Abstract
Unravelling the timing of the metazoan radiation is crucial for elucidating the macroevolutionary processes associated with the Cambrian explosion. Because estimates of metazoan divergence times derived from molecular clocks range from quite shallow (Ediacaran) to very deep (Mesoproterozoic), it has been difficult to ascertain whether there is concordance or quite dramatic discordance between the genetic and geological fossil records. Here, we show using a range of molecular clock methods that the major pulse of metazoan divergence times was during the Ediacaran, which is consistent with a synoptic reading of the Ediacaran macrobiota. These estimates are robust to changes in priors, and are returned with or without the inclusion of a palaeontologically derived maximal calibration point. Therefore, the two historical records of life both suggest that although the cradle of Metazoa lies in the Cryogenian, and despite the explosion of ecology that occurs in the Cambrian, it is the emergence of bilaterian taxa in the Ediacaran that sets the tempo and mode of macroevolution for the remainder of geological time.
Keywords: molecular clock, Cambrian explosion, fossil calibration, Ediacaran
1. Introduction
Central to unravelling the causality and biological significance of the Cambrian explosion is accurately and precisely elucidating the origination times of the metazoan phyla. Despite the fact that the Cambrian explosion is geologically obvious (Darwin 1859), it has long been argued that this same geological record, owing to its incompleteness, might be misleading when considering metazoan origins (Runnegar 1982b). As Runnegar (1986) recognized, a second ‘fossil record’, the genetic record written in the DNA of all living organisms, could be used to test hypotheses about the completeness of the geological record (Peterson et al. 2007), and initial attempts at using a molecular clock strongly suggested that metazoans had a deep and cryptic Precambrian history (Runnegar 1982a, 1986; Wray et al. 1996; reviewed recently by Conway Morris 2006). Nonetheless, several palaeontologists have cogently argued that the fossil record provides positive evidence for the absence of Early Neoproterozoic and Mesoproterozoic animals, casting doubt on the veracity of these molecular clock estimates (Budd & Jensen 2000, 2003; Jensen et al. 2005; Conway Morris 2006; Butterfield 2007). Hence, comparisons between the genetic and geological fossil records of early animal evolution, as currently understood, suggest that either the geological record is woefully incomplete or that there is something seriously awry with our reading of the genetic record (Bromham 2006).
To explore the apparent incongruity between the known fossil record and the very deep estimates of metazoan diversification as revealed by molecular clocks, Peterson and colleagues (Peterson et al. 2004; Peterson & Butterfield 2005) assembled the largest novel dataset yet, showing that the two records were remarkably concordant: metazoans originated sometime during the Cryogenian, and bilaterians arose during the Ediacaran. Part of the reason for the prior discrepancy concerned the use of vertebrate divergence times. Peterson et al. (2004) discovered that there was an approximately twofold rate reduction across the vertebrate protein-coding genome as compared with the three invertebrate lineages examined (echinoderms, molluscs and insects), consistent with total genome comparisons between vertebrates and dipteran insects (Zdobnov et al. 2002). However, some studies using invertebrate calibrations have also inferred divergence times consistent with a cryptic Precambrian history of Metazoa (Pisani et al. 2004; Regier et al. 2005), suggesting that the twofold rate reduction across the vertebrate genome is only one of many factors influencing the estimation of divergence times (Linder et al. 2005; Peterson & Butterfield 2005).
In addition, Peterson et al.'s (2004) estimates and explanations were called into question by several workers, notably Blair & Hedges (2005) who argued that Peterson et al. (2004) used palaeontologically derived calibration points as maxima as opposed to minima, which generated spuriously shallow estimates for metazoan divergences. Although false, as Peterson et al. (2004) stated explicitly (see also Peterson & Butterfield 2005), this criticism highlights an important issue surrounding the use of molecular clocks, namely the proper way to incorporate calibration points into molecular clock analyses (Benton & Donoghue 2007). Recent experimental analyses have shown the importance of numerous, well-constrained calibration points for returning accurate and precise estimates of divergence times, and thus highlighting the need to pay particular attention to this aspect of molecular dating (Roger & Hug 2006; Hug & Roger 2007). Nonetheless, difficulties arise when incorporating fossils into a molecular clock analysis: unlike the establishment of a minimal divergence time for any two taxa, which is simply the first appearance of either one of the taxa, estimating the maximum divergence time is much more difficult (Benton & Donoghue 2007). Two types of maxima have been proposed: a ‘hard’ maximum proposes an absolute value for the oldest possible date of divergence; whereas a ‘soft’ maximum treats a divergence as having some chance of being older than a particular date, depending on a probability distribution used to describe the calibration point (Hedges & Kumar 2004; Yang & Rannala 2006; Benton & Donoghue 2007).
