET•Ca2+ grew in 20% PEG8000, 0.1 M HEPES pH 7.5, 1.6 M ammonium sulfate, and 1 mM CaCl2 at 4°C. The reservoir solution for crystallization of dimeric GCaMP2•Ca2+ consisted of 0.2 M sodium formate, 20% PEG 3350, 10 mM MgCl2, and 1 mM CaCl2 (dimer 1), or 0.2 M MgCl2, 0.1 M Bis-Tris pH 6.5, 25% PEG3350, and 1 mM CaCl2 (dimer 2). During preparation of this manuscript a third crystallization condition has been reported, producing crystals within the same space group and similar unit cell constants (Rodriguez Guilbe et al., 2008). Crystals producing the structure of dimer 1 were grown from partially dimeric protein (as judged by size exclusion chromatography and analytical ultracentrifugation). The structure of dimer 2 was obtained from EGTA-treated, monomeric protein. All crystals were cryo-protected using crystallization solutions supplemented with 20% xylitol, frozen in liquid nitrogen, and kept at 100 K during data collection.
Crystallographic statistics for data collection are shown in Table 1. Data sets were collected using synchrotron radiation at the Cornell High Energy Synchrotron Source (CHESS, Ithaca, beamline A1, wavelength 0.977Å). Data reduction was carried out with the software package HKL2000 (Otwinowski and Minor, 1997). Phases were obtained from molecular replacement using the software package PHENIX (Adams et al., 2002) with structures of GFP-S65T (PDB code: 1EMA) (Ormo et al., 1996) and Ca2+-bound calmodulin (PDB code: 1MXE) (Clapperton et al., 2002) as search models. Manual refinement in COOT (Emsley and Cowtan, 2004) and minimization using PHENIX (Adams et al., 2002) yielded the final models with good geometry (with all residues being in allowed regions of the respective Ramachandran plots). Illustrations were made in Pymol (DeLano Scientific). Buried surface areas were calculated using the program CNS (Brunger et al., 1998). Electrostatic potentials were calculated using the program APBS (Baker et al., 2001).
Table 1.
Data collection and refinement statistics
| cpEGFP | GCaMP2ΔRSET Ca2+-bound | GCaMP2 dimer Ca2+-bound (1) | GCaMP2 dimer Ca2+-bound (2) | |
|---|---|---|---|---|
| Data collectiona, b | ||||
| Space group | P212121 | P41212 | C2 | C2 |
| Cell dimensions | ||||
| a, b, c (Å) | 51.1, 62.2, 69.5 | 122.0, 122.0, 97.8 | 127.9, 47.3, 69.6 | 127.5, 47.0, 67.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 100.7, 90.0 | 90.0, 100.9, 90.0 |
| Resolution (Å) | 50-1.45 (1.50-1.45) | 50-2.00 (2.07-2.00) | 50-1.75 (1.81-1.75) | 50-2.60 (2.69-2.60) |
| Rsym | 4.2 (13.2) | 12.7 (71.0) | 4.0 (21.1) | 14.2 (44.1) |
| I / σI | 43.3 (9.9) | 23.6 (3.7) | 35.5 (5.0) | 8.0 (2.3) |
| Completeness (%) | 99.3 (93.2) | 100.0 (100.0) | 100.0 (99.9) | 98.0 (85.5) |
| Measured reflections | 302,513 | 881,335 | 188,674 | 41,030 |
| Unique reflections | 39,659 | 50,531 | 41,444 | 12,458 |
| Redundancy | 7.6 (4.1) | 17.4 (15.2) | 4.6 (4.3) | 3.3 (2.6) |
| Refinementc | ||||
| Resolution (Å) | 50-1.45 | 50-2.00 | 50-1.75 | 50-2.60 |
| Rwork / Rfreed | 13.2/17.5 | 16.3/18.7 | 16.8/21.4 | 22.4/27.0 |
| Free R test set size (#/%) | 3171 / 8.1 | 4821 / 10.0 | 4076 / 10.1 | 1167 /10.1 |
| No. atoms | ||||
| Protein | 1851 | 3194 | 3158 | 3054 |
| Ligand/ion | - | 4 (Ca2+) | 4 (Ca2+) | 4 (Ca2+) |
| Water | 383 | 453 | 518 | 55 |
| B-factors | ||||
| Protein | 13.40 | 28.19 | 23.95 | 28.91 |
| Ligand/ion | n.a. | 28.93 | 16.29 | 44.65 |
| Water | 34.70 | 34.11 | 38.78 | 24.18 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.011 | 0.008 | 0.019 | 0.003 |
| Bond angles (°) | 1.510 | 1.126 | 1.664 | 0.739 |
Values as defined in SCALEPACK (Otwinowski and Minor, 1997).
Highest resolution shell is shown in parenthesis.
Values as defined in PHENIX (Adams et al., 2002). All structures were solved by Molecular Replacement.
No sigma cutoffs.
Small angle X-ray scattering (SAXS) and SAXS-based shape reconstruction
SAXS data were collected at the Cornell High Energy Synchrotron Source (CHESS, Ithaca, beamline G1) at an electron energy of 8KeV covering the range of momentum transfer 0.019 < S < 0.22 Å-1. Buffer exchange into Ca2+-free (gel filtration buffer supplemented with 10 mM EGTA) and Ca2+-saturated (gel filtration buffer supplemented with 1 mM CaCl2) buffer was carried out by size exclusion chromatography. Protein samples and buffer controls were centrifuged at 13,200 × g for 20 min before data acquisition. Scattering data were collected in triplicates at protein concentrations between 5-20 mg/ml with exposure times of 5 seconds. Buffer background scattering was collected from gel filtration buffers used during buffer exchange. Background-subtraction, averaging, and scaling were carried out using the program PRIMUS (Konarev et al., 2003), and data was analyzed using the programs GNOM and CRYSOL (Svergun et al., 1995; Svergun, 1992). Only scattering data with Smax*Rg<1.3, computed from Guinier plots at low angle regions, was considered for further analysis. Kratky plots, experimental molecular weight determinations, and normalized scattering intensities were used to assess the folded-state of the proteins and overall data quality using the program IGOR PRO (version 5.04B). Distance distribution functions (P[r]) and Dmax were determined using the program GNOM (Svergun, 1992).
Ab initio free atom modeling was performed using the program DAMMIN (Svergun, 1999) with the distance distribution function as the fitting target. Forty independent simulations are carried out for each protein. Superposition, averaging, and filtering using the program DAMAVER (Volkov and Svergun, 2003) yielded shape reconstructions. Calculated scattering curves and goodness of fit (χ) were calculated using the program CRYSOL (Svergun et al., 1995).
Analytical ultracentrifugation
Sedimentation velocity experiments were carried out using an XL-I analytical ultracentrifuge (Beckman Coulter) equipped with an AN-60 Ti rotor. Proteins (5-15 μM) were diluted in AUC buffer (100 mM NaCl, 25 mM HEPES pH 7.4, and 1 mM TCEP) and were analyzed at a centrifugation speed of 40,000 rpm. Data collection was carried out at 280 nm, followed by data analysis using the program SedFit (version 11.0) using default parameters.
Absorbance and fluorescence spectroscopy
Absorbance spectra (320-500 nm; 10 mm path length) of purified proteins (20 μM) were recorded in duplicate on a DU730 UV/Vis spectrophotomer (Beckman Coulter) at 25°C. Emission spectra (495-600 nm) were recorded in triplicate on a fluorescence spectrophotomer (Photon Technology International) at a protein concentration of 100 nM and an excitation wavelength of 488 nm. The Ca2+ calibration buffer kit (Biotium, Inc) was used to create accurate assay conditions with regard to Ca2+ concentration. Ca2+ sensitivity of purified proteins was determined by measuring the fluorescence intensity at 508 nm under Ca2+-free (10 mM EGTA) and Ca2+-saturated conditions (40 μM Ca2+). Sensitivity to changes in pH was measured at a protein concentration of 10 μM using buffers containing sodium acetate (pH 4.5-5.0), MES (pH 5-6.5), HEPES (pH 6.5-8), or CHES (pH 8.5-10). pKa values were calculated by fitting the experimental data to the Hill equation.
Supplementary Material
Acknowledgments
We are grateful to the scientists and staff members at CHESS for assistance with synchrotron data collection, and to the scientist at the protein facility at Cornell for assistance with analytical ultracentrifugation experiments. We thank Gary Peng for his assistance with plasmid preparation and Lei Tan for her contribution of MATLAB code used in the distance matrix analysis.
Abbreviations
- CaM
calmodulin
- cpEGFP
circularly permutated enhanced green fluorescent protein
- EGTA
ethylene glycol tetraacetic acid
- GECI
genetically encoded calcium indicator
- SAXS
small-angle X-ray scattering
- SEC
size exclusion chromatography
Footnotes
Accession Numbers Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank under ID code XXXX, XXXX, XXXX, and XXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams P, Grosse-Kunstleve R, Hung L, Ioerger T, McCoy A, Moriarty N, Read R, Sacchettini J, Sauter N, Terwilliger T. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci U S A. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Clapperton JA, Martin SR, Smerdon SJ, Gamblin SJ, Bayley PM. Structure of the complex of calmodulin with the target sequence of calmodulin-dependent protein kinase I: studies of the kinase activation mechanism. Biochemistry. 2002;41:14669–14679. doi: 10.1021/bi026660t. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hendel T, Mank M, Schnell B, Griesbeck O, Borst A, Reiff DF. Fluorescence changes of genetic calcium indicators and OGB-1 correlated with neural activity and calcium in vivo and in vitro. J Neurosci. 2008;28:7399–7411. doi: 10.1523/JNEUROSCI.1038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries CM, Whitten AE, Harris SP, Trewhella J. Small-angle X-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. J Mol Biol. 2008;377:1186–1199. doi: 10.1016/j.jmb.2008.01.080. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- Kotlikoff MI. Genetically encoded Ca2+ indicators: using genetics and molecular design to understand complex physiology. J Physiol. 2007;578:55–67. doi: 10.1113/jphysiol.2006.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux J, Taylor MS, Bonev AD, Hannah RM, Solodushko V, Shui B, Tallini Y, Kotlikoff MI, Nelson MT. Functional architecture of inositol 1,4,5-trisphosphate signaling in restricted spaces of myoendothelial projections. Proc Natl Acad Sci U S A. 2008;105:9627–9632. doi: 10.1073/pnas.0801963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M, Griesbeck O. Genetically encoded calcium indicators. Chem Rev. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- Mao T, O’Connor DH, Scheuss V, Nakai J, Svoboda K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS ONE. 2008;3:e1796. doi: 10.1371/journal.pone.0001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Nausch LW, Ledoux J, Bonev AD, Nelson MT, Dostmann WR. Differential patterning of cGMP in vascular smooth muscle cells revealed by single GFP-linked biosensors. Proc Natl Acad Sci U S A. 2008;105:365–370. doi: 10.1073/pnas.0710387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Joint use of small-angle X-ray and neutron scattering to study biological macromolecules in solution. Eur Biophys J. 2006;35:567–576. doi: 10.1007/s00249-006-0063-9. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porod G. General Theory. In: Kratky O, editor. Small-Angle X-Ray Scattering. Academic Press; London: 1982. pp. 17–51. [Google Scholar]
- Reiff DF, Ihring A, Guerrero G, Isacoff EY, Joesch M, Nakai J, Borst A. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Guilbe MM, Alfaro Malave EC, Akerboom J, Marvin JS, Looger LL, Schreiter ER. Crystallization and preliminary X-ray characterization of the genetically encoded fluorescent calcium indicator protein GCaMP2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:629–631. doi: 10.1107/S1744309108016059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roell W, Lewalter T, Sasse P, Tallini YN, Choi BR, Breitbach M, Doran R, Becher UM, Hwang SM, Bostani T, et al. Engraftment of connexin 43-expressing cells prevents post-infarct arrhythmia. Nature. 2007;450:819–824. doi: 10.1038/nature06321. [DOI] [PubMed] [Google Scholar]
- Souslova EA, Belousov VV, Lock JG, Stromblad S, Kasparov S, Bolshakov AP, Pinelis VG, Labas YA, Lukyanov S, Mayr LM, Chudakov DM. Single fluorescent protein-based Ca2+ sensors with increased dynamic range. BMC Biotechnol. 2007;7:37. doi: 10.1186/1472-6750-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Brekke JF, Shui B, Doran R, Hwang SM, Nakai J, Salama G, Segal SS, Kotlikoff MI. Propagated endothelial Ca2+ waves and arteriolar dilation in vivo: measurements in Cx40BAC GCaMP2 transgenic mice. Circ Res. 2007;101:1300–1309. doi: 10.1161/CIRCRESAHA.107.149484. [DOI] [PubMed] [Google Scholar]
- Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci U S A. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraban M, Zhan H, Whitten AE, Langley DB, Matthews KS, Swint-Kruse L, Trewhella J. Ligand-induced conformational changes and conformational dynamics in the solution structure of the lactose repressor protein. J Mol Biol. 2008;376:466–481. doi: 10.1016/j.jmb.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.