Abstract
The recombinational environment influences patterns of molecular evolution through the effects of Hill–Robertson interference. Here, we examine genome-wide patterns of gene expression with respect to recombinational environment in Drosophila melanogaster. We find that regions of the genome lacking crossing over exhibit elevated levels of expression, and this is most pronounced for genes on the entirely non-crossing over fourth chromosome. We find no evidence for differences in the patterns of gene expression between regions of high, intermediate and low crossover frequencies. These results suggest that, in the absence of crossing over, selection to maintain control of expression may be compromised, perhaps due to the accumulation of deleterious mutations in regulatory regions. Alternatively, higher gene expression may be evolving to compensate for defective protein products or reduced translational efficiency.
Keywords: recombination, crossing over, Drosophila, gene expression
1. Introduction
The recombinational environment is known to influence patterns of molecular evolution through the effects of Hill–Robertson interference (Hill & Robertson 1966; Felsenstein 1974), such that regions of the genome with no recombination or low rates of recombination experience a reduced effective population size (Ne). This leads to reduced rates of adaptive evolution and a lower efficacy of selection against deleterious mutations (Kimura 1983). In Drosophila, several studies have examined the effects of recombination on patterns of evolution, and showed that linkage impedes the ability of natural selection both to incorporate beneficial mutations and remove deleterious ones (Betancourt & Presgraves 2002; Marais et al. 2004; Presgraves 2005; Bartolomé & Charlesworth 2006; Bachtrog et al. 2008; Larracuente et al. 2008).
In a genome-wide comparison of Drosophila melanogaster and Drosophila yakuba, Haddrill et al. (2007) found that regions of the genome that completely lacked crossing over exhibited elevated rates of non-synonymous and long intron divergence, as well as reduced codon usage bias, and attributed this to a severe reduction in the efficacy of selection, particularly on the fourth (dot) chromosome, which lacks crossing over under normal conditions (Ashburner et al. 2005). Consistent with previous studies (Betancourt & Presgraves 2002; Marais et al. 2004), Haddrill et al. (2007) also noted a negative relationship between the rate of non-synonymous divergence (measured using both the codon-based measure dN (Yang 1997) and the nucleotide-based measure KA (Comeron 1995); see Bierne & Eyre-Walker (2003) and Haddrill et al. (2007) for discussions of the two measures) and the level of codon usage bias (measured by Fop, the frequency of optimal codons).
There is, however, a well-established positive correlation between Fop for a gene and its level of expression (Duret & Mouchiroud 1999; Marais et al. 2001, 2004) and a negative correlation between the level of expression and non-synonymous divergence (Marais et al 2004; Larracuente et al. 2008). This raises an interesting alternative possible explanation for the results of Haddrill et al. (2007). If the genes found in the non-crossover regions of the Drosophila genome are a group of genes that are expressed at a very low level, the elevated levels of non-synonymous divergence and low codon usage bias observed on the D. melanogaster/D. yakuba fourth chromosome (and other non-crossover regions) could simply be explained by the relationships between gene expression, non-synonymous divergence and codon usage bias.
Here, we report a re-analysis of the dataset analysed by Haddrill et al. (2007) combined with genome-wide expression data. Surprisingly, we find that expression levels are elevated in non-crossing over regions, and this pattern is particularly strong for the fourth chromosome genes. Consistent with Haddrill et al. (2007), we find no evidence for differences in gene expression between regions of high, intermediate and low crossing over. The effects of gene expression cannot, therefore, explain the elevated levels of non-synonymous divergence and low codon usage bias in the non-crossover regions of the Drosophila genome.