Most modern molecular clock methods (e.g. Sanderson 1997, 2002; Thorne et al. 1998; Drummond et al. 2006) allow constraining, as well as fixing, the age of a calibration point, so that every fossil divergence can be defined using a minimum and a maximum. This is a significant improvement over older molecular clock approaches (e.g. Kumar & Hedges 1998) because it allows the integration of palaeontological uncertainty in the estimation of divergence times. However, most existing molecular clock software including ‘r8s’ (Sanderson 2004) and ‘Multidivtime’ (Thorne & Kishino 2002), do not distinguish between hard and soft maxima, instead treating all maxima as hard. The difficulty here is that divergence times estimated with uncertain maxima treated as if they were hard can only give minimum estimates for the true divergence time, as the soft maxima might significantly underestimate the true age of the calibration points. Nonetheless, Drummond et al. (2006) have now implemented Bayesian relaxed molecular clock methods (in the software package Beast) where soft maxima can be properly modelled using a probability distribution, and can thus be older than their proposed fossil date.
Here, we set out to explore the diversification of animal phyla in the Neoproterozoic using alternative relaxed molecular clock approaches while testing the stability of our results to the choice of different priors and to the deletion of palaeontologically derived maxima, and modelling soft maxima using the most appropriate probability distribution. We find that, although deleting or relaxing maxima tends to push divergence times towards the past (as expected), all estimates are largely congruent between algorithms. We conclude that a synoptic reading of both the geological and genetic fossil records demonstrates that the Ediacaran was the time of major diversification of most higher-level animal taxa and set the stage for Phanerozoic-like macroecology and macroevolution.
2. Material and methods
(a) Molecular characters
All taxa are taken from Sperling et al. (2007) where a concatenated alignment of seven different housekeeping genes, for a total of 2059 amino acid positions and 44 representative species (see Peterson et al. 2004; Peterson & Butterfield 2005), was analysed using Bayesian methods (MrBayes v. 3.1.2; Ronquist & Huelsenbeck 2003; see Sperling et al. 2007 for details).
(b) Molecular clock calibration
Calibration points were taken from Peterson et al. (2004) except for the minimum estimate for crown-group Eleutherozoa, which was adjusted from 475 to 480 Myr ago in light of the discovery of a slightly older asterozoan (Blake & Guensberg 2005), and the minimum and the maximum for crown-group Diptera were taken from Benton & Donoghue (2007). Several new maxima and minima were incorporated into this analysis. First, the maximum for the origin of crown-group echinoderms was set at 520 Myr ago, the first appearance of stereom in the fossil record. Because stereom is a highly distinctive skeletal material, and its presence in numerous stem-group taxa (Smith 2005) demonstrates that stereom is a total-group echinoderm character, it must have evolved before the origin of the crown group. Indeed, if stereom had evolved much earlier (i.e. before the Tommotian, ˜525 Myr ago), then one would expect that stereom would be aragonitic as opposed to calcitic, given that mineral choice seems dictated by the sea water chemistry at the time the skeleton was first acquired (Porter 2007). Second, this same time point also sets the minimum for Ambulacraria (Echinodermata+Hemichordata), as echinoderms appear before hemichordates in the rock record (Budd & Jensen 2003). Third, because ambulacrarians are characterized by the possession of four to six coeloms in each animal (Peterson et al. 2000; Smith et al. 2004), and because coeloms cannot predate the first appearance of bilaterian traces (Budd & Jensen 2000, 2003), the first appearance of traces sets the maximum age for crown-group Ambulacraria, ca 555 Myr ago (Martin et al. 2000; Jensen et al. 2005). Fourth, the maximum for the origin of Gastropoda+Bivalvia, is the first appearance of skeletons in the fossil record, ca 542 Myr ago (Bengtson 1994; Amthor et al. 2003). Fifth, the maximum for the origin of crown-group demosponges, is the first appearance of demosponge-specific biomarkers (McCaffrey et al. 1994; Love et al. 2006; see Peterson et al. 2007 for discussion), sometime after the Sturtian, ca 657 Myr ago (Kendall et al. 2006). Finally, the maximum for the origin of crown-group Eumetazoa, which was only used in the Beast analyses, is argued to be 635 Myr ago based on palaeoecological observations (Peterson & Butterfield 2005).
Newly incorporated minima include the first appearance of arthropod traces 525 Myr ago (Budd & Jensen 2003) as a minimum for the divergence between insects and the priapulid, the first appearance of medusozoans 500 Myr ago (Hagadorn et al. 2002) as a minimum for the origin of the crown-group Cnidaria, and the first appearance of vertebrates 520 Myr ago, as the minimum for the origin of crown-group chordates (Benton & Donoghue 2007).
(c) Molecular estimates of divergence times
Molecular estimates of divergence times were obtained using the Bayesian methods of Thorne et al. (1998) as implemented in Multidivtime (Thorne & Kishino 2002), and Drummond et al. (2006) as implemented in Beast v. 1.4.2 (Drummond & Rambaut 2007). All divergence times were calculated assuming the tree topology of figure 1, which was derived from MrBayes (see above and Sperling et al. 2007). For the Multidivtime analyses, branch lengths were estimated using the Estbranches program from the Multidivtime package, under the WAG model. For Beast analyses, starting branch lengths were assigned arbitrarily to match the constraints imposed by the calibrations.
