Abstract
In the face of ever-increasing threats to coral reef ecosystems, it is essential to understand the impact of natural predators in order to devise appropriate management strategies. Destructive population explosions of the crown-of-thorns starfish Acanthaster planci have devastated coral reefs throughout the Indo-Pacific for decades. But despite extensive research, the causes of outbreaks are still unclear. An important consideration in this research is that A. planci has been regarded as a single taxonomic entity. Using molecular data from its entire distribution, we find that A. planci is in fact a species complex. This discovery has important consequences for future coral reef research, and might prove critical for successful reef conservation management.
Keywords: species complex, sibling species, Acanthaster planci, crown-of-thorns starfish
1. Introduction
Coral reefs, the most species-rich marine ecosystems, are subjected to growing anthropogenic pressure, limiting their resilience to natural threats such as corallivorous predators (Bellwood et al. 2004). Among those, the crown-of-thorns starfish (COTS) Acanthaster planci is infamous for its dramatic population explosions (called outbreaks) that have devastated coral reefs throughout the Indo-Pacific for decades, making it a major management issue (Birkeland & Lucas 1990; Veron 2008). But despite extensive research into COTS biology, the causes of outbreaks are still not clear; they probably involve a variable set of interacting natural and anthropogenic factors that lead to increased recruitment (Engelhardt & Lassig 1997). An important consideration in both COTS research and management is that A. planci has been regarded as a single species throughout its distribution, and therefore the same ecological and behavioural traits are assumed worldwide.
Acanthaster planci's long-lived pelagic larva—surviving from three to four weeks in normal conditions (Yamaguchi 1973) to about seven weeks in marginal food regimes as found in oceanic conditions (Lucas 1982)—would be expected to promote genetic homogeneity. But this species appears to be highly structured (Benzie 1999), in line with other recent studies of widespread marine invertebrates (e.g. Becker et al. 2007). Using sequences of the mitochondrial cytochrome oxidase subunit I gene (COI) from samples covering its entire distribution, we show that A. planci consists of four deeply diverged clades that form a pan-Indo-Pacific species complex (as identified by DNA taxonomy; Vogler & Monaghan 2007).
2. Material and methods
DNA was extracted using a Qiagen MagAttract 96 DNA Plant Core Kit from 237 A. planci and two Acanthaster brevispinus tissue samples, collected by SCUBA diving and snorkel from 1987 to 2008 (table 1 in the electronic supplementary material). A fragment of COI, corresponding to the ‘barcoding’ fragment, was amplified and sequenced with the following primers: COTS_COI_F4734 5′-GCCTGAGCAGGAATGGTTGGAAC-3′ and COTS_COI_R5433 5′-CGTGGGATATCATTCCAAATCCTGG-3′. Sequences were assembled using CodonCode Aligner (http://www.codoncode.com/aligner), and the 632 bp remaining after quality-based end-clipping were aligned in SeaView (Galtier et al. 1996) with Patiria pectinifera (accession number: D16378) as an outgroup. All sequences were deposited in EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/: accession numbers: FM174472–174675, FM177190–177203, FM202070–202090).
Genetic distances between and within clades were calculated with Mega4 (Tamura et al. 2007) using the Kimura two-parameter model of sequence evolution (K2P), to enable comparisons with other asteroid datasets (Waters et al. 2004). There are no fossil data or geological calibration points available to date the separation between the four clades, so divergence times were approximated by applying the most accurate COI divergence rates available for echinoderms to the K2P distances (2.9–4.5% Myr−1; Lessios 2008). To test the 95 per cent connectivity limit as a species threshold (Hart & Sunday 2007), a parsimony haplotype network was built using TCS 1.2.1 (Clement et al. 2000). A neighbour-joining (NJ) analysis and NJ bootstrap analysis (1000 replicates) were carried out in PAUP* v. 4.0.b10 (Swofford 2003). After inferring the best-fit nucleotide evolution model using the Akaike information criterion as implemented in Modeltest v. 3.7 (Posada & Crandall 1998), we estimated the maximum-likelihood tree under a GTR+Γ+I model in PhyML v. 2.4.4, including 1000 bootstrap replicates (Guindon & Gascuel 2003).
