Abstract
Inflammasomes are cytosolic multiprotein complexes that sense microbial infection and trigger cytokine production and cell death. However, the molecular components of inflammasomes, and what they sense, remain poorly defined. Here we demonstrate that 35 amino acids from the C-terminus of flagellin triggered inflammasome activation in the absence of bacterial contaminants or secretion systems. To further elucidate the host flagellin-sensing pathway, we generated mice deficient in Naip5. Naip5-deficient mice failed to activate the inflammasome in response to the 35 amino acids of flagellin or in response to Legionella pneumophila infection. Taken together, these data clarify the molecular basis for the cytosolic response to flagellin.
Inflammasomes are cytosolic multiprotein complexes that are critical regulators of inflammation, and are required for proteolytic activation of the cysteine protease caspase-1 (refs. 1–3). Caspase-1 (A000492; http://www.signaling-gateway.org/molecule/query?afcsid=A000492) is itself required for the proteolytic processing and release of inflammatory cytokines such as interleukin 1β (IL-1β) and IL-18, as well as for induction of a necrotic-like cell death called pyroptosis1–3. The molecular components and structures of inflammasomes remain poorly defined. It is believed that multiple distinct inflammasomes may exist, each containing a key scaffold protein of the NLR (nucleotide-binding domain, leucine-rich repeat) superfamily that confers specificity for particular microbial products. For example, NLR proteins of the NLRP1 family (also called NALP1) appear to activate the inflammasome in response to anthrax lethal toxin4 and bacterial muramyl dipeptide5. In contrast, the NLR protein NLRP3 (also called NALP3 or cryopyrin) has been proposed to sense a wide range of stimuli including bacterial RNA6, viral DNA7, uric acid crystals8, muramyl dipeptide9, 10, nigericin11, amyloid-beta12, and other irritants13–16. There is at present no molecular explanation for how a single NLR protein can be activated by all these microbial products and the precise molecular nature of what is sensed by any inflammasome remains undefined.
The inflammasome containing the NLR protein IPAF (also called NLRC4) is one of the best characterized inflammasomes, and has been proposed by several groups to respond to the presence of flagellin in the cytosol17–19. Flagellin-deficient mutants of Salmonella typhimurium and Legionella pneumophila are defective in IPAF-dependent inflammasome activation, and flagellin, purified from or expressed in bacteria, triggers IPAF-dependent caspase-1 activation when delivered to the cytosol of macrophages by use of a pore-forming toxin (listeriolysin O (LLO)) or transfection reagents17–21. It was proposed that during natural infections, flagellin triggers inflammasome activation upon secretion into the host cytosol via bacterial type III/IV secretion systems17–21. However, doubts have been expressed as to whether flagellin is indeed sensed cytosolically22, 23 since none of the existing studies eliminated the possibility that bacterial secretion systems, LLO, transfection reagents, and/or copurifying bacterial contaminants, contributed to activation of caspase-1. Moreover, the role of IPAF in sensing flagellin remains unclear. It has been reported that IPAF-deficient (Nlrc4−/−) macrophages also fail to activate caspase-1 in response to non-flagellated pathogens, including flagellin-deficient Pseudomonas aeruginosa24 and Shigella flexneri25. In addition, IPAF has been proposed to play a key role in caspase-1 activation in response to aerolysin, a pore-forming toxin26. Thus it has been proposed that IPAF may not in fact respond to flagellin but instead may respond to bacterial contaminants, membrane pores or type III and/or IV secretion systems22, 23, which appear to be required for inflammasome activation by all bacterial pathogens tested.
Even less clear is the role of NLR apoptosis inhibitory protein 5 (Naip5, Birc1e), a protein that hetero-oligomerizes with IPAF27, 28, and has also been proposed to be involved in cytosolic sensing of flagellin20, 21. However, the role of Naip5 in inflammasome activation remains unclear29, 30. Like IPAF, Naip5 contains a nucleotide binding domain and leucine rich repeats, but Naip5 lacks a caspase recruitment domain (CARD). Instead, Naip5 contains baculovirus inhibitor of apoptosis repeats (BIRs), and is thus classified as a member of the inhibitor of apoptosis (IAP) protein family31. Indeed, it was reported that the BIRs from human NAIP are capable of binding and inhibiting the apoptotic caspases, caspase-3, -7 and -9 (refs. 32, 33) though these findings have been questioned31. Macrophages from the A/J mouse strain carry a defective Naip5 allele, which (like IPAF deficiency) results in permissiveness to L. pneumophila replication34, 35, but A/J macrophages fully or almost fully activated caspase-1 in response to L. pneumophila20, 21, 28, 29. Several groups have proposed that the role of Naip5 in resistance to L. pneumophila is unrelated to inflammasome activation but is instead due to modulation of the intracellular fate of the legionella-containing vacuole17, 29, 36–38. However, the absence of Naip5 ‘knockout’ mice, and the reliance on the A/J allele of Naip5, which may not be a null allele, has led to much uncertainty as to the role of Naip5 in inflammasome activation.
In this report we clarify many of the above issues. We define a short peptide domain from the C-terminus of flagellin that appears sufficient to activate the inflammasome in the absence of bacterial contaminants, pores or secretion systems. Importantly, we also generated Naip5-deficient (Naip5−/−) mice, and demonstrate a requirement for Naip5 in inflammasome activation in response to the C-terminus of flagellin and L. pneumophila infection. Our results provide an example of an ‘inhibitor of apoptosis’ protein being required for inflammasome activation rather than for apoptosis inhibition. Taken together, our results clarify the molecular basis of inflammasome activation.