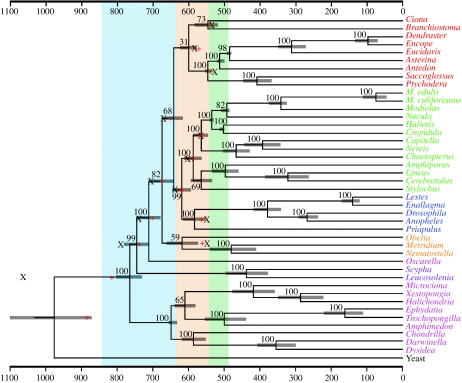
Figure 1.
The timing of the metazoan radiation according to the molecular clock. The phylogenetic tree for 41 metazoan taxa rooted on the yeast Saccharomyces cerevisiae as determined by Bayesian phylogenetic analysis (see text) is shown. The deuterostomes are shown in red, spiralian protostomes in green, ecdysozoan protostomes in blue, cnidarians in orange, the homoscleromorph Oscarella in salmon pink, calcisponges in purple and demosponges in magenta. The nodes of the tree are positioned according to the optimum as determined from the Bayesian autocorrelated method of Thorne et al. (1998), as implemented in the software package ‘Multidivtime’ (Thorne & Kishino 2002) using a root prior of 1000 Myr ago (s.d.=500 Myr ago). The 95% HPD credibility intervals are shown in brackets. The red crosses are the estimates for clades with internal calibration points as determined by Bayesian algorithm Beast (Drummond et al. 2006) using uniform priors and an exponential rate distribution; black Xs are the estimates using exponential priors and the same rate distribution. Note that much of the metazoan diversification occurs during the Ediacaran (brown), which lies between the Cryogenian (ice blue) and the Cambrian (green).
For the Multidivtime analyses, a prior age for the root node (in our case the Fungi–Metazoa split) must be specified. We assumed a 1000 Myr ago prior for this node (Knoll 1992; Douzery et al. 2004), and then tested whether this choice affected our results by performing analyses in which this age was changed to 100 Myr ago (s.d.=500 Myr ago), 1500 Myr ago (s.d.=500 Myr ago) and 2000 Myr ago (s.d.=750 Myr ago). Other priors used in Multidivtime analyses include the mean and standard deviation of the prior distribution at the root node, and ‘Minab’ (parameter for beta prior on proportional node depth). The mean and standard deviation of the prior distribution of the rate at the root node were set to 0.039, as estimated from the data following the procedure outlined in the Multidivtime manual, and the effect that 100-fold changes to this parameter had on the results were assessed. The Minab parameter affects the distribution of the nodes through time—Minab values greater than 1 will cause the nodes to repel each other, while values smaller than 1 will cause the nodes to attract each other. This parameter was set to 1 for our analyses, but we assessed how changing the Minab parameter from 0.6 to 1.4 affected our results.
Beast implements uncorrelated relaxed clock methods, which assumes an overall distribution of rates across branches, but does not assume that the rates on adjacent branches are autocorrelated. We used both the exponential and lognormal rate distributions with two different calibration schemes: one with hard maxima in which most calibrations were treated as uniform priors on clade ages, and a second with only soft maxima, in which all calibrations were treated as exponential priors, with 95% of their density lying between the uniform maximum and minimum. In both schemes, the maximum at 635 Myr ago was treated as an exponential prior, with 90% of its density lying below 635 Myr ago, giving a 10% prior chance that this calibration point is incorrect. All other priors and operators were kept at default settings, except that all operators that alter the tree topology were disabled.
Ninety-five per cent highest posterior density (HPD) credibility intervals are automatically calculated by Multidivtime, and were calculated using the program Tracer for the Beast analyses. To test whether our priors dominated the posterior distribution, all our Beast and Multidivtime analyses were also performed without data and the results obtained in these runs were compared with those obtained when the data were actually analysed.
3. Results
Molecular divergence times were estimated using the topology shown in figure 1. Support for Cnidaria and Deuterostomia was low (67 and 33%, respectively), probably owing to long-branch artefacts (Pisani 2004) associated with Ciona and Obelia in particular (indeed the value for Deuterostomia increases to more than 90% with the removal of Ciona), but given the clear monophyly of the phyla Chordata and Cnidaria constraining these nodes should not generate spurious molecular divergence estimates. Most of the other nodes were strongly supported, including Calcispongia+Eumetazoa and Eumetazoa, supporting the results of Peterson & Butterfield (2005), and contra the conclusions of Rokas and colleagues (Rokas et al. 2005; see also Baurain et al. 2006). Indeed, within Protostomia, for example, all but one node (Stylochus+Nemertea) have posterior probability values above 80%, and both Lophotrochozoa and Ecdysozoa, as well as Annelida+Mollusca, have clade credibility values of 100%. In addition, we find strong support for the node Homoscleromorpha+Eumetazoa, which indicates that there are at least three independent extant sponge lineages (Sperling et al. 2007).