We used a method separating species diversification from coalescent processes in a phylogenetic tree by comparing two models describing the likelihood of branching patterns (Pons et al. 2006). The null model assumes that the entire sample derives from a single population undergoing a single coalescent process, whereas the general mixed Yule coalescent (GMYC) model classifies the observed branching time intervals into two categories, as the result of either inter- or intraspecific processes of lineage sorting. A log-likelihood ratio test is then used to assess which model provides a better fit. The GMYC model additionally integrates scaling parameters for both the diversification (pk+1) and coalescent (pj) processes, which allow departures from strict assumptions of constant population size and rates of cladogenesis. The models were fitted using an R script provided by T. Barraclough (Pons et al. 2006) to an ultrametric tree obtained by non-parametric rate smoothing of the NJ tree (Sanderson 1997), as implemented in the R package APE (Paradis et al. 2004).
3. Results
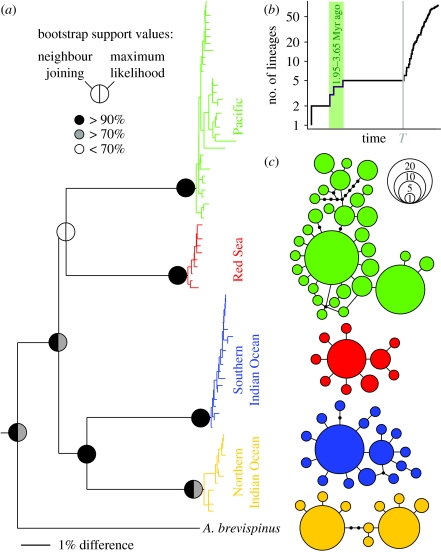
Evidence for species status of the clades comes, first, from the extent of the genetic distances between them. These ranged from 8.8 to 10.6% (as opposed to <0.7% within clades), equivalent to the distances between sibling species in other starfish (Waters et al. 2004). According to these distances, the four clades are estimated to have diverged between 1.95 and 3.65 Myr ago. Second, the COI haplotypes grouped into four disconnected statistical parsimony networks at the 95 per cent connection limit (figure 1c) suggested as a species delimitation threshold (Hart & Sunday 2007). Third, using a method that differentiates (i) interspecific from (ii) intraspecific diversification processes through a phylogenetic approach (Pons et al. 2006), we identified the same four clusters, corresponding to the putative sibling species (figure 1a). Indeed, the GMYC model, which assumes a steep increase in branching rates from (i) to (ii) at a threshold T, was preferred over the null model of uniform branching rates (log LGMYC=432.6, 2ΔL=31.1, Χ32-test, p<0.001; figure 1b). Both of the scaling parameters for the diversification (pk+1=−0.27) and coalescent (pj=0.04) processes were smaller than 1.
Figure 1.
Pan-Indo-Pacific Acanthaster species complex (a) Acanthaster COI NJ tree (rooted with Patiria pectinifera, not shown), GMYC clusters in colour, bootstrap support values for both the NJ and maximum-likelihood analyses depicted on main nodes only; scale bar, 1% difference). (b) Lineages-through-time plot based on the ultrametric tree obtained by non-parametric rate smoothing of the phylogeny depicted in (a); grey line is branching rate threshold T; green shaded area highlights the timing of the diversification events, which at a COI divergence rate of 2.9–4.5% Myr−1 (Lessios 2008) corresponds to the Pliocene–early Pleistocene (between 1.95 and 3.65 Myr ago). (c) Four disconnected statistical parsimony networks at the 95 per cent connectivity limit, corresponding to the putative species; same colours as in (a).
4. Discussion
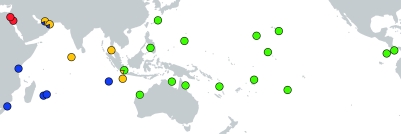
We find that A. planci consists of four strongly differentiated and highly supported mitochondrial clades, from the Red Sea, the Pacific (Pac), the Northern (NIO) and the Southern Indian Ocean (SIO) (figures 1a and 2), that together form a species complex. Although cryptic speciation is a widespread phenomenon in the marine realm, this finding is quite surprising for an organism as extensively studied as A. planci over the past decades.