2. Material and methods
The Haddrill et al. (2007) dataset was combined with genome-wide expressed sequence tag (EST) data from D. melanogaster, downloaded from the NCBI UniGene database (http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=&db=unigene), which contains counts of the number of ESTs associated with each gene in a number of different tissue types. We used the total number of EST counts across all tissue types as the measure of gene expression. We were able to extract expression profiles for 7398 of the 7612 genes in the Haddrill et al. (2007) dataset, including 3752, 2477, 1082 and 87 genes in regions of high, intermediate, low and no crossing over, respectively, thereby including all non-crossover genes analysed by Haddrill et al. (2007). The non-crossover category was further subdivided into 67 fourth chromosome and 20 non-fourth chromosome genes. We refer to crossing over rather than recombination, because there is evidence that gene conversion occurs in the regions of the D. melanogaster genome with very low or zero frequencies of crossing over (Langley et al. 2000; Jensen et al. 2002; Gay et al. 2007).
3. Results
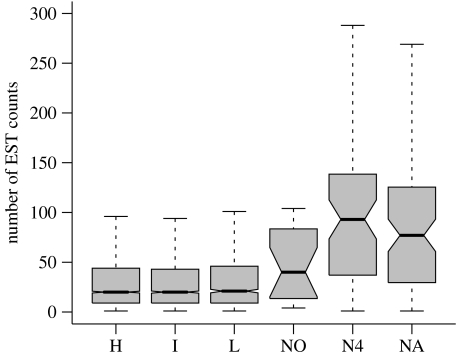
The recombinational environment had a highly significant effect on the levels of gene expression (Kruskal–Wallis test: H=69.12, p<10−5), and this effect was entirely due to the non-crossover regions, since no significant differences in expression levels were found between high, intermediate and low crossover regions (H=2.28, p=0.31). Figure 1 shows that non-crossover regions exhibit elevated levels of expression compared with the rest of the genome, with median number of EST counts almost four times higher for the non-crossing over region (median=77) than the high (20), intermediate (20) and low (21) crossing over regions.
Figure 1.
Notched boxplot of the number of EST counts for each recombination region, high (H), intermediate (I) and low (L) frequency of crossing over, and regions of no crossing over, divided into non-fourth chromosome genes (NO), fourth chromosome genes (N4) and all no crossover genes (NA). The box extends from the lower to the upper quartile, with a line in the middle at the median. The dotted bars represent the 5th and 95th percentiles.
Since Haddrill et al. (2007) found differences in non-synonymous divergence between genes on the fourth chromosome and other non-crossover genes, we examined the level of expression in these groups separately. Figure 1 shows that within the non-crossing over genes, non-fourth chromosome genes show levels of expression intermediate between the fourth chromosome and the rest of the genome (median EST counts: non-fourth chromosome=40, fourth chromosome=93), and are significantly different from both these groups (Wilcoxon signed-rank test: non-fourth versus fourth, W=893.5, p=0.024; non-fourth versus high, intermediate and low regions combined, W=53480.5, p=0.037). This is consistent with the results of Haddrill et al. (2007) for intron divergence and codon usage bias in non-fourth chromosome genes, which were intermediate between the fourth chromosome genes and the rest of the genome.
Marais et al. (2004) and Larracuente et al. (2008) reported a negative relationship between non-synonymous divergence and expression level, and we see the same relationship here (Spearman's rank correlation, Rs, between EST count and KA, with 95% confidence intervals obtained by bootstrapping across genes: high crossover region Rs=−0.40 (−0.43 to −0.37); intermediate crossover region Rs=−0.42 (−0.45 to −0.39); low crossover region Rs=−0.36 (−0.41 to −0.31); and non-crossover region Rs=−0.24 (−0.42 to −0.03)). However, within the non-crossover region, this relationship is significantly different from zero only for the non-fourth chromosome genes (Rs=−0.58 (−0.82 to −0.17)) but not the fourth chromosome genes (Rs=−0.19 (−0.41 to 0.05)); the differences between the categories are, however, not significant. A positive relationship between expression level and the codon usage bias measure, Fop, has also been reported in Drosophila (Duret & Mouchiroud 1999; Marais et al. 2001, 2004). Consistent with this, we find a significantly positive relationship between EST count and Fop for all crossing over regions (high crossover region Rs=0.33 (0.31 to 0.37); intermediate crossover region Rs=0.32 (0.28 to 0.35); low crossover region Rs=0.16 (0.10 to 0.22)), but not for any of the non-crossover regions (all non-crossover genes Rs=−0.16 (−0.38 to 0.06); fourth chromosome genes Rs=−0.02 (−0.28 to 0.24); non-fourth chromosome genes Rs=−0.11 (−0.48 to 0.30)).
