Abstract
The brown alga Fucus serratus is a key foundation species on rocky intertidal shores of northern Europe. We sampled the same population off the coast of southern Norway in 2000 and 2008, and using 26 microsatellite loci, we estimated the changes in genetic diversity and effective population size (Ne). The unexpectedly low Ne (73–386) and Ne/N ratio (10−3–10−4), in combination with a significant decrease (14%) in allelic richness over the 8-year period, suggests an increased local extinction risk. If small Ne proves to be a common feature of F. serratus, then being abundant may not be enough for the species to weather future environmental changes.
Keywords: effective population size, Fucus serratus, genetic diversity, Ne/N
1. Introduction
Several recent experiments have demonstrated that the genetic diversity of a population is positively correlated with its ability to maintain fitness in the face of environmental changes (e.g. Ehlers et al. 2008). Genetic diversity is a function of a population's effective size (Ne, the size of an ideal population undergoing the same amount of genetic drift as the real population or, roughly, the number of individuals that successfully contribute to the next generation). Natural populations, however, are rarely ideal and Ne is usually smaller than N (see Palstra & Ruzzante 2008). Consequently, even populations with a large census size may face a loss of genetic diversity. Clearly, small values of Ne can constrain a population's evolutionary response to environmental change much more than the actual census size (Frankham et al. 2002).
Classically, temporal measurements of demographic parameters (growth, sex ratio and mating system) have been used to estimate Ne (Frankham 1995), but each parameter is difficult to measure directly in natural populations. An alternative approach uses genetic data to assess temporal changes in allele frequency within a given population (Waples 1989). A DNA-based temporal method has been used for several animal species, but for only a very few plant species (see Palstra & Ruzzante 2008), and to our knowledge, for no species of marine macroalgae.
Intertidal shores throughout the North Atlantic are characterized by Fucus spp., with Fucus serratus dominant from the mid-intertidal to lower subtidal (Lüning 1990). The species is dioecious, reproduces annually with a generation time of 1–2 years (Coyer et al. 2007), and microsatellite analyses suggest a panmictic unit of 0.5–2 km (Coyer et al. 2003). We sampled the same population of F. serratus off the coast of southern Norway in 2000 and 2008, and using 26 microsatellite loci, we estimated Ne and temporal changes in genetic diversity.
2. Material and methods
(a) Sampling and DNA extraction
Slåttholmen is a small islet directly offshore from the Espeland Marine Biological Station (University of Bergen), Norway. Census size (N) of the F. serratus population was obtained in May 2008 using a systematic sampling approach: a 50 m transect line was established at the upper extent of F. serratus, along which 3–5, 0.25 m2 quadrats were placed perpendicular and contiguously (to the lowest extent) at 1.5 m intervals (175 quadrats). Three patches separated from Slåttholmen by approximately 250, 500 and 1000 m were censused in the same manner with transects varying from 30 to 50 m (depending upon topology); mean densities of the four areas were then combined for a grand mean density. The total census size of F. serratus was estimated by determining the range of suitable habitat within a 1 km radius of Slåttholmen using aerial maps and the software ImageJ (Abramoff et al. 2004), assuming a width of 2 m (determined from the contiguous quadrats) and using the grand mean density.
Tissue samples were collected from 64 individuals spaced at 1 m intervals along a linear transect. Exactly the same area was sampled in June 2000 and April 2008. Tissue was blotted dry and placed in silica crystals for dehydration and storage; DNA was extracted and purified as described by Hoarau et al. (2007) and modified by Coyer et al. (in press).
(b) Microsatellite genotyping
A total of 26 microsatellite loci were genotyped: FsA198, FsB113, FsB128, FsE6, FsF4 (Coyer et al. 2002); L20, L38, L58, L94 (Engel et al. 2003); and F9, F12, F14, F17, F19, F21, F22, F36, F37, F45, F47, F49, F50, F58, F65, F69, F72 (Coyer et al. in press). The genotypes were visualized on an ABI 3730 automatic sequencer (Applied Biosystems) and analysed with GeneMapper v. 4.0 software (Applied Biosystems).
(c) Data analysis
The average number of alleles per locus (α), Hexp (non-biased; Nei 1978) and θ estimator of FST (2000 replications; Weir & Cockerham 1984) were estimated with Genetix v. 4.02 (Belkhir et al. 2001). Ne was estimated using a 1- and 2-yr generation time with the software MLNe (Wang & Whitlock 2003). As the model assumes no selection, we also tested for neutrality using the FST-outlier test (Beaumont & Nichols 1996), which revealed locus F65 as the only significant outlier (p<0.01). Therefore, we estimated Ne with and without locus F65. An estimate of temporal changes in Ne was obtained using the software TMVP, which estimates Ne at the time of the oldest (2000) and most recent sample (2008), assuming a model of exponential growth or decline during the interval (Beaumont 2003).