Figure 2.
Geographical distribution of COI haplotypes from the four putative COTS species. Pie charts indicate relative frequency of each species per sampling location. Colours are the same as in figure 1.
Assuming a COI divergence rate of 2.9–4.5% Myr−1 (Lessios 2008), the four clades are estimated to have diverged in the Pliocene–Early Pleistocene (1.95–3.65 Myr ago). The speciation process was probably driven by sea-level changes (Pillans et al. 1998), isolating populations between major oceans (e.g. Pac versus NIO; Voris 2000). Additionally, restricted circulation patterns could have reduced larval interchange between populations (e.g. SIO versus NIO; Pollock 1993). Furthermore, the strong patterns of regional differentiation may have been enhanced by ecological differences among lineages (Reid et al. 2006). The populations of all four sibling species appear to be expanding, as supported by both the GMYC scaling parameter for the coalescent process (pj<1; Pons et al. 2006) and the overall star shape of each species' haplotype network (figure 1c; Avise 2000).
Our discovery of four highly differentiated clades in one of the world's most destructive coral predators has significant conservation implications. Identifying cryptic speciation is essential to adequately study and contain species that require management (Bickford et al. 2007). Although the status of A. planci is relatively poorly documented from the Indian Ocean and the Red Sea, outbreaks there do not appear to be as massive and widespread as in the Pacific (Zann 2000), suggesting that outbreak patterns might vary between the different sibling species. Up to now, however, the overwhelming majority of COTS research has been performed in the Pacific. Failure to recognize the existence of the sibling species could have contributed to a lack of understanding of the processes that lead to outbreaks in the different COTS lineages, by extrapolating results obtained from the Pacific studies to A. planci's entire distribution for both research and management purposes.
Future research will be required to investigate whether the life history, behavioural patterns and/or ecological requirements that may affect the outbreak dynamics of these four independent evolutionary COTS lineages have diverged sufficiently to necessitate lineage-specific management. This could prove to be crucial for the design of appropriate management strategies to minimize the impact of future catastrophic COTS outbreaks in different regions of the world.
Acknowledgments
Funded by the EU Marie-Curie Early Stage Research Training HOTSPOTS MEST-CT-2005-020561. We thank Tim Barraclough, Imperial College, London, for R scripts, Alfried Vogler for ideas, Oliver Voigt and Daniel Jackson for their helpful comments, Kerry Roper, Bastian Bentlage, Bernie Degnan for assistance, and the Universita¨tsstiftung Go¨ttingen for funding. We also thank Lyndon de Vantier, Elisabeth Illidge-Evans, Alexander Keck, Gordon Kirkwood, Gustav Paulay, Serge Planes, Peter Schupp, Molly Timmers and Sven Uthicke for providing samples, as well as all the institutions and individuals who supported the authors of this study during fieldwork. For recent samples, we acknowledge the Egyptian Environmental Affairs Agency, the Fujeirah Municipality (UAE), the Oman Ministry of Environment and Climate Affairs, Five Oceans LLC (Oman) and CORDIO Kenya. All experiments comply with current German laws.
Supplementary Material
Sampling location information, number of sequences per location, collection reference, accession numbers
References
- Avise J.C. Harvard University Press; Cambridge, MA: 2000. Phylogeography: the history and formation of species. [Google Scholar]
- Becker B.J, Levin L.A, Fodrie F.J, McMillan P.A. Complex larval connectivity patterns among marine invertebrate populations. Proc. Natl Acad. Sci. USA. 2007;104:3267–3272. doi: 10.1073/pnas.0611651104. doi:10.1073/pnas.0611651104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood D.R, Hughes T.P, Folke C, Nyström M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. doi:10.1038/nature02691 [DOI] [PubMed] [Google Scholar]
- Benzie J.A.H. Major genetic differences between crown-of-thorns starfish (Acanthaster planci) populations in the Indian and Pacific Oceans. Evolution. 1999;53:1782–1795. doi: 10.1111/j.1558-5646.1999.tb04562.x. doi:10.2307/2640440 [DOI] [PubMed] [Google Scholar]
- Bickford D, Lohman D, Sodhi N, Ng P, Meier R, Winker K, Ingram K, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. doi:10.1016/j.tree.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Birkeland C.E, Lucas J.S. CRC Press; Boca Raton, FL: 1990. Acanthaster planci: major management problem of coral reefs. [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Engelhardt, U. & Lassig, B. 1997 A review of the possible causes and consequences of outbreaks of the crown-of-thorns starfish (Acanthaster planci) on the great barrier reef—an Australian perspective. In The Great Barrier Reef Science, Use and Management a National Conf. Proceedings, pp. 243–259. Townsville, Australia: Great Barrier Reef Marine Park Authority.