4. Discussion
The main conclusion of this study is that the elevated level of non-synonymous divergence previously observed in the non-crossing over regions of the Drosophila genome (Haddrill et al. 2007) cannot simply be explained by the established negative correlation between non-synonymous divergence and expression level. Genes in the non-crossover regions actually experience higher levels of expression than the rest of the genome, opposite to what is expected on this basis. Within the non-crossover regions, the fourth chromosome genes show the most extreme elevations in expression, with median EST counts double those in other non-crossover regions. This pattern of the fourth chromosome showing the most extreme deviations from the rest of the genome is consistent with previous findings, and may reflect enhanced Hill–Robertson effects on a larger region of zero recombination than elsewhere, and/or a long history of no crossing over on the fourth chromosome, which may not apply to other non-crossover genes (Haddrill et al. 2007).
Since Haddrill et al. (2007) concluded that the efficacy of selection was severely reduced in the non-crossing over regions, especially on the fourth chromosome, these results may suggest that selection has lost control of expression in the non-crossover regions, which would imply that these genes are being expressed at a higher level than optimal. If these regions are accumulating deleterious mutations, these mutations are likely to affect sequences important for regulating expression patterns as well as coding regions. Indeed, Haddrill et al. (2007) found that divergence in long introns was close to that of short introns in non-crossover regions, despite evidence for strong selective constraints in long introns in the rest of the genome (Bergman & Kreitman 2001; Andolfatto 2005; Haddrill et al. 2005; Halligan & Keightley 2006); introns are likely to contain regulatory elements important for the control of expression (Casillas et al 2007), and evidence from the non-recombining neo-Y chromosome of Drosophila miranda suggests that regulatory mutations that have accumulated on this chromosome have increased expression levels, as well as decreased them (Bachtrog 2006).
In addition to this, the genome-wide relationships observed between non-synonymous divergence/Fop and expression level may have broken down in the non-crossover regions, further suggesting that, while selection normally maintains tight control over expression patterns, this regulation appears to have deteriorated in the absence of crossing over. However, relaxation of selective constraints might be expected to increase the variance in expression level between genes, and there is some evidence for this (Rifkin et al. 2005). After transformation of our data to a log scale, however, there is no statistical support for higher variance in expression level in the non-crossover regions (data not shown), suggesting that other mechanisms may be influencing the patterns observed.
Alternatively, it is possible that the effects observed are a result of the evolution of a form of dosage compensation. If genes in non-crossover regions are accumulating deleterious mutations that reduce the function of their proteins, selection may favour mutations that increase expression to compensate for this, similar to what has been proposed for the evolution of X and Y chromosomes (e.g. Charlesworth & Charlesworth 2000; Bachtrog 2006). Similarly, if the presence of unpreferred codons in the fourth chromosome/other non-crossover genes results in a reduction in translational efficiency or accuracy, the higher expression level observed may represent a means of compensating for this. Interestingly, it has been reported that the protein ‘Painting of fourth’ (POF) exclusively binds to the fourth chromosome in Drosophila (Larsson et al. 2004). The extent of POF binding to fourth chromosome genes is correlated with their transcription levels, and mutational knockouts of POF lead to reduced gene expression on the fourth chromosome (Johansson et al. 2007). This suggests that it is involved in chromosome 4-specific gene regulation, perhaps even having a dosage compensation-like role.
Acknowledgements
We are very grateful to Daniel Halligan for assistance with the statistical analysis package R, Beatriz Vicoso for assistance with the EST database, Andrea Betancourt for drawing our attention to POF and two anonymous reviewers for useful comments on the manuscript. P.R.H. was supported by a grant from NERC to B.C., F.M.W. by a Marie Curie Early Stage Fellowship and B.C. by the Royal Society.