3. Results and discussion
Our estimate of Ne was between 73 and 386 (table 1) comparable to the average Ne of 260 obtained from temporal studies of 65 diverse species (Palstra & Ruzzante 2008). The Ne/N ratio was unexpectedly low, 1.8×10−3–3.5×10−4, based on an estimated census size of 208 000 individuals within a 1 km radius of Slåttholmen (grand mean density (s.d.) from four areas=10.13 m−2 (3.86); potential Fucus habitat=10.28×0.002 km). For land plants, Ne/N ratios of approximately 10−1 have been estimated with temporal analyses (see Siol et al. 2007). The low Ne/N ratio for F. serratus, therefore, appears to be more similar to the 10−3–10−6 differences observed among species with a type III survivorship curve (high fecundity and high juvenile mortality) such as bony fishes (e.g. Hoarau et al. 2005) or shrimp (Ovenden et al. 2007). In F. serratus, female fecundity is also very high (approx. 106 eggs per female; Knight & Parke 1950; J. A. Coyer, G. Hoarau, W. T. Stam & J. L. Olsen, unpublished data) and high juvenile mortality is probable given the low dispersal of gametes (1–2 m; Serrão et al. 1997) and the relatively low density of adults compared with gamete production. Similar to fishes and shrimp, F. serratus appears to have a high variance in individual reproductive success, which probably leads to the low Ne/N ratio we estimated.
Table 1.
Estimation of Ne in a population of F. serratus collected in 2000 and 2008. (Ne was estimated (95% CI) with the software MLNe over a generation time interval of 1 or 2 yrs using all 26 loci and without the EST-derived microsatellite locus F65 a significant outlier, see §2c).
| Ne | |
|---|---|
| generation time=2 yrs | |
| all loci | 99 (73–193) |
| without F65 | 125 (87–200) |
| generation time=1 yr | |
| all loci | 188 (136–279) |
| without F65 | 241 (168–386) |
Gene flow can significantly bias estimates of Ne (Fraser et al. 2007). Immigration from highly differentiated populations will lead to changes in allele frequencies larger than expected by genetic drift alone and a subsequent underestimation of Ne. This scenario is very unlikely in F. serratus, as gene flow is very restricted (Coyer et al. 2003) and such immigrants would have highly divergent genotypes, a pattern not revealed using a factorial correspondence analysis (electronic supplementary material). Immigration from weakly differentiated neighbouring populations cannot be excluded, although it would result in an overestimation of Ne (Fraser et al. 2007). Additionally, sampling bias due to small-scale family structure can be excluded as no kinship was detected within 1–100 m (encompassing our sampling scale; Coyer et al. 2003). Thus, our estimate of Ne is likely to be reasonably unbiased.
Populations with small Ne may suffer from inbreeding depression and loss of genetic diversity, potentially leading to a reduced capacity to respond to changing environmental conditions and to increased extinction risk (Frankham 2005). An Ne of at least 500 appears necessary to minimize inbreeding depression and to maintain gene diversity (Frankham et al. 2002). Consequently, populations with small Ne and limited dispersal, such as F. serratus, could face localized extinction in the face of any environmental perturbation that reduces census size and/or constrains the population.