Using this topology as a constraint tree, divergence times were estimated using the Bayesian autocorrelated method of Thorne et al. (1998), as implemented in the software package Multidivtime (Thorne & Kishino 2002). These Bayesian estimates are robust to changes in the age of the root prior as the estimates are essentially the same whether the age is 100 Myr ago (s.d.=500 Myr ago) or 2000 Myr ago (s.d.=750 Myr ago; table 1), suggesting that the age of the root prior is not biasing the analyses. Also, changing the value of Minab, or the mean rate of evolution of the root node, did not change our results (not shown). Running the analyses without data confirmed that our results were not dominated by our choice of priors (not shown). The suggestion that fungi diverged from animals ca 1000 Myr ago (Knoll 1992; Douzery et al. 2004) was confirmed by all our analyses that did not assume a particular age for the root node. Thus, we used the values derived from the 1000 Myr ago (s.d.=500 Myr ago) prior on figure 1.
Table 1.
Optima (maxima and minima) in millions of years derived from Multidivtime (M) and Beast (B) for five key metazoan divergences.
| method | Metazoa | Eumetazoa | Bilateria | Protostomia | Deuterostomia |
|---|---|---|---|---|---|
| M-1000a | 766 (803,731) | 676 (709,645) | 643 (671,617) | 619 (648,594) | 601 (625,579) |
| M-100b | 760 (798,725) | 672 (706,642) | 641 (669,615) | 618 (645,592) | 600 (624,578) |
| M-2000c | 774 (812,739) | 679 (712,648) | 645 (674,619) | 622 (649,595) | 602 (626,580) |
| M-1000-Dd | 904 (997,825) | 743 (798,694) | 686 (727,649) | 653 (689,619) | 624 (655,596) |
| B-UCEX uniforme | 815 (1621,625) | 676 (849, 579) | 652 (764,570) | 620 (692,556) | 572 (614,537) |
| B-UCEX expf | 1067 (2358,612) | 707 (985, 581) | 669 (870,566) | 638 (784,556) | 582 (695,529) |
| B-UCLN uniformg | 891 (995,640) | 739 (822,607) | 699 (768,588) | 660 (715,572) | 640 (706,559) |
| B-UCLN exph | 953 (1093,821) | 779 (869, 694) | 733 (808,663) | 688 (751,629) | 677 (746,607) |
Age of the root prior is 1000 Myr ago (s.d.=500 Myr ago);
Age of the root prior is 100 Myr ago (s.d.=500 Myr ago);
Age of the root prior is 2000 Myr ago (s.d.=750 Myr ago);
Age of the root prior is 1000 Myr ago (s.d.=500 Myr ago) and estimates derived without considering the demosponge maximum of 657 Myr ago;
Estimates derived using an exponential rate distribution and uniform priors.
Estimates derived using an exponential rate distribution and exponential priors;
Estimates derived using a lognormal rate distribution and uniform priors;
Estimates derived using a lognormal rate distribution and exponential priors.
The removal of the deeper calibration point, namely the maximum age of 657 Myr ago for the origin of crown-group demosponges, resulted in increasing the estimate for the age of crown-group Metazoa by approximately 18% (from 766 to 904 Myr ago; table 1). Nonetheless, the age for both crown-group Protostomia and crown-group Deuterostomia increased by only approximately 4–5%, suggesting that the results derived with the use of this maximum are generally robust. Given its position in the tree, the geological depth of the divergence, and the unique nature of the evidence (biomarkers), this maximum is most likely adding both accuracy and precision to the clock estimates.
We next explored these same divergence times using the models implemented in Beast (Drummond et al. 2006). In general, the estimates derived from Beast using an exponential rate distribution and uniform priors (red crosses in figure 1) are similar to those derived from Multidivtime (table 1). The analyses that use exponential priors are somewhat deeper than those that use uniform priors (black Xs in figure 1), and those using a lognormal rate distribution are deeper than those derived from an exponential rate distribution (table 1), presumably because the exponential distribution on rates is leading to greater autocorrelation between rates. Analyses without data again confirmed that the priors were not dominating the data (results not shown).
4. Discussion
(a) Concordance between the genetic and geological fossil records
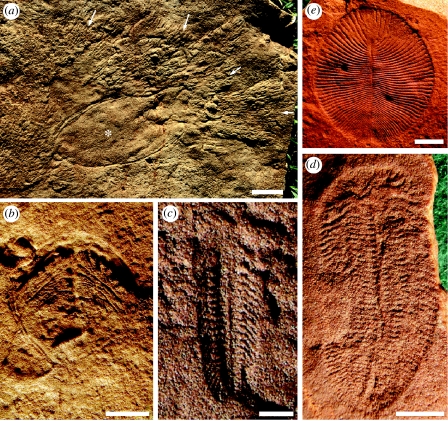
Here we have shown, using a variety of analyses and appropriately testing for biases that may have been introduced by the use of palaeontologically derived maxima, that the genetic fossil record strongly supports the notion that the diversification of metazoans in general, and bilaterian metazoans in particular, occurred during the Ediacaran Period, 635–542 Myr ago (Knoll et al. 2004, 2006). How do these molecular estimates compare with the known geological record? Macroscopic fossils of the Ediacara biota span the upper half of the Ediacaran Period, from 575 to 542 Myr ago (Grotzinger et al. 1995; Martin et al. 2000; Bowring et al. 2003; Condon et al. 2005). Since most of these fossils occur as soft-bodied impressions in relatively coarse-grained siliciclastic sedimentary rocks, a comprehensive array of palaeobiological interpretations of the Ediacara biota has been put forth. Nonetheless, a few taxa stand out as potential candidates for affinities within Metazoa. One taxon in particular, Kimberella, has generated much discussion as a possible triploblastic metazoan. Not only does it compare well in external form to molluscs (Fedonkin & Waggoner 1997), in a few cases an everted proboscis is preserved (Gehling et al. 2005) that is inferred to contain a radula-like organ given the association between specimens of Kimberella (figure 2a, asterisk) and aligned sets of paired scratch marks (figure 2a, arrows; Gehling et al. 2005). These findings suggest that Kimberella was preserved in place while grazing on substrate microbial mats (Seilacher 1999; Gehling et al. 2005). Given that we estimated the divergence between annelids and molluscs to be ca 570 Myr ago (figure 1), it is possible, if not probable, that Kimberella is allied with modern molluscs.