References
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. doi:10.1038/nature04107 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic K.G, Hawley R.S. Cold Spring Harbor Laboratory Press; New York, NY: 2005. Drosophila A laboratory handbook. [Google Scholar]
- Bachtrog D. Expression profile of a degenerating neo-Y chromosome in Drosophila. Curr. Biol. 2006;16:1694–1699. doi: 10.1016/j.cub.2006.07.053. doi:10.1016/j.cub.2006.07.053 [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Hom E, Wong K.M, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. doi:10.1186/gb-2008-9-2-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C, Charlesworth B. Evolution of amino acid sequences and codon usage on the Drosophila miranda neo-sex chromosomes. Genetics. 2006;174:2033–2044. doi: 10.1534/genetics.106.064113. doi:10.1534/genetics.106.064113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman C.M, Kreitman M. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 2001;11:1335–1345. doi: 10.1101/gr.178701. doi:10.1101/gr.178701 [DOI] [PubMed] [Google Scholar]
- Betancourt A.J, Presgraves D.C. Linkage limits the power of natural selection in Drosophila. Proc. Natl Acad. Sci. USA. 2002;99:13 616–13 620. doi: 10.1073/pnas.212277199. doi:10.1073/pnas.212277199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics. 2003;165:1587–1597. doi: 10.1093/genetics/165.3.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas S, Barbadilla A, Bergman C.M. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol. Biol. Evol. 2007;24:2222–2234. doi: 10.1093/molbev/msm150. doi:10.1093/molbev/msm150 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. doi:10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 1995;41:1152–1159. doi: 10.1007/BF00173196. doi:10.1007/BF00173196 [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl Acad. Sci. USA. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. doi:10.1073/pnas.96.8.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay J, Myers S, McVean G. Estimating meiotic gene conversion rates from population genetic data. Genetics. 2007;177:881–894. doi: 10.1534/genetics.107.078907. doi:10.1534/genetics.107.078907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P.R, Charlesworth B, Halligan D.L, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. doi:10.1186/gb-2005-6-8-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P.R, Halligan D.L, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. doi:10.1186/gb-2007-8-2-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D.L, Keightley P.D. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. doi:10.1101/gr.5022906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G, Robertson A. The effect of linkage on the limits of artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Jensen M.A, Charlesworth B, Kreitman M. Patterns of genetic variation at a chromosome 4 locus of Drosophila melanogaster and D. simulans. Genetics. 2002;160:493–507. doi: 10.1093/genetics/160.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A.-M, Stenberg P, Bernhardsson C, Larsson J. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 2007;26:2307–2316. doi: 10.1038/sj.emboj.7601604. doi:10.1038/sj.emboj.7601604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Cambridge University Press; Cambridge, UK: 1983. The neutral theory of molecular evolution. [Google Scholar]
- Langley C.H, Lazzaro B.P, Phillips W, Heikkinen E, Braverman J.M. Linkage disequilibria and the site frequency spectrum in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics. 2000;156:1837–1852. doi: 10.1093/genetics/156.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A.M, Sackton T.B, Greenberg A.J, Wong A, Singh N.D, Sturgill D, Zhang Y, Oliver B, Clark A.G. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. doi:10.1016/j.tig.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Larsson J, Svensson M.J, Stenberg P, Mäkitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc. Natl Acad. Sci. USA. 2004;101:9728–9733. doi: 10.1073/pnas.0400978101. doi:10.1073/pnas.0400978101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl Acad. Sci. USA. 2001;98:5688–5692. doi: 10.1073/pnas.091427698. doi:10.1073/pnas.091427698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Domazet-Loso T, Tautz D, Charlesworth B. Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J. Mol. Evol. 2004;59:771–779. doi: 10.1007/s00239-004-2671-2. doi:10.1007/s00239-004-2671-2 [DOI] [PubMed] [Google Scholar]
- Presgraves D.C. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. doi:10.1016/j.cub.2005.07.065 [DOI] [PubMed] [Google Scholar]
- Rifkin S.A, Houle D, Kim J, White K.P. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. doi:10.1038/nature04114 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]