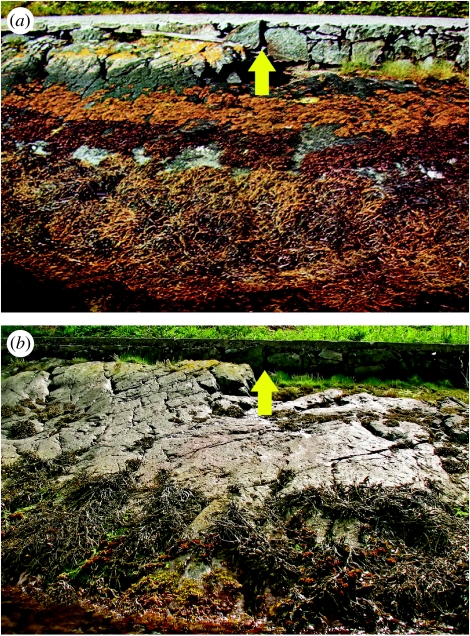
Considerable differences in fucoid abundance are noted when comparing photographs taken at Slåttholmen in 1968 (Lewis 1968) and 2008: two species (Pelvetia canaliculata and Fucus spiralis) were absent and another (Ascophyllum nodosum) was significantly reduced in 2008 (figure 1). Fucus serratus also has declined (as it has in the general area; K. Sjøtun 2004–2008, unpublished data), although it still remains widespread. Observed changes may be due to the increased frequency of exceptionally warm summers/mild winters observed in southern Norway during the past decade (Husa et al. 2007) and/or the 10-fold increase of herbivores relative to nearby areas (E. Heggøy 2001–2007, personal communication). Genetic data also revealed that from 2000 to 2008, Ne declined nearly threefold, allelic richness decreased significantly (14%; p<0.05, 10 000 permutations, FSTAT; Goudet 2002) and pairwise FST was highly significant (table 2).
Figure 1.
Photograph of the Slåttholmen site in (a) 1968 (Lewis 1968) and (b) 2008. Arrows indicate a common location.
Table 2.
Genetic changes in a population of F. serratus over eight generations. (Abbreviations: average number of alleles−locus (α), Hexp (non-biased), θ estimator of FST (reflecting temporal differences in allelic frequencies between 2000 and 2008) and estimates of temporal changes in Ne. Parenthetical values of Ne represent estimations without locus F65 (see table 1 legend); n.s., non-significant; *p<0.05; **p<0.001.)
| 2000 | 2008 | |
|---|---|---|
| α* | 5.7143 | 4.8929 |
| Hexpn.s. | 0.4602 | 0.4570 |
| Ne | 437 (392) | 152 (230) |
| θ** | 0.017 | |
The Slåttholmen population may be at risk and its decline will lead to significant changes in the intertidal community structure. If the very small Ne and Ne/N ratio estimated here are more generally characteristic of F. serratus (and possibly other fucoids), then the species may be less resilient to environmental change than assumed. Being abundant does not guarantee local survival in the long term.
Acknowledgments
This study was supported by NWO-ALW 2006 (contract number 815.01.011 to J.A.C. and 863.05.008 to G.H.), Schure-Beijerinck-Popping 2008-22 and the Directorate for Nature Management (Norway).
Supplementary Material
References
- Abramoff M.D, Magelhaes P.J, Ram S.J. Image processing with image. J. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- Beaumont M.A. Estimation of population growth or decline in genetically monitored populations. Genetics. 2003;164:1139–1160. doi: 10.1093/genetics/164.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont M.A, Nichols R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B. 1996;263:1619–1626. doi:10.1098/rspb.1996.0237 [Google Scholar]
- Belkhir K, Borsa P, Goudet J, Raufaste N, Bonhomme F. Laboratoire Genome et populations; Montpellier, France: 2001. Genetix, logiciel sous Windows pour la genetique des populations. [Google Scholar]
- Coyer J.A, Veldsink J.H, Stam W.T, Serrão E.A, Stam W.T, Olsen J.L. Characterization of microsatellite loci in the marine rockweeds, Fucus serratus and F. evanescens (Heterokontophyta; Fucaceae) Mol. Ecol. Notes. 2002;2:35–37. doi:10.1046/j.1471-8286.2002.00138.x [Google Scholar]
- Coyer J.A, Peters A.F, Stam W.T, Olsen J.L. Post-ice age recolonization and differentiation of Fucus serratus L. (Fucaceae: Phaeophyta) populations in northern Europe. Mol. Ecol. 2003;12:1817–1829. doi: 10.1046/j.1365-294x.2003.01850.x. doi:10.1046/j.1365-294X.2003.01850.x [DOI] [PubMed] [Google Scholar]
- Coyer J.A, Hoarau G, Stam W.T, Olsen J.L. Hybridization and introgression in a mixed population of the intertidal seaweeds Fucus distichus and F. serratus. J. Evol. Biol. 2007;16:3606–3616. doi: 10.1111/j.1420-9101.2007.01411.x. [DOI] [PubMed] [Google Scholar]
- Coyer, J. A. et al In press. Expressed sequence tag derived polymorphic SSR markers for Fucus serratus and amplification in other species of Fucus Mol. Ecol. Resour. [DOI] [PubMed]