Figure 2.
Putative Ediacaran metazoans. (a) Natural cast on bed base of Kimberella resting trace (asterisk) and Radulichnus radular feeding trace fans (arrows); scale bar, 1 cm. (b)Parvancorina minchami; scale bar, 1 cm. (c) Spriggina floundersi; scale bar, 10 mm. (d) Marywadea ovata; scale bar, 10 mm. (e) Dickinsonia costata; scale bar, 2 cm.
What about other higher-level clades? Our estimates suggest that arthropods diverged from priapulids ca 575 Myr ago, suggesting that stem-group panarthropods (Nielsen 2001) should be present in Upper Ediacaran rocks. Interestingly, several taxa compare favourably with a panarthropod interpretation. For example, large specimens of Parvancorina show lateral structures originating on either side of the medial ridge that might be characterized as appendages (figure 2b). In fact, in external form, Parvancorina bears a striking resemblance to the unmineralized kite-shaped Cambrian arthropod Skania (Lin et al. 2006). Spriggina (figure 2c) also preserves large numbers of appendage-like structures, and still others like Marywadea (figure 2d) show apparent cephalic branching structures that resemble digestive caecae in arthropods. Importantly (see below), all of these taxa were no larger than 10 cm in maximum dimension (Gehling 1999; Fedonkin 2003; see figure 2), and appear simultaneously with the first demonstrable trace fossils (Droser et al. 2005; Jensen et al. 2005). The absence of arthropod scratch marks (Seilacher 1999), though, is not too worrisome given that such traces would demand the presence of sclerotized appendages to cut through the ubiquitously present microbial mats, a character not necessitated by the presence of stem-group panarthropods, or even deeply nested stem-group arthropods, in Ediacaran-aged sediments.
Indeed, the distinct possibility remains that this fauna preserves numerous stem-group forms ranging from basal triploblasts up through basal ecdysozoans, spiralians and possibly even deuterostomes. Given the enigmatic nature of some very prominent taxa like Dickinsonia (figure 2e), a taxon that appears capable of some form of limited motility (Gehling et al. 2005), a position for Dickinsonia within total-group Eumetazoa is not out of the question. In fact, mobile but saprophytic feeding without the use of a gut would be compelling evidence that some form of ectomesoderm predates the advent of endoderm.
(b) Discordance between the genetic and geological fossil records
Of course, many others have addressed these questions using a similar approach, and it is worth comparing our results against not only the fossil record but also with other molecular clock estimates as well. It compares well with some molecular analyses, notably Peterson et al. (2004) and Peterson & Butterfield (2005), all of whom argued that the last common ancestor of protostomes and deuterostomes evolved not more than 635 Myr ago. But recently, Blair & Hedges (2005), argued for much deeper divergences, based on a series of penalized likelihood (Sanderson 2002) analyses using r8s (Sanderson 2004) in which every calibration point was treated as a minimum. They suggested that the divergence between ambulacrarian and chordate deuterostomes was 896 Myr ago (with the 95% CI spanning from 832 to 1022 Myr ago). They further argued that the divergence between hemichordates and echinoderms was 876 Myr ago (725 and 1074 Myr ago), and the origin of crown-group echinoderms was 730 Myr ago. Finally, they estimated that the divergence between starfish and sea urchins was 580 Myr ago. Unfortunately, their results are most likely spurious because as Sanderson (2004) pointed out, r8s cannot converge on a unique solution if only minima are used to calibrate penalized likelihood analyses, which is supported by the fact that their estimate for the origin of a mineralized, coelomate taxon like crown-group Echinodermata precedes their appearance in the fossil record by some 200 Myr ago.
Of course, neither the genetic nor the geological fossil record has a monopoly on historical accuracy, and as much as molecular evolutionists need to keep in mind the relevant palaeontological data, palaeontologists need to keep in mind estimates derived from molecular clocks (Donoghue & Benton 2007). For example, Budd & Jensen (2000, 2003) argued that bilaterians could not have had an extensive Precambrian history, as suggested by almost all molecular clocks, as the trace fossil record, and the inferred morphology of these animals, is not consistent with an origin much before 555 Myr ago. They observed that possession of coelom(s) and a blood vascular system (BVS) is inconsistent with a meiofaunal origin, as tiny organisms would have had no need for a transport system like the BVS, and are only consistent with a size large enough to be detected in the geological record. In general, we agree with their arguments, and use their insights to set a maximum age for crown-group Ambulacraria (see above).
