Abstract
Phylogeography has recently become more abundant in studies of demographic history of both wild and domestic species. A single nucleotide polymorphism (SNP) in the intron of the Y-chromosomal gene UTY19 displays a north–south gradient in modern cattle. Support for this geographical distribution of haplogroups has previously also been seen in ancient cattle from Germany. However, when analysing 38 historic remains of domestic bulls and three aurochs from northern Europe for this SNP we found no such association. Instead, we noted extensive amounts of temporal variation that can be attributed to transportation of cattle and late breed formation.
Keywords: ancient DNA, Bos taurus, single nucleotide polymorphism, Y chromosome
1. Introduction
Recent decades have seen an increase in the number of phylogeographic studies of populations and species, including domestication studies (Emerson & Hewitt 2005). Along with the increasing number of publications, the need for proper sampling and temporal data (Ramakrishnan et al. 2005) has become evident. For example, when modern European brown bears (Ursus arctos) were investigated, the genetic pattern indicated an obvious case of climatic dependence upon southern refugias (Taberlet et al. 1998). Ancient samples, on the contrary, indicate a continuity of prehistoric gene flow (Valdiosera et al. 2007, 2008). Ancient samples from other species have also contradicted analyses based solely on modern samples, as seen with the language-related FOXP2 gene in humans. Based on contemporary samples, it was suggested that the non-synonymous mutations unique in present-day Homo sapiens evolved after the split with Neandertals (Enard et al. 2002). But additional ancient samples showed that it evolved before the speciation of H. sapiens (Krause et al. 2007).
In modern European cattle (Bos taurus), a north–south gradient of genetic diversity has been observed in various genetic systems (Jann et al. 2004; Cymbron et al. 2005; Li et al. 2007), including the Y chromosome. One single nucleotide polymorphism (SNP) in the 19th intron of the Y-chromosomal gene UTY (UTY19) and one 2 bp insertion–deletion polymorphism in ZFY intron 5 distinguishes two haplogroups: Y1, primarily found in northern Europe, and Y2, mainly distributed in southern Europe (Götherström et al. 2005). When the same geographical pattern was observed in ancient cattle and aurochs (Bos primigenius), it was interpreted as a sign of potential hybridization in Europe between domestic cows and aurochs bulls (Götherström et al. 2005). Although most studies of mitochondrial DNA from ancient and modern cattle show no sign of local contributions by European aurochs to the modern gene pool (Bollongino et al. 2006; Edwards et al. 2007), some but very limited cattle–aurochs hybridizations in Europe cannot be ruled out (Beja-Pereira et al. 2006; Achilli et al. 2008). Nevertheless, whether any local introgression between aurochs bulls and domestic cattle occurred in Europe is yet to be determined.
In order to explore how a temporal dataset, spanning from the Bronze Age to the present day, relates to the previous results, we use the UTY19 SNP that is known to be alternatively fixed in the two haplogroups. SNP status was studied in 38 historic cattle remains mainly from Scandinavia, as well as in three additional aurochs specimens. A strong geographical signal in the ancient samples, with the majority of samples belonging to haplogroup Y1, would support the previous indications of cattle–aurochs hybridization in Europe. Temporal variation of allele frequency, on the contrary, would not be consistent with a conserved signal from the time of domestication, and would require other explanations.
2. Material and methods
(a) Samples
The majority of the samples (all dated with the archaeological context) were Swedish mediaeval material (n=21) collected from the following places: the fortified village of Eketorp (Öland), Stora Kålltorp, Lödöse, Skara and Visby. The post-mediaeval historic material (n=15) consists of bones from the harbour in Marstrand and was dated to the eighteenth century. The English material consists of one mediaeval bone from York and one undated aurochs sample found on the coast close to Southampton. From Hungary (Szazhalombatta), two aurochs and one domestic specimen dated to 1600–1700 BC were analysed. All samples were identified as male (data not shown) using a previously described protocol (Svensson et al. 2008). The material is summarized in table 1 (and electronic supplementary material, table 1). To further investigate the pattern in modern animals, DNA from 27 Lithuanian bulls (table 1) was analysed. Data from Götherström et al. (2005) were included in the statistical analysis.
Table 1.
Location, date and the number of animals assigned to haplogroups Y1 and Y2 for the domestic cattle included in the statistical analysis.
| location | date | Y1/Y2a |
|---|---|---|
| Hungary | 1600–1700 BC | 0/1 |
| Eketorp/Sweden | AD 1100 | 0/8 |
| Visby/Sweden | AD 1100 | 0/2 |
| Skara/Sweden | AD 1200 | 0/1 |
| Marstrand/Sweden | AD 1700 | 8/6 |
| Lithuania | AD 2000 | 26/1 |
Number of individuals assigned to each haplogroup.
(b) Sample extraction and amplification
Ancient DNA was extracted according to a previously published protocol (Svensson et al. 2007). DNA was amplified with primers targeting 133 bp of UTY19 starting from nucleotide position 378 in the reference sequence AY936543. For details on DNA extraction, PCR conditions and SNP typing, see the electronic supplementary material.
(c) Statistical analyses
Allelic dropout is a frequent problem when working with degraded material (Taberlet & Waits 1998). Since the Y chromosome is a haploid system, the frequency bias due to allelic dropout is not possible. Therefore, a minimum of two successful SNP identifications for each historic sample were considered sufficient for inclusion in the statistical analysis. The Fisher exact probability two-tailed test, as implemented in Statisticav. 8, was used to test for differentiation between the different geographical and temporal groups. We also calculated the allele frequency needed for statistical significance, using the binomial distribution, for the mediaeval and modern samples.
3. Results
Twenty eight out of the 41 samples were successfully typed more than once, and were thus included in the statistical calculations. The three aurochs samples from northern Europe all displayed allele A, corresponding with the southern haplogroup Y2 (table 1 and electronic supplementary material, table 1).
All but one of the modern Lithuanian cattle displayed allele C, analogous to the northern haplogroup Y1 (table 1). No significant difference was found between Lithuanian and modern bulls from Sweden and Finland (p=0.1927), while they were significantly different compared with animals from central and southern Europe (p=0 in both comparisons). Therefore, the bulls from Lithuania, Sweden and Finland were treated as one group (northern Europe).
Since the previously observed north–south geographical pattern did not apply to our new data, we conducted statistical tests to investigate both the temporal and geographical differences between all geographical and temporal groups. The allele frequency in Swedish mediaeval bulls was significantly different (p=0.0029) compared with that of eighteenth century bulls. It was also significantly different from that of contemporary northern European cattle (p=0; figure 1). We detected no difference between the mediaeval bulls compared with modern southern European cattle (p=0.5688). Also, the eighteenth century material from Marstrand did not indicate any significant difference (p=1) when compared with modern cattle from Britain, Holland, Germany and Switzerland. Cattle from these groups display both alleles at almost equal frequency.
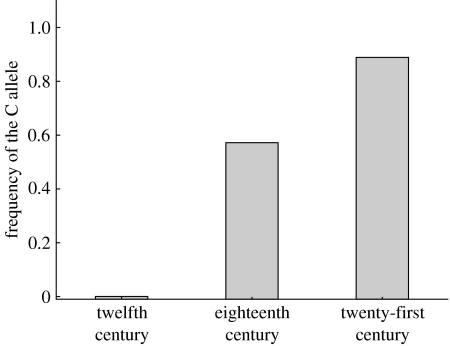
Figure 1.
The frequency of the C allele, corresponding to haplogroup Y1 in the animals from Eketorp, Visby and Skara (twelfth century), Marstrand (eighteenth century) and modern animals from Sweden, Finland and Lithuania (twenty-first century). Note that a binomial distribution alone allows for a frequency of below 0.23 of the C allele in the mediaeval material and, similarly, a frequency of above 0.79 in the modern material.
4. Discussion
We did not observe a geographical pattern of variation in ancient cattle similar to the one observed in modern animals. Instead, the allele frequency fluctuated over time, often from fixation of one allele at one time point to fixation of the other allele at another time. Our data suggest that there was fixation for (or at least a major representation of) the A allele in Southern Scandinavia some 900 years ago. Since then there has been a shift; both alleles were well represented in Scandinavia 300 years ago, but the C allele was almost completely fixed in modern samples. This variation suggests that Scandinavia was similar to modern central Europe concerning the frequencies of the UTY19 SNP during the eighteenth century, and also that the fixation for the C allele in northern Europe only occurred after the last known aurochs had been killed (1627 in Poland). Fluctuations over time can also be seen in the German material (Götherström et al. 2005 and electronic supplementary material), yet another indication of the fact that present-day frequencies of the UTY19 SNP have little to do with early cattle domestication. A better explanation for the fluctuation of allele frequency and eventual fixation is an increased interest in specialized breeds. In mediaeval and post-mediaeval Europe, with improved possibilities for significant levels of long-distance trade, cattle could easily have been moved from one part of Europe to another. With the increased interest in breeding, as well as elevated breeding sophistication, combined with the use of only a few bulls to create modern breeds during the nineteenth and twentieth centuries, the present-day frequency of the UTY19 SNP is more likely to be an effect of the most recent century's cattle keeping rather than events that possibly took place in central Europe some 5–8 Myr ago.
Acknowledgments
Samples were kindly provided by Maria Vretemark (Hungary, Skara, Stora Kålltorp, Saleby and Marstrand), Ylva Telldahl (Eketorp), Gustav Malmborg (Visby), Terry O'Connor (York), Jaco Weinstock (English aurochs), Per Persson (Lödöse) and Jurgita Baubliene (Modern DNA). We thank Judith Mank, Cecilia Anderung and two anonymous reviewers for their helpful comments on the manuscript. A.G. is a Research Fellow of the Royal Swedish Academy of Science.
Supplementary Material
References
- Achilli A, et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr. Biol. 2008;18:R157–R158. doi: 10.1016/j.cub.2008.01.019. doi:10.1016/j.cub.2008.01.019 [DOI] [PubMed] [Google Scholar]
- Beja-Pereira A, et al. The origin of European cattle: evidence from modern and ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:8113–8118. doi: 10.1073/pnas.0509210103. doi:10.1073/pnas.0509210103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollongino R, Edwards C.J, Alt K.W, Burger J, Bradley D.G. Early history of European domestic cattle as revealed by ancient DNA. Biol. Lett. 2006;2:155–159. doi: 10.1098/rsbl.2005.0404. doi:10.1098/rsbl.2005.0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymbron T, Freeman A.R, Malheiro M.I, Vigne J.D, Bradley D.G. Microsatellite diversity suggests different histories for Mediterranean and Northern European cattle populations. Proc. R. Soc. B. 2005;272:1837–1843. doi: 10.1098/rspb.2005.3138. doi:10.1098/rspb.2005.3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C.J, et al. Mitochondrial DNA analysis shows a Near Eastern Neolithic origin for domestic cattle and no indication of domestication of European aurochs. Proc. R. Soc. B. 2007;274:1377–1385. doi: 10.1098/rspb.2007.0020. doi:10.1098/rspb.2007.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B.C, Hewitt G.M. Phylogeography. Curr. Biol. 2005;15:R367–R371. doi: 10.1016/j.cub.2005.05.016. doi:10.1016/j.cub.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher S.E, Lai C.S.L, Wiebe V, Kitano T, Monaco A.P, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. doi:10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- Götherström A, Anderung C, Hellborg L, Elburg R, Smith C, Bradley D.G, Ellegren H. Cattle domestication in the Near East was followed by hybridization with aurochs bulls in Europe. Proc. R. Soc. B. 2005;272:2345–2350. doi: 10.1098/rspb.2005.3243. doi:10.1098/rspb.2005.3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jann O.C, Ibeagha-Awemu E.M, Ozbeyaz C, Zaragoza P, Williams J.L, Ajmone-Marsan P, Lenstra J.A, Moazami-Goudarzi K, Erhardt G. Geographic distribution of haplotype diversity at the bovine casein locus. Genet. Select. Evol. 2004;36:243–257. doi: 10.1186/1297-9686-36-2-243. doi:10.1051/gse:2003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J, et al. The derived FOXP2 variant of modern humans was shared with neandertals. Curr. Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. doi:10.1016/j.cub.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Li M.H, et al. The genetic structure of cattle populations (Bos taurus) in northern Eurasia and the neighbouring Near Eastern regions: implications for breeding strategies and conservation. Mol. Ecol. 2007;16:3839–3853. doi: 10.1111/j.1365-294X.2007.03437.x. doi:10.1111/j.1365-294X.2007.03437.x [DOI] [PubMed] [Google Scholar]
- Ramakrishnan U, Hadly E.A, Mountain J.L. Detecting past population bottlenecks using temporal genetic data. Mol. Ecol. 2005;14:2915–2922. doi: 10.1111/j.1365-294X.2005.02586.x. doi:10.1111/j.1365-294X.2005.02586.x [DOI] [PubMed] [Google Scholar]
- Svensson E.M, Anderung C, Baubliene J, Persson P, Malmström H, Smith C, Vretemark M, Daugnora L, Götherström A. Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim. Genet. 2007;38:378–383. doi: 10.1111/j.1365-2052.2007.01620.x. doi:10.1111/j.1365-2052.2007.01620.x [DOI] [PubMed] [Google Scholar]
- Svensson E.M, Götherström A, Vretemark M. A DNA test for sex identification in cattle confirms osteometric results. J. Archaeol. Sci. 2008;35:942–946. doi:10.1016/j.jas.2007.06.021 [Google Scholar]
- Taberlet P, Waits L.P. Non-invasive genetic sampling. Trends Ecol. Evol. 1998;13:26–27. doi: 10.1016/s0169-5347(97)01276-7. doi:10.1016/S0169-5347(97)01276-7 [DOI] [PubMed] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A.G, Cosson J.F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. doi:10.1046/j.1365-294x.1998.00289.x [DOI] [PubMed] [Google Scholar]
- Valdiosera C.E, et al. Staying out in the cold: glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol. Ecol. 2007;16:5140–5148. doi: 10.1111/j.1365-294X.2007.03590.x. doi:10.1111/j.1365-294X.2007.03590.x [DOI] [PubMed] [Google Scholar]
- Valdiosera C.E, et al. Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos) Proc. Natl Acad. Sci. USA. 2008;13:5123–5128. doi: 10.1073/pnas.0712223105. doi:10.1073/pnas.0712223105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.