- Ehlers A, Worm B, Reusch T.B.H. Importance of genetic diversity in eelgrass Zostera marina for its resilience to climate warming. Mar. Ecol. Prog. Ser. 2008;355:1–7. doi:10.3354/meps07369 [Google Scholar]
- Engel C.R, Brawley S, Edwards K.J, Serrão E. Isolation and cross-species amplification of microsatellite loci from the fucoid seaweeds Fucus vesiculosus, F. serratus, and Ascophyllum nodosum (Heterokontophyta, Fucaceae) Mol. Ecol. Notes. 2003;3:180–182. doi:10.1046/j.1471-8286.2003.00390.x [Google Scholar]
- Frankham R. Effective population size-adult population size ratios in wildlife: a review. Genet. Res. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Frankham R. Genetics and extinction. Biol. Conserv. 2005;126:131–140. doi:10.1016/j.biocon.2005.05.002 [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Cambridge University Press; Cambridge, UK: 2002. Introduction to conservation genetics. [Google Scholar]
- Fraser D.J, Hansen M.M, Østergaard S, Tessier N, Legault M, Bernatchez L. Comparative estimation of effective population sizes and temporal gene flow in two contrasting population systems. Mol. Ecol. 2007;16:3866–3889. doi: 10.1111/j.1365-294X.2007.03453.x. doi:10.1111/j.1365-294X.2007.03453.x [DOI] [PubMed] [Google Scholar]
- Goudet, J. 2002 FSTAT 2.9.3.2, a program to estimate and test gene diversities and fixation indices. See http://www2.unil.ch/popgen/softwares/fstat.htm
- Hoarau G, Boon E, Jongma D.N, Ferber S, Palsson J, Van der Veer H.W, Rijnsdorp A.D, Stam W.T, Olsen J.L. Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleronectes platessa L.) Proc. R. Soc. B. 2005;272:497–503. doi: 10.1098/rspb.2004.2963. doi:10.1098/rspb.2004.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G, Coyer J.A, Stam W.T, Olsen J.L. A fast and inexpensive DNA extraction/purification method for brown macroalgae. Mol. Ecol. Notes. 2007;7:191–193. doi:10.1111/j.1471-8286.2006.01587.x [Google Scholar]
- Husa V, Steen H, Åsen P.A. Hvordan vil makroalgesamfunnene langs norskekysten påvirkes av økt sjøtemperatur? In: Dahl E, Hansen P.K, Haug T, Karlsen Ø, editors. Kyst og havbruk 2007, Fisken og havet, særnr. 2-2007. 2007. pp. 23–27. [Google Scholar]
- Knight M, Parke M. A biological study of Fucus vesiculosus L. and F. serratus L. J. Mar. Biol. Assoc. UK. 1950;29:439–514. [Google Scholar]
- Lewis J.R. Water movements and their role in rocky shore ecology. 2nd European Symposium on Marine Biology. Sarsia. 1968;34:13–26. [Google Scholar]
- Lüning K. J. Wiley; New York, NY: 1990. Seaweeds: their environment, biogeography and ecophysiology. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovenden J.R, Peel D, Street R, Courtney A, Hoyle S, Peel S, Podlich H. The genetic effective and adult census size of an Australian population of tiger prawns (Penaeus escullentus) Mol. Ecol. 2007;16:127–138. doi: 10.1111/j.1365-294X.2006.03132.x. doi:10.1111/j.1365-294X.2006.03132.x [DOI] [PubMed] [Google Scholar]
- Palstra F, Ruzzante D.E. Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 2008;15:3428–3447. doi: 10.1111/j.1365-294x.2008.03842.x. doi:10.1111/j.1365-294X.2008.03842.x [DOI] [PubMed] [Google Scholar]
- Serrão E.A, Kautsky L, Lifvergren T, Brawley S.H. Gamete dispersal and pre-recruitment mortality in Baltic Fucus vesiculosus. Phycologia Suppl. 1997;36:101–102. [Google Scholar]
- Siol M, Bonnin I, Olivieri I, Prosperi J.M, Ronfort J. Effective population size associated with self-fertilization: lessons from temporal changes in allele frequencies in the selfing annual Medicago truncatula. J. Evol. Biol. 2007;20:2349–2360. doi: 10.1111/j.1420-9101.2007.01409.x. doi:10.1111/j.1420-9101.2007.01409.x [DOI] [PubMed] [Google Scholar]
- Wang J.L, Whitlock M.C. Estimating effective population size and migration rates from genetic samples over space and time. Genetics. 2003;161:429–446. doi: 10.1093/genetics/163.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R.S. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics. 1989;121:379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.