However, the same argument cannot be extended to many other parts of the bilaterian tree. Contra Budd & Jensen (2000), there is no evidence for homology of coeloms either between protostomes and deuterostomes or even within both protostomes and deuterostomes. Since the coelom is, by definition, just a mesodermally lined cavity (Ruppert 1991; Nielsen 2001), the possession of the space itself cannot be used as an argument of similarity. Instead, topological similarity must be used, and when it is, it strongly suggests homology, for example, within Ambulacraria (Peterson et al. 2000; Smith et al. 2004), but not homology between any other higher taxa (Ruppert 1991; Nielsen 2001). Thus, outside of Ambulacraria, the trace fossil record cannot be used to set a maximum for most bilaterian divergences. In fact, the small size of many putative Ediacaran bilaterians (figure 2), and the fact that acoel flatworms are now recognized as the sister group to the remaining bilaterians (Baguñà & Riutort 2004; Peterson et al. 2005; Sempere et al. 2007), is consistent with an argument that small size and absence of a coelom are primitive for Bilateria. This then removes the final obstacle to a pre-555 Myr ago origin for Bilateria, which is consistent with both the appearance of many different bilaterian lineages in the Ediacaran (figure 2) and the molecular clock (figure 1).
Despite the presence of many different stem-group taxa, the Ediacaran is still a transitional ecology, with these organisms confined to a two-dimensional mat world. This stands in dramatic contrast to the Early Cambrian where the multi-tiered food webs that so typify the Phanerozoic were established with the eumetazoan invasion of both the pelagos and the infaunal benthos (Butterfield 1997, 2001; Vannier & Chen 2000, 2005; Dzik 2005; Peterson et al. 2005; Vannier et al. 2007). Hence, although the Ediacaran is an apparent quantum leap in ecological complexity as compared with the ‘boring billions’ that characterize Earth before the Ediacaran, it is still relatively simple when compared with the Cambrian, yet another quantum leap in organismal and ecological evolution. Thus, the Ediacaran stands as the transition interval between the ‘Precambrian’ and the Phanerozoic (Butterfield 2007). Whether the Ediacaran transition was triggered by the introduction of eumetazoans, as argued by Peterson & Butterfield (2005), or by the introduction of mobile, macrophagous triploblasts, as is suggested by our analyses reported here (figure 1), or some other factor or combination of factors, remains to be more fully studied through continued exploration of the relevant rock sections throughout the world, and continued improvements in molecular clock methods.
5. Conclusions
Both the genetic and geological fossil records, each with their own inherent biases and artefacts, are largely congruent with one another, and for historical disciplines congruence of independent datasets is the strongest argument one can make for historical accuracy (Pisani et al. 2007). Thus, our analyses suggest that while the cradle of metazoan life is in the Cryogenian, and the explosion of metazoan ecology occurred in the Cambrian, it is the emergence of bilaterians in the Ediacaran that established the ecological and evolutionary rules that largely govern Earth's macrobiota for the remainder of geological time.
Acknowledgments
K.J.P. was supported by the National Science Foundation; J.A.C. was supported by an RCUK Academic Fellowship and J.G.G. was supported by the Australian Research Council Discovery Project (DG0453393), the A.R.C. Linkage Project LP0774959 including the South Australian Museum and Beach Petroleum Pty Ltd, and the SA Museum Waterhouse Club. We would like to thank P. Donoghue (U. Bristol) for his usual perspicacity, two anonymous reviewers for their helpful comments on an earlier version of this paper, and T. Littlewood (NHM) and M. Telford (UCL) for inviting us to contribute to this symposium volume. Finally, K.J.P. would like to thank all of the students who have come through the lab and contributed data to this project, and the South Australian Museum for a very enlightening visit.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Evolution of the animals: a Linnean tercentenary celebration’.
References
- Amthor J.E, Grotzinger J.P, Schröder S, Bowring S.A, Ramezani J, Martin M.W, Matter A. Extinction of Cloudina and Namacalathus at the Precambrian–Cambrian boundary in Oman. Geology. 2003;31:431–434. doi:10.1130/0091-7613(2003)031<0431:EOCANA>2.0.CO;2 [Google Scholar]
- Baguñà J, Riutort M. The dawn of bilaterian animals: the case of acoelomorph flatworms. Bioessays. 2004;26:1046–1057. doi: 10.1002/bies.20113. doi:10.1002/bies.20113 [DOI] [PubMed] [Google Scholar]
- Baurain D, Brinkman H, Philippe H. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors. Mol. Biol. Evol. 2006;24:6–9. doi: 10.1093/molbev/msl137. doi:10.1093/molbev/msl137 [DOI] [PubMed] [Google Scholar]
- Bengtson, S. 1994 The advent of animal skeletons. In Early life on Earth. Nobel Symposium no. 84 (ed. S. Bengtson), pp. 412–425. New York, NY: Columbia University Press.