- Galtier N, Gouy M, Gautier C. SeaView and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. doi:10.1093/bioinformatics/12.6.543 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. PhyML—a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hart M.W, Sunday J. Things fall apart: biological species form unconnected parsimony networks. Biol. Lett. 2007;3:509–512. doi: 10.1098/rsbl.2007.0307. doi:10.1098/rsbl.2007.0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios H.A. The great American schism: divergence of marine organisms after the rise of the central American isthmus. Annu. Rev. Ecol. Evol. Syst. 2008;36:63–91. doi:10.1146/annurev.ecolsys.38.091206.095815 [Google Scholar]
- Lucas J.S. Quantitative studies of feeding and nutrition during larval development of the coral reef asteroid Acanthaster planci (L.) J. Exp. Mar. Biol. Ecol. 1982;65:173–193. doi:10.1016/0022-0981(82)90043-0 [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. doi:10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Pillans B, Chappell J, Naish T.R. A review of the Milankovitch climatic beat: template for Plio-Peistocene sea-level changes and sequence stratigraphy. Sediment. Geol. 1998;122:5–21. doi:10.1016/S0037-0738(98)00095-5 [Google Scholar]
- Pollock D.E. Speciation in spiny lobsters—clues to climatically-induced changes in ocean circulation patterns. Bull. Mar. Sci. 1993;53:937–944. [Google Scholar]
- Pons J, Barraclough T, Gomez-Zurita J, Cardoso A, Duran D, Hazell S, Kamoun S, Sumlin W, Vogler A. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. doi:10.1080/10635150600852011 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Reid D, Lal K, Mackenzie-Dodds J, Kaligis F, Littlewood D, Williams S. Comparative phylogeography and species boundaries in Echinolittorina snails in the central Indo-West Pacific. J. Biogeogr. 2006;33:990–1006. doi:10.1111/j.1365-2699.2006.01469.x [Google Scholar]
- Sanderson M.J. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 1997;14:1218–1231. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software v. 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. doi:10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Veron J.E.N. Harvard University Press; Cambridge, MA: 2008. A reef in time: the great barrier reef from beginning to end. [Google Scholar]
- Vogler A.P, Monaghan M.T. Recent advances in DNA taxonomy. J. Zool. Syst. Evol. Res. 2007;45:1–10. doi:10.1111/j.1439-0469.2006.00384.x [Google Scholar]
- Voris H.K. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 2000;27:1153–1167. doi:10.1046/j.1365-2699.2000.00489.x [Google Scholar]
- Waters J, O'Loughlin P.M, Roy M.S. Cladogenesis in a starfish species complex from southern Australia: evidence for vicariant speciation? Mol. Phylogenet. Evol. 2004;32:236–245. doi: 10.1016/j.ympev.2003.11.014. doi:10.1016/j.ympev.2003.11.014 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M. 1973 Early life-histories of coral reef asteroids, with special reference to Acanthaster planci (L.). In Biology and geology of coral reefs: 2, biology 1 (eds O. A. Jones & R. Endean), pp. 369–387. New York, NY: Academic Press.
- Zann, L. P. 2000 Status of the crown-of-thorns starfish in the Indian Ocean. In Coral reefs of the Indian Ocean: their ecology and conservation (eds T. R. McClanahan, C. R. C. Sheppard, D. O. Obura), pp. 59–63. New York, NY: Oxford University Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling location information, number of sequences per location, collection reference, accession numbers