Results
Flagellin alone triggers IPAF and caspase-1
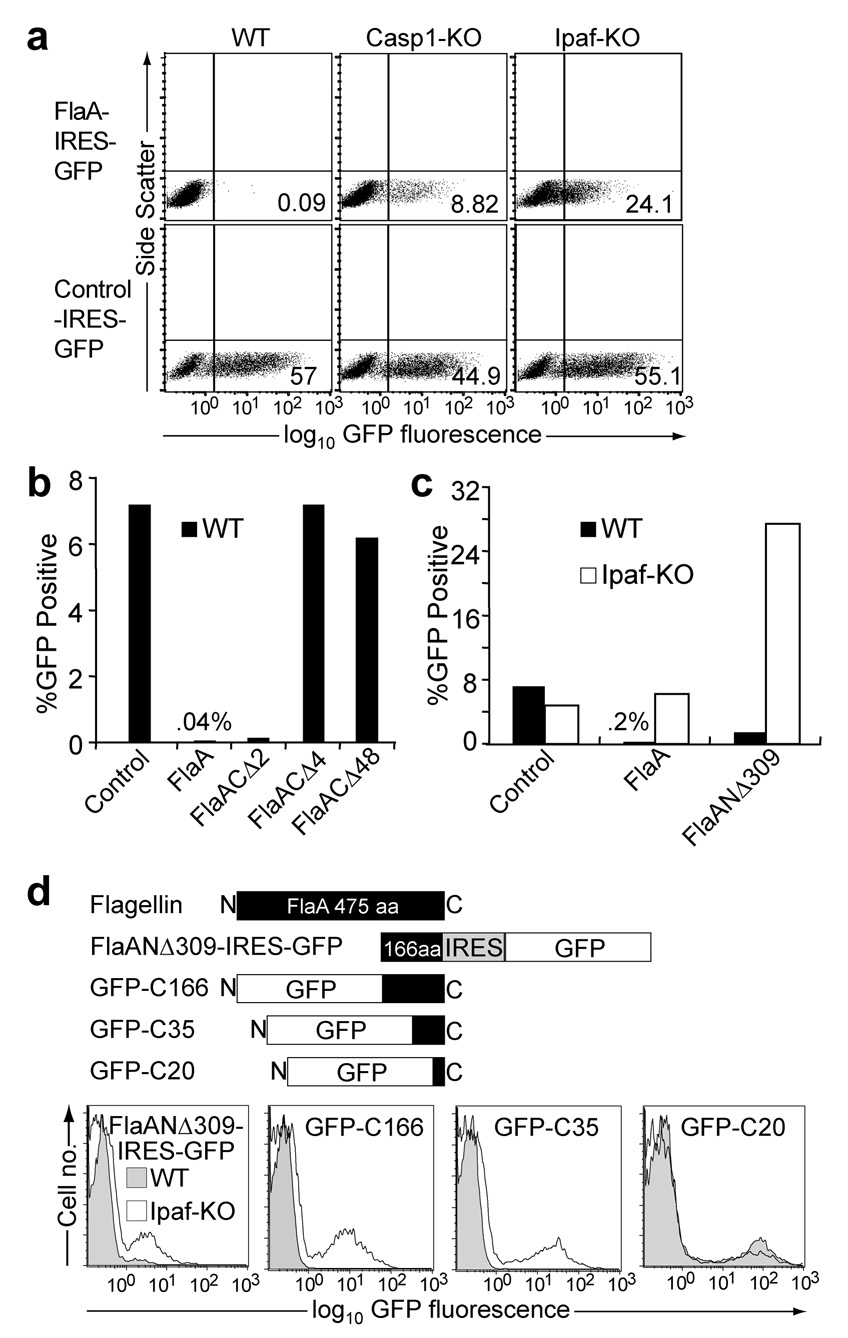
To determine whether flagellin itself, in the absence of bacterial modifications, contaminants, secretion systems or transfection reagents, is sufficient to trigger IPAF and macrophage death, we used retroviral transduction to express L. pneumophila flagellin (FlaA) directly in the macrophage cytosol from a eukaryotic promoter. The retroviral expression constructs also contained an internal ribosomal entry site (IRES) and a green fluorescent protein (GFP) gene to permit identification of positively transduced cells. Primary bone marrow cells were transduced, differentiated into macrophages in the presence of macrophage colony stimulating factor, and resulting macrophages were analyzed for GFP expression four days after transduction. Although wildtype B6 macrophages were efficiently transduced with a control retrovirus, we were unable to recover B6 macrophages transduced with a FlaA-expressing retrovirus when analyzed four days after transduction (Fig. 1a). By contrast, caspase-1-deficient and IPAF-deficient macrophages were transduced with FlaA-expressing retrovirus (Fig. 1a). Thus, the inability to transduce B6 macrophages with FlaA is due to an activity of IPAF and caspase-1 (presumably induction of pyroptotic cell death, see below). We conclude that flagellin itself is sensed in the cytosol in the absence of other bacterially-derived signals.
Figure 1. The C-terminus of flagellin is necessary and sufficient to trigger IPAF- and caspase-1-dependent macrophage death.
(a) Flow cytometry of wild-type, caspase-1-deficient and IPAF-deficient macrophages transduced with retroviruses expressing flagellin from L. pneumophila (FlaA) or a control gene (Irgb10) followed by an internal ribosomal entry site (IRES) and GFP. (b,c) Graphs of flow cytometry data on wild-type or IPAF-deficient macrophages transduced with retroviruses expressing a control protein, full length FlaA, or flagellin lacking the C-terminal 2, 4 or 48 amino acids (FlaACΔ2, FlaACΔ4, FlaACΔ48) (b) or flagellin lacking the N-terminal 309 amino acids (FlaANΔ309) (c). (d) Flow cytometry of wild-type or IPAF-deficient macrophages transduced with retroviruses expressing FlaANΔ309-IRES-GFP (left) or fusions of GFP and the following truncations of the C-terminal end of flagellin: C166 (GFP-C166), C35 (GFP-C35) and C20 (GFP-C20). Results are representative of 2–5 independent experiments.
Flagellin C-terminus triggers IPAF-dependent pyroptosis
In order to identify the region of flagellin that is sensed cytosolically, we transduced macrophages with retroviruses expressing a series of C- and N-terminal deletion mutants of flagellin from L. pneumophila. Flagellins lacking 4 or 48 C-terminal amino acids (FlaACΔ4, FlaACΔ48) were not cytotoxic and efficiently transduced macrophages (Fig. 1b). In contrast, a deletion mutant lacking the N-terminal two-thirds of flagellin (FlaANΔ309) still triggered IPAF-dependent cell death (Fig. 1c). These results implied that the C-terminal third of flagellin is necessary and sufficient to trigger IPAF. To define further the portion of flagellin that is sensed by IPAF, macrophages were transduced with retroviruses that express GFP fused to the C-terminal 20 (GFP-C20), 35 (GFP-C35) or 166 (GFP-C166) amino acids of flagellin. Although transduction of B6 macrophages with the GFP-C20 construct was not cytotoxic and resulted in significant green fluorescence, the GFP-C35 and GFP-C166 expression constructs failed to transduce wild-type, but not IPAF-deficient macrophages (Fig. 1d). Thus, we conclude that the C-terminal 35 amino acids of flagellin fused to GFP are sufficient to activate IPAF in the cytosol.
Although the simplest interpretation of our data is that cytosolic expression of flagellin triggers an IPAF- and capsase-1-dependent pyroptotic cell death of B6 macrophages, we considered the alternative possibility that flagellin-expressing retrovirus selectively failed to transduce B6 macrophages. Both possibilities are formally consistent with the lack of GFP+ macrophages four days after transduction. To demonstrate that B6 macrophages were first being transduced by flagellin-expressing retrovirus, and then dying, we analyzed bone marrow cells at early timepoints after transduction. At 6 hours after transduction, we were able to detect both B6 and IPAF-deficient bone marrow cells expressing the GFP-C35 flagellin construct. We found that B6 cells transduced with the GFP-C35 flagellin construct gradually disappeared from the culture, whereas IPAF-deficient cells were maintained (Supplementary Fig. 1a). The large number of spontaneously dying cells in primary bone marrow cell cultures precluded an analysis of whether GFP+ B6 cells subsequently become permeable to DNA-intercalating dyes (e.g., 7AAD), a late measure of cell death. However, we were able to observe 7AAD+ cells accumulate among immortalized bone marrow-derived macrophages that were transduced with the cytotoxic GFP-C35 construct (Supplementary Fig. 1b). By contrast, 7AAD+ cells did not accumulate among cells transduced with the noncytotoxic GFP-C20 construct. Although permeability to 7AAD does not distinguish late apoptotic from pyroptotic cells, the observed dependence of the cell death on caspase-1 (Fig. 1a) and a lack of dependence on caspase-3 (Supplementary Figure 1c) are consistent only with pyroptosis. We conclude that cytosolic expression of the C-terminal 35 amino acids of flagellin, fused to GFP, induces IPAF-capsase-1-inflammasome-dependent pyroptotic cell death.
IPAF and TLR5 respond to distinct regions of flagellin
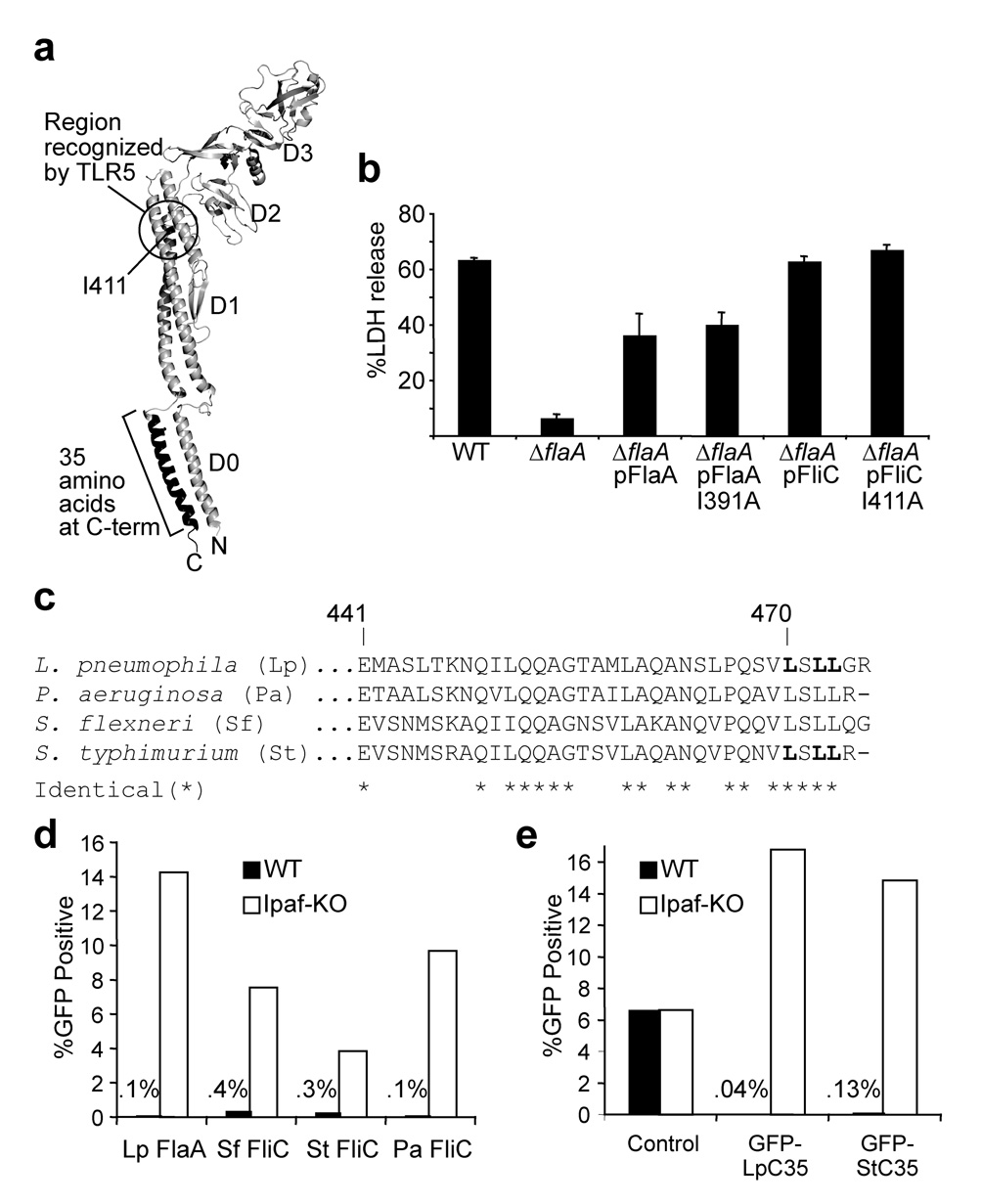
Previous studies demonstrated that sensing of flagellin by Toll-like receptor 5 (TLR5) requires amino acids in the D1 region of flagellin (e.g., isoleucine 411)39, whereas the above studies indicate it is the C-terminal D0 region that is sensed cytosolically (Fig. 2a). When expressed from S. typhimurium, an I411A flagellin mutant also reportedly failed to activate caspase-1, but this may have resulted from a failure of the I411A flagellin to be translocated into host cells by S. typhimurium18. We circumvented this difficulty by expressing salmonella flagellin (FliC) in flagellin-deficient (ΔflaA) L. pneumophila. As we previously showed, salmonella flagellin (fliC) is able to restore cytotoxicity to the ΔflaA L. pneumophila mutant21. Importantly, both wild-type and I411A FliC were equally able to restore cytotoxicity to the ΔflaA mutant (Fig. 2b), implying that amino acids required for TLR5 sensing are not required for inflammasome activation. Our results therefore establish that there are two distinct innate immune pathways for detecting flagellin: a cell-surface TLR5-dependent pathway that senses the D1 region of flagellin and triggers NF-κB activation and a cytosolic IPAF-dependent pathway that senses the C-terminal D0 region and then activates the inflammasome.
Figure 2. The cytotoxic C-terminus of flagellin is highly conserved and is distinct from the region sensed by TLR5.
(a) Model of the salmonella flagellin monomer as it appears within the assembled flagellar filament40. Isoleucine 411 is critical for ‘sensing’ of flagellin by TLR539, and corresponds to I391 in L. pneumophila flagellin. (b) Cell death assayed by release of lactate dehydrogenase (LDH) from B6 macrophages infected with indicated L. pneumophila strains (MOI = 1). Error bars represent s.d. of triplicate samples. (c) Alignment of the C-terminal 35 amino acids of flagellin; bolded leucines in Lp and St flagellin were mutated to alanine in the experiments in Fig. 3. (d,e) Graphs of flow cytometry data on wild-type (WT) or IPAF-deficient (IPAF-KO) macrophages transduced with retroviruses expressing either full-length flagellin from L. pneumophila (Lp-FlaA), S. typhimurium (St-FliC), P. aeruginosa (Pa-FliC) or S. flexneri (Sf-FliC) upstream of IRES-GFP (d) or the C-terminal 35 amino acids (C35) of L. pneumophila (Lp) or S. typhimurium (St) flagellin fused to the C-terminus of GFP (e). The percentage of macrophages that were GFP+ is indicated. Results are representative of 2–5 independent experiments.
Essential amino acids in flagellin are sensed in the cytosol
The C-terminal 35 amino acids of flagellin are essential for flagellum filament assembly40 and are highly conserved (Fig. 2c). Because of this conservation, we predicted that cytosolic expression of flagellins from S. typhimurium, P. aeruginosa and S. flexneri should be sufficient to trigger IPAF, as was indeed observed (Fig. 2d). Importantly, the C-terminal 35 amino acids of salmonella flagellin, fused to GFP, were also sufficient to trigger activation of IPAF (Fig. 2e). Thus, the minimal domain within flagellin sensed by IPAF appears to be conserved among bacterial species. However, flagellin is not necessarily involved in inflammasome activation for all these bacterial species24, 25; indeed, many bacterial species may evade detection by failing to express or secrete potentially cytotoxic flagellins into the host cytosol.
We further refined the region of flagellin sensed by IPAF by mutating several conserved C-terminal leucines to alanines. Mutation of L470 slightly reduced the cytotoxicity of flagellin, whereas mutation of L472 and L473 had a greater effect, and mutation of all three leucines abolished the ability of the GFP-C35 flagellin fusion to trigger IPAF-dependent cell death (Fig. 3a). Importantly, immunoblot analysis suggested that the mutations did not adversely affect the abundance or stability of the GFP-C35 fusion (Fig. 3b). Introduction of the same point mutations into a copy of flagellin on the chromosome of L. pneumophila abolished motility of L. pneumophila (data not shown), indicating that the amino acids sensed by IPAF are critical for function of the flagellum. As expected, the point mutations also abolished the ability of L. pneumophila to trigger macrophage pyroptosis (Fig. 3c), IL-1β release (Fig. 3d) or caspase-1 cleavage (Supplementary Fig. 4), and also allowed L. pneumophila to evade IPAF-mediated growth restriction and replicate in B6 macrophages (Fig. 3e).
Figure 3. Leucines in the C-terminus of L. pneumophila flagellin are critical for activation of the inflammasome.
(a) Flow cytometry of wild-type and IPAF-deficient macrophages transduced with GFP-C35 harboring the mutations L470A (GFP-C35A), L472A and L473A (GFP-C35AA), or L470A, L472A and L473A (GFP-C35AAA). (b) Immunoblot of IPAF-deficient macrophages expressing the constructs in a with anti-GFP. (c) Cell death indicated by release of lactate dehydrogenase (LDH) ±s.d. from wild-type macrophages infected with wild-type (WT), flagellin-deficient (ΔflaA) or ΔflaA L. pneumophila complemented with wild-type flagellin (ΔflaA ::flaA) or the leucine to alanine mutants L470A (ΔflaA::flaA-A), L472A and L473A (ΔflaA::flaA-AA), all three mutations (ΔflaA::flaA-AAA), salmonella flagellin ((ΔflaA ::fliC), or the fliI mutant (fliI::Cm), which is non-flagellated but still expresses flaA21, 53. Macrophages were assayed for release of LDH after 4 hours of infection. (d) ELISA assay for release of IL-1β from Pam3CSK4-primed bone-marrow-derived macrophages infected at an MOI of 1 with the indicated strains. (e) Replication of the indicated L. pneumophila strains, in wild-type and IPAF-deficient macrophages, measured by colony forming units. Results are representative of 2–3 independent experiments.
Although the C-terminus of flagellin is not known to direct its translocation into host cells, our results may be explained by a failure of the mutant flagellins to be translocated into the macrophage cytosol. Unfortunately, there is no assay sensitive enough to measure translocation of legionella flagellin into host cells, likely because the amounts of legionella flagellin translocated are very low. Recently, however, type III-dependent translocation of salmonella flagellin (FliC) into host cells was detected by use of a flagellin-β-lactamase fusion reporter protein41. We therefore decided to use the salmonella system to ascertain whether the conserved C-terminal leucines were required for cytotoxicity and translocation of salmonella flagellin. We found that S. typhimurium expressing a mutant FliC (in which the three C-terminal leucines (Fig. 2c) were changed to alanine) was much less cytotoxic to macrophages than was S. typhimurium expressing wild-type FliC (Supplementary Fig. 2a). Importantly, translocation of the mutant flagellin into macrophages was not impaired by the leucine-to-alanine mutations, as measured by the β-lactamase fusion assay (Supplementary Fig. 2b). These data establish that conserved amino acids at the C-terminus of flagellin are required for triggering the IPAF inflammasome, independent of the requirements for translocation into host cells. The salmonella data also strongly validate our retroviral transduction approach for mapping the regions within flagellin that trigger inflammasome activation.
A recent report suggested that amino acids at the N-terminus of pseudomonas flagellin may contribute to IPAF activation42. This study did not distinguish whether the N-terminal mutations in flagellin affected recognition by IPAF or translocation of flagellin into host cells. Future studies will therefore be required to reconcile these results with our findings. It should also be emphasized that in addition to the C-terminal leucines we identified, there may be other regions within the minimal 35 amino acids that are required for IPAF activation. For example, the GFP-C20 construct contains the C-terminal leucines but is not cytotoxic, implying that amino acids outside the C-terminal 20 amino acids but within the C-terminal 35 amino acids may also play a role. Future studies will be required to determine whether these amino acids (i.e., 21–35 from the C-terminus) are directly sensed or whether they merely physically extend the C-terminus so that it is available to be recognized.
Inflammasome activation by L. pneumophila requires Naip5
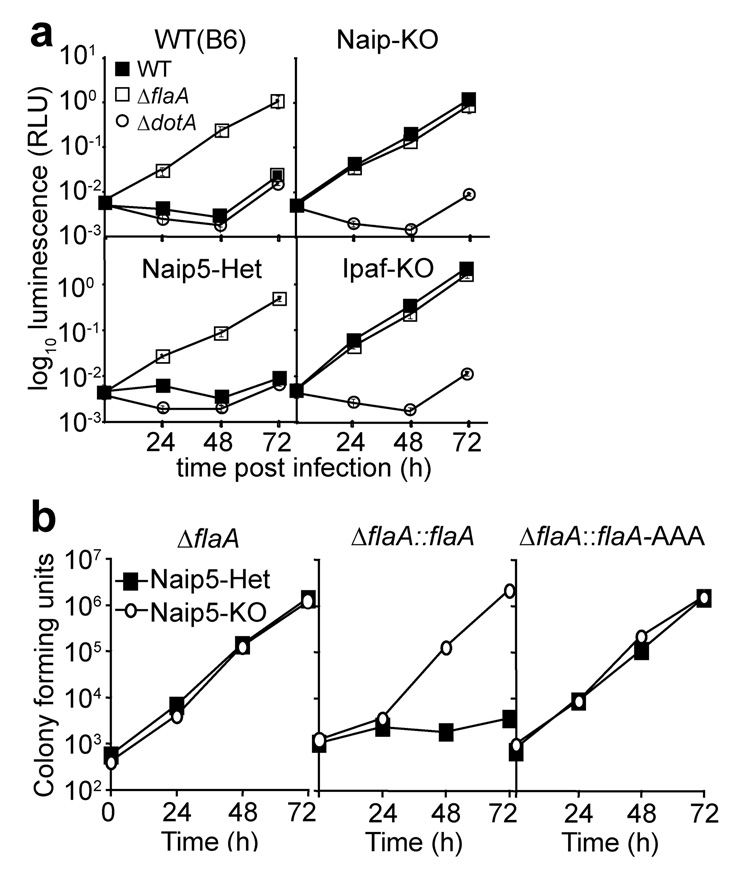
To clarify the role of Naip5 in inflammasome activation, we generated Naip5-deficient mice on a pure C57BL/6 genetic background (Supplementary Fig. 3). We found that Naip5-deficient macrophages completely failed to activate caspase-1, release IL-1β or undergo pyroptotic cell death in response to infection with L. pneumophila (Fig. 4a–c). Naip5-deficient macrophages were also highly permissive to replication of L. pneumophila (Fig. 5a,b). As also observed previously20, 21, we found that B6.A-Chr13 macrophages (which carry the A/J Naip5 allele) exhibit only a small defect in pyroptotic cell death and caspase-1 activation in response to L. pneumophila (Fig. 4a,b). Thus, it appears that the A/J allele of Naip5 is indeed partially functional.
Figure 4. Naip5 is required for activation of caspase-1 and release of IL-1β induced by L. pneumophila.
(a) Cell death assayed by release of lactate dehydrogenase (LDH) from peritoneal macrophages infected by wild-type (WT) or flagellin-deficient (ΔflaA) L. pneumophila (MOI = 1). Chr13-AJ mice carry chromosome 13 and the Naip5 locus from A/J mice on the B6 background. (b) Immunoblot of processed p10 of active caspase-1 detected in supernatants of bone-marrow-derived macrophages infected with wild-type (WT) or flagellin-deficient (ΔflaA) L. pneumophila at an MOI of 1. (c) Sandwich ELISA assay for release of IL-1β ±s.d. from Pam3CSK4-primed wild-type (B6), Naip5-deficient (Naip5-KO) or Naip5-heterozygous (Naip5-het) bone-marrow-derived macrophages infected with wild-type (WT) or flagellin-deficient (ΔflaA) L. pneumophila at an MOI of 1. Results are representative of 2–5 independent experiments.
Figure 5. Naip5 is required for restriction of L. pneumophila replication in macrophages.
(a) Luminescence assay50 to measure replication of wild-type (WT), ΔflaA or type IV secretion-deficient ΔdotA L. pneumophila in bone-marrow-derived macrophages (MOI = 0.01). Values represent mean ± s.d. of triplicate samples. (b) Replication measured by colony forming units of ΔflaA, or ΔflaA complemented with either wild-type flaA (ΔflaA::flaA) or with the triple leucineto-alanine mutant (ΔflaA::flaA-AAA, see Fig. 3); colony forming units were evaluated at daily intervals after infection. Results are representative of 2–3 independent experiments.
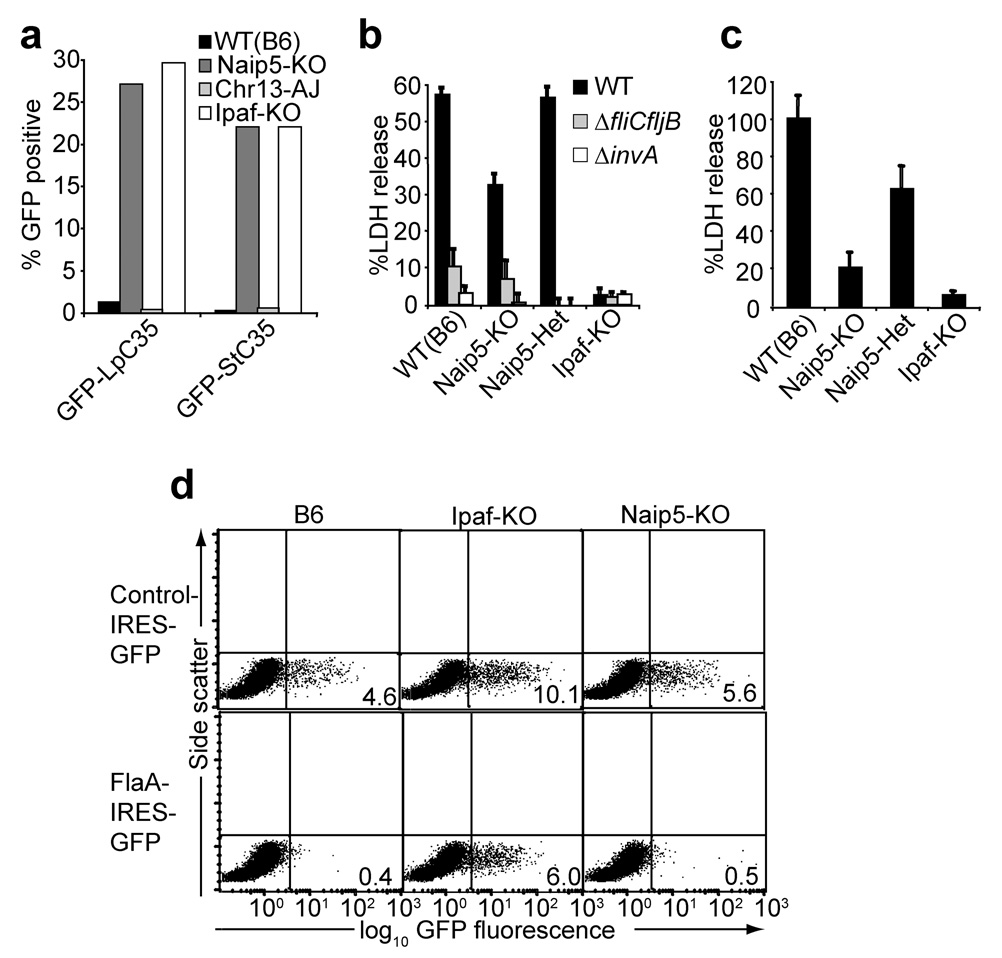
We sought to determine whether Naip5 is required for inflammasome activation in response to the same 35 amino acid flagellin-derived peptide that activates IPAF. Macrophages were transduced with retroviral constructs that express the C-terminal 35 amino acid peptide from L. pneumophila and S. typhimurium flagellin fused to GFP. These constructs were cytotoxic to B6 macrophages, but were transduced as efficiently into Naip5-deficient macrophages as into IPAF-deficient macrophages (Fig. 6a). Thus, Naip5 is required for inflammasome activation in response to the same region of flagellin that activates IPAF.
Figure 6. Role of Naip5 in recognition of L. pneumophila, S. typhimurium and P. aeruginosa.
(a) Graph of flow cytometry data on macrophages from the indicated strains transduced with retroviruses expressing GFP fused to the C-terminal 35 amino acids of flagellin from L. pneumophila (Lp-C35) or S. typhimurium (St-C35). (b,c) Cell death indicated by release of lactate dehydrogenase (LDH) ±s.d. from bone marrow-derived macrophages infected with wild-type (WT), SPI-1 type III secretion system-deficient (ΔinvA) or flagellin-deficient (ΔfliCfljB) S. typhimurium (MOI = 2) (b) or in bone marrow-derived macrophages by wild-type P. aeruginosa (MOI = 10) (c). (d) Flow cytometry of macrophages transduced with retroviruses expressing a control protein or full-length L. pneumophila flagellin (FlaA). Results are representative of at least 3 independent experiments.
Not all IPAF functions require Naip5
Although our results demonstrate an unequivocal requirement for Naip5 in inflammasome activation in response to L. pneumophila, we were interested to determine if Naip5 was required for all known functions of IPAF. For instance, we hypothesized that closely related Naip proteins (e.g., Naip6 has 95% amino acid identity to Naip5) might circumvent the requirement for Naip5 in particular instances. Indeed, we observed that P. aeruginosa and S. typhimurium triggered pyroptotic cell death that was IPAF-dependent but only partly Naip5-dependent (Fig. 6b,c). In the case of infection with S. typhimurium, Naip5-independent IPAF activation was still flagellin-dependent, since a flagellin-deficient fliCfljB mutant was not cytotoxic (Fig. 6a). In addition, we observed that retroviral-mediated overexpression of full-length flagellin from L. pneumophila was cytotoxic to Naip5-deficient but not IPAF-deficient macrophages (Fig. 6d). Since the 35 amino acid C-terminal region of flagellin is sensed in a strictly Naip5-dependent manner (Fig. 6a), these results may suggest that regions of flagellin outside of the C-terminal 35 amino acids can contribute to IPAF activation in the absence of Naip5. Regardless of how other regions of flagellin might contribute to sensing in overexpression scenarios, it remains clear that flagellin-dependent activation of the inflammasome upon natural infection with L. pneumophila is strictly Naip5-dependent (Fig. 4).
Discussion
The specific molecular determinants capable of activating inflammasomes are in general poorly defined. Here we have specified particular amino acids at the C-terminus of flagellin that are required to trigger a cytosolic inflammasome comprised of IPAF and Naip5. Our results significantly clarify the molecular nature of the trigger of the Naip5-IPAF inflammasome, and argue against contributions from membrane damage, bacterial secretion systems, or other bacterial contaminants. Since the region of flagellin that triggers the Naip5-IPAF inflammasome appears to be a relatively short peptide, our findings may be valuable in the design of peptide mimetic compounds that could have therapeutic application. For example, small peptide-based compounds that mimic the activity of the pro-apoptotic protein Smac have shown promise as anti-tumor drugs43.
Naip5 has previously been proposed to be dispensable for inflammasome activation in response to legionella flagellin29. However, previous studies have relied on the use of the defective A/J allele of Naip5, which our data (Fig. 4) indicate is not a null allele. Our results with Naip5-deficient mice provide new and unequivocal evidence that Naip5 functions upstream of caspase-1 activation. However, the question as to whether the permissiveness of A/J macrophages to L. pneumophila is related to defective caspase-1 activation, or to another activity17, 29, 36–38 downstream of Naip5, remains to be settled definitively. The use of quantitative single-cell assays21, 28, as opposed to qualitative assays29, reveals clear defects in caspase-1 activation in A/J macrophages. Moreover, caspase-1-deficient macrophages exhibit increased permissiveness to L. pneumophila17, 28. Thus we tend to favor a simple model in which L. pneumophila replication in macrophages is restricted as a consequence of Naip5-IPAF-dependent caspase-1 activation, though more complex models can also be envisaged.
It is interesting that an inhibitor of apoptosis (IAP) gene, Naip5, appears to be required for activation of the inflammasome. Other IAP family members, such as XIAP and cIAP-1/2, were previously described to function as apoptosis inhibitors44. Our data, and other recent data43, 45, 46, suggest that IAPs may have broader functions within cells than previously assumed. It remains to be determined whether the BIR domains in Naip5 are critical for regulation of inflammasome activation.
A major outstanding question is whether inflammasomes are triggered directly by microbial products, or alternatively, whether microbial products have indirect effects on host cells that ultimately result in inflammasome activation47. Our results do not settle this question. The fact that C-terminal leucine-to-alanine point mutations can abolish the ability of flagellin to trigger the inflammasome is strongly suggestive of a direct physical interaction between flagellin and a host sensor. However, it remains possible that the flagellin peptide indirectly triggers the inflammasome. For example, the flagellin peptide might adversely interfere with some host cellular process, and this physiological disruption might be what activates the inflammasome. These questions will be settled in future studies aimed at determining whether a host protein, possibly Naip5 or IPAF, directly binds the region of flagellin we have identified. These studies are likely to be challenging given that direct binding between a microbial ligand and a cytosolic sensor has yet to be unequivocally demonstrated.
One particularly surprising result was our observation that transduction of macrophages with a construct expressing the C-terminal 35 amino acids of flagellin fused to GFP resulted in Naip5-dependent cell death, whereas full-length flagellin triggered Naip5-independent (but IPAF-dependent) cell death. This difference does not appear to be related to fusion of flagellin to GFP, since full-length flagellin fused to GFP triggers Naip5-independent cell death (J.P. and R.E.V., unpublished results). Instead, it appears that regions of flagellin outside the C-terminal 35 amino acids can contribute, directly or indirectly, to Naip5-independent IPAF-activation, at least in overexpression scenarios. Given that Ipaf is also required for inflammasome activation in response to certain non-flagellated bacteria24, 25, we favor a model in which Ipaf provides a flexible scaffold for the assembly of multiple distinct inflammasomes, only a subset of which require Naip5. Future studies are required to address the specificity of alternative Ipaf-containing inflammasomes and the possible involvement of other Naip paralogs.
Our data indicate that the Naip5-IPAF inflammasome responds to a region of flagellin that is distinct from that which is sensed by TLR5 at the cell surface. This raises the interesting question as to why two independent innate immune pathways have convergently evolved to recognize the same microbial molecule. Naip5-IPAF and TLR5 are localized to distinct subcellular compartments and trigger markedly different signalling outcomes – inflammasome activation and cell death versus NF-κB activation. Thus, it seems likely that the innate immune system is not simply geared to detect the mere presence or absence of microbial ligands. Instead, pathogen sensors are distributed to distinct subcellular compartments in order to discriminate and respond appropriately to different classes of microbes, e.g., those that access the cytosol from those that do not. The importance of subcellular localization of pathogen sensors is important in other contexts as well48, and is likely a feature of general importance for appropriate immune responses.
Methods
Mice
Wild-type C57BL/6 (B6) mice and B6 mice harboring A/J chromosome 13 (B6.A-Chr13) were obtained from the Jackson Labs (Bar Harbor, ME). IPAF-deficient (Nlrc4−/−) mice49 were obtained from S. Mariathasan and V. Dixit (Genentech). Caspase-1-deficient (Casp1−/−)3 were the gift of A. Van der Velden and M. Starnbach. Naip5-deficient mice (Naip5−/−) were generated by gene targeting in Bruce4 C57BL/6-derived ES cells (Supplementary Fig. 3) and maintained on a pure B6 background. Animal experiments were approved by the UC Berkeley Animal Care and Use Committee.
Bacterial Strains
LP02 is a streptomycin-resistant thymidine auxotroph derived from L. pneumophila LP01. An unmarked deletion of flaA was described previously21. The ΔflaA strain was complemented by pBBR1-MCS2 plasmids expressing flagellin from L. pneumophila or S. typhimurium (or point mutants thereof). The ΔflaA strain was also complemented by reintroducing a copy of flagellin (or point mutants thereof) onto the chromosome under the control of the endogenous L. pneumophila flaA promoter. FlaA and its promoter were cloned into the suicide vector pSR47S and introduced onto the chromosome by a single crossover and selection with kanamycin. Salmonella typhimurium LT2 and isogenic mutants were the gift of A. Van der Velden and M. Starnbach. Pseudomonas aeruginosa strain PAK was the gift of T. Machen. S. typhimurium was grown overnight in Luria-Bertani broth and reinoculated at a 1:100 dilution and grown to mid-exponential phase (3h) to induce SPI-I. Overnight cultures of P. aeruginosa were diluted 1:10 and grown 3h.
Growth Curves
Growth curves were performed as previously described50. Briefly, macrophages were plated in 96 or 24 well tissue culture plates at 5 × 10 5 /mL, allowed to adhere, and then infected with stationary phase L. pneumophila at an MOI of 0.01. Growth of luminescent L. pneumophila strains was assessed in an LmaxII plate-reading luminometer (Molecular Devices). Growth of non-luminescent strains was assessed by plating for colony forming units on BCYE plates.
Cytotoxicity Assays
Cytotoxicity was measured by evaluating the activity of lactate dehydrogenase (LDH) released from cells51. Overnight cultures of L. pneumophila in stationary phase were added at an MOI of 1 to a confluent monolayer of macrophages, and plates were spun at 400 ×g for 10 minutes to ensure comparable infectivity of motile and non-motile strains. Culture supernatants were assayed for LDH activity after 4h. S. typhimurium (LT2 background) was added at an MOI of 2, and P. aeruginosa at an MOI of 10. Plates were spun as described above, and gentamicin (100ug/ml) was added 30 min post infection (PI) to kill extracellular bacteria. LDH release was measured after 4h and 2h PI, respectively. Specific lysis was calculated as a percentage of detergent lysed macrophages. The cytotoxicity experiments in Supplementary Fig. 2 were performed as previously described41.
Retroviral Constructs and Production
Genes encoding flagellin or control protein were cloned into a replication defective mouse stem-cell retroviral construct (pMSCV2.2). Retroviral particles were generated using Phoenix-eco packaging cells, and were used to transduce bone marrow cells after 48h and 72h culture in macrophage colony stimulating factor. Cells were typically analyzed 3–4 days after the first transduction. In the experiment in Supplementary Fig. 1b, v-raf/v-myc immortalized B6-derived bone marrow macrophages52 (gift of D. Golenbock and K. Fitzgerald) were transduced with VSV pseudotyped virus.
Western blotting
Bone marrow-derived macrophages (106) were seeded in a 6-well plate and infected at an MOI of 1. At 1h post infection, the bacterial suspension was replaced with fresh media without serum, and infection was allowed to continue for 2 hours. Subsequently, supernatants were harvested and precipitated with 10% trichloroacetic acid (TCA). Precipitated proteins were separated on 12% gels (Invitrogen), blotted onto Immobilon-P transfer membrane (Millipore) and probed with rabbit polyclonal anti-caspase-1 p10 antibody (Santa Cruz Biotechnology; antibody sc-514).
Cytokine ELISA
Bone-marrow derived macrophages were primed with 0.5 µg/ml Pam3CSK4 (gift of G. Barton) for 4 hours prior to infection. Supernatants from infected macrophages were analyzed by sandwich ELISA. IL-1β was captured on a monoclonal hamster anti-mouse IL-1β antibody (eBioscience) and detected using biotinylated polyclonal rabbit anti-mouse IL-1β (eBioscience). Recombinant mouse IL-1β (R&D Systems) was used as a standard.
Supplementary Material
Acknowledgements
We thank S. Goodart and M. Michelman for help in generation of the Naip5-deficient mice, V. Dixit and S. Mariathasan for IPAF-deficient mice, K. Fitzgerald and D. Golenbock for immortalized B6 macrophages, C. Roy and C. Case for Caspase-3-deficient femurs and D. Raulet, G. Barton, and the Barton and Vance labs for discussions. R. Vance is a recipient of a Cancer Research Institute Investigator Award and acknowledges funding support from NIH grants AI075039 and AI070739. J. Persson is funded by Stiftelsen Olle Engkvist Byggmästare through the Swedish Research Council.
References and Notes
- 1.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 3.Li P, et al. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 4.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 5.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 7.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 8.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 9.Marina-Garcia N, et al. Pannexin-1-Mediated Intracellular Delivery of Muramyl Dipeptide Induces Caspase-1 Activation via Cryopyrin/NLRP3 Independently of Nod2. J Immunol. 2008;180:4050–4057. doi: 10.4049/jimmunol.180.6.4050. [DOI] [PubMed] [Google Scholar]
- 10.Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 11.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 12.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008 doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dostert C, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008 doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host IPAF. J Biol Chem. 2006 doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 18.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 19.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 20.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- 23.Sutterwala FS, Ogura Y, Flavell RA. The inflammasome in pathogen recognition and inflammation. J Leukoc Biol. 2007 doi: 10.1189/jlb.1206755. [DOI] [PubMed] [Google Scholar]
- 24.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 27.Damiano JS, Oliveira V, Welsh K, Reed JC. Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses. Biochem J. 2004;381:213–219. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 29.Lamkanfi M, et al. The Nod-Like Receptor Family Member Naip5/Birc1e Restricts Legionella pneumophila Growth Independently of Caspase-1 Activation. J Immunol. 2007;178:8022–8027. doi: 10.4049/jimmunol.178.12.8022. [DOI] [PubMed] [Google Scholar]
- 30.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci U S A. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davoodi J, Lin L, Kelly J, Liston P, MacKenzie AE. Neuronal apoptosis-inhibitory protein does not interact with Smac and requires ATP to bind caspase-9. J Biol Chem. 2004;279:40622–40628. doi: 10.1074/jbc.M405963200. [DOI] [PubMed] [Google Scholar]
- 33.Maier JK, et al. The neuronal apoptosis inhibitory protein is a direct inhibitor of caspases 3 and 7. J Neurosci. 2002;22:2035–2043. doi: 10.1523/JNEUROSCI.22-06-02035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diez E, et al. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet. 2003;33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- 35.Wright EK, et al. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol. 2003;13:27–36. doi: 10.1016/s0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- 36.Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol. 2005;7:765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortier A, de Chastellier C, Balor S, Gros P. Birc1e/Naip5 rapidly antagonizes modulation of phagosome maturation by Legionella pneumophila. Cell Microbiol. 2006 doi: 10.1111/j.1462-5822.2006.00839.x. [DOI] [PubMed] [Google Scholar]
- 38.Watarai M, et al. Legionella pneumophila is internalized by a macropinocytotic uptake pathway controlled by the Dot/Icm system and the mouse Lgn1 locus. J Exp Med. 2001;194:1081–1096. doi: 10.1084/jem.194.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith KD, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 40.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 41.Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype typhimurium. J Biol Chem. 2007 doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 42.Franchi L, et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 43.Wu H, Tschopp J, Lin SC. Smac mimetics and TNFalpha: a dangerous liaison? Cell. 2007;131:655–658. doi: 10.1016/j.cell.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- 45.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 46.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 47.Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 49.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 50.Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- 51.Decker T, Lohmann-Matthes ML. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J Immunol Methods. 1988;115:61–69. doi: 10.1016/0022-1759(88)90310-9. [DOI] [PubMed] [Google Scholar]
- 52.Blasi E, et al. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature. 1985;318:667–670. doi: 10.1038/318667a0. [DOI] [PubMed] [Google Scholar]
- 53.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.