- Benton M.J, Donoghue P.C.J. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. doi:10.1093/molbev/msl150 [DOI] [PubMed] [Google Scholar]
- Blair J.E, Hedges S.B. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. doi:10.1093/molbev/msi225 [DOI] [PubMed] [Google Scholar]
- Blake D.B, Guensberg T.E. Implications of a new Early Ordovician asteroid (Echinodermata) for the phylogeny of asterozoans. J. Paleontol. 2005;79:395–399. doi:10.1666/0022-3360(2005)079<0395:IOANEO>2.0.CO;2 [Google Scholar]
- Bowring S, Myrow P, Landing E, Ramezani J, Grotzinger J. Geochronological constraints on terminal Neoproterozoic events and the rise of metazoans. Geophys. Res. Abs. 2003;5:13219. [Google Scholar]
- Bromham L. Molecular dates for the Cambrian explosion: is the light at the end of the tunnel an oncoming train? Palaeontol. Electron. 2006;9:2E. [Google Scholar]
- Budd G.E, Jensen S. A critical reappraisal of the fossil record of the bilaterian phyla. Biol. Rev. Camb. Philos. Soc. 2000;75:253–295. doi: 10.1017/s000632310000548x. doi:10.1017/S000632310000548X [DOI] [PubMed] [Google Scholar]
- Budd G.E, Jensen S. The limitations of the fossil record and the dating of the origin of the Bilateria. In: Donoghue P.C.J, Smith M.P, editors. Telling evolutionary time: molecular clocks and the fossil record. Taylor & Francis; London, UK: 2003. pp. 166–189. [Google Scholar]
- Butterfield N.J. Plankton ecology and the Proterozoic–Phanerozoic transition. Paleobiology. 1997;23:247–262. [Google Scholar]
- Butterfield N.J. Ecology and evolution of Cambrian plankton. In: Zhuravlev A.Y, Riding R, editors. The ecology of the Cambrian radiation. Columbia University Press; New York, NY: 2001. pp. 200–216. [Google Scholar]
- Butterfield N.J. Macroevolution and macroecology through deep time. Palaeontology. 2007;50:41–55. doi:10.1111/j.1475-4983.2006.00613.x [Google Scholar]
- Condon D, Zhu M, Bowring S, Wang W, Yang A, Jin Y. U–Pb ages from the Neoproterozoic Doushantuo Formation, China. Science. 2005;308:95–98. doi: 10.1126/science.1107765. doi:10.1126/science.1107765 [DOI] [PubMed] [Google Scholar]
- Conway Morris S. Darwin's Dilemma: the realities of the Cambrian ‘explosion’. Phil. Trans. R. Soc. B. 2006;361:1069–1083. doi: 10.1098/rstb.2006.1846. doi:10.1098/rstb.2006.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. John Murray; London, UK: 1859. On the origin of species. [Google Scholar]
- Donoghue P.C.J, Benton M.J. Rocks and clocks: calibrating the tree of life using fossils and molecules. Trends Ecol. Evol. 2007;22:424–431. doi: 10.1016/j.tree.2007.05.005. doi:10.1016/j.tree.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Douzery E.J.P, Snell E.A, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils. Proc. Natl Acad. Sci. USA. 2004;101:15 386–15 391. doi: 10.1073/pnas.0403984101. doi:10.1073/pnas.0403984101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droser M.L, Gehling J.G, Jensen S.R. Ediacaran trace fossils: true and false. In: Briggs D.E.G, editor. Evolving form and function: fossils and development. Peabody Museum of Natural History, Yale University; New Haven, CT: 2005. pp. 125–138. [Google Scholar]
- Drummond A.J, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. doi:10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J, Ho S.Y.W, Phillips M.J, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzik J. Behavioral and anatomical unity of the earliest burrowing animals and the cause of the “Cambrian explosion”. Paleobiology. 2005;31:503–521. doi:10.1666/0094-8373(2005)031[0503:BAAUOT]2.0.CO;2 [Google Scholar]
- Fedonkin M.A. The origin of the Metazoa in light of the Proterozoic fossil record. Paleontol. Res. 2003;7:9–41. doi:10.2517/prpsj.7.9 [Google Scholar]
- Fedonkin M.A, Waggoner B.M. The Late Precambrian fossil Kimberella is a mollusc-like bilaterian organism. Nature. 1997;388:868–871. doi:10.1038/42242 [Google Scholar]
- Gehling J.G. Microbial mats in terminal Proterozoic siliciclastics: Ediacaran death masks. Palaios. 1999;14:40–57. doi:10.2307/3515360 [Google Scholar]
- Gehling J.G, Droser M.L, Jensen S.R, Runnegar B.N. Ediacara organisms: relating for to function. In: Briggs D.E.G, editor. Evolving form and function: fossils and development: Proceedings of a symposium honoring Adolf Seilacher for his contributions to paleontology, in celebration of his 80th birthday. Peabody Museum of Natural History, Yale University; New Haven, CT: 2005. pp. 43–66. [Google Scholar]
- Grotzinger J.P, Bowring S.A, Saylor B.Z, Kaufman A.J. Biostratigraphic and geochronologic constraints on early animal evolution. Science. 1995;270:598–604. doi:10.1126/science.270.5236.598 [Google Scholar]
- Hagadorn J.W, Dott R.H.J, Damrow D. Stranded on a Late Cambrian shoreline: medusae from central Wisconsin. Geology. 2002;30:147–150. doi:10.1130/0091-7613(2002)030<0147:SOALCS>2.0.CO;2 [Google Scholar]
- Hedges S.B, Kumar S. Precision of molecular time estimates. Trends Genet. 2004;20:242–247. doi: 10.1016/j.tig.2004.03.004. doi:10.1016/j.tig.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Hug L.A, Roger A.J. The impact of fossils and taxon sampling on ancient molecular dating analyses. Mol. Biol. Evol. 2007;24:1889–1897. doi: 10.1093/molbev/msm115. doi:10.1093/molbev/msm115 [DOI] [PubMed] [Google Scholar]
- Jensen S, Droser M.L, Gehling J.G. Trace fossil preservation and the early evolution of animals. Palaeogeogr. Palaeoclim. Palaeoecol. 2005;220:19–29. doi:10.1016/j.palaeo.2003.09.035 [Google Scholar]
- Kendall B, Creaser R.A, Selby D. Re–Os geochronology of postglacial black shales in Australia: constraints on the timing of “Sturtian” glaciation. Geology. 2006;34:729–732. doi:10.1130/G22775.1 [Google Scholar]
- Knoll A.H. The early evolution of eukaryotes: a geological perspective. Science. 1992;256:622–627. doi: 10.1126/science.1585174. doi:10.1126/science.1585174 [DOI] [PubMed] [Google Scholar]
- Knoll A.H, Walter M.R, Narbonne G.M, Christie-Blick N. A new period for the geologic time scale. Science. 2004;305:621–622. doi: 10.1126/science.1098803. doi:10.1126/science.1098803 [DOI] [PubMed] [Google Scholar]
- Knoll A.H, Walter M.R, Narbonne G.M, Christie-Blick N. The Ediacaran period: a new edition to the geologic time scale. Lethaia. 2006;39:13–30. doi:10.1080/00241160500409223 [Google Scholar]
- Kumar S, Hedges S.B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. doi:10.1038/31927 [DOI] [PubMed] [Google Scholar]
- Lin J.-P, et al. A Parvancorina-like arthropod from the Cambrian of South China. Hist. Biol. 2006;18:33–45. [Google Scholar]
- Linder H, Hardy C.R, Rutschmann F. Taxon sampling effects in molecular clock dating: an example from the African Restionaceae. Mol. Phylogenet. Evol. 2005;35:569–582. doi: 10.1016/j.ympev.2004.12.006. doi:10.1016/j.ympev.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Love G.D, Fike D.A, Grosjean E, Stalvies C, Grotzinger J, Bradley A.S, Bowring S, Condon D, Summons R.E. Constraining the timing of basal metazoan radiation using molecular biomarkers and U–Pb isotope dating. Geochim. Cosmochim. Acta. 2006;70:A371. doi:10.1016/j.gca.2006.06.748 [Google Scholar]
- Martin M.W, Grazhdankin D.V, Bowring S.A, Evans D.A.D, Fedonkin M.A, Kirschvink J.L. Age of Neoproterozoic bilaterian body and trace fossils, White Sea, Russia: implications for metazoan evolution. Science. 2000;288:841–845. doi: 10.1126/science.288.5467.841. doi:10.1126/science.288.5467.841 [DOI] [PubMed] [Google Scholar]
- McCaffrey M.A, Moldowan J.M, Lipton P.A, Summons R.E, Peters K.E, Jeganathan A, Watt D.S. Paleoenvironmental implications of novel C30 steranes in Precambrian to Cenozoic age petroleum and bitumen. Geochim. Cosmochim. Acta. 1994;58:529–532. doi:10.1016/0016-7037(94)90481-2 [Google Scholar]
- Nielsen C. Oxford University Press; Oxford, UK: 2001. Animal evolution: interrelationships of the living phyla. [Google Scholar]
- Peterson K.J, Butterfield N.J. Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc. Natl Acad. Sci. USA. 2005;102:9547–9552. doi: 10.1073/pnas.0503660102. doi:10.1073/pnas.0503660102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K.J, Arenas-Mena C, Davidson E.H. The A/P axis in echinoderm ontogeny and evolution: evidence from fossils and molecules. Evol. Dev. 2000;2:93–101. doi: 10.1046/j.1525-142x.2000.00042.x. doi:10.1046/j.1525-142x.2000.00042.x [DOI] [PubMed] [Google Scholar]
- Peterson K.J, Lyons J.B, Nowak K.S, Takacs C.M, Wargo M.J, McPeek M.A. Estimating metazoan divergence times with a molecular clock. Proc. Natl Acad. Sci. USA. 2004;101:6536–6541. doi: 10.1073/pnas.0401670101. doi:10.1073/pnas.0401670101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K.J, McPeek M.A, Evan D.A.D. Tempo and mode of early animal evolution: inferences from rocks, Hox, and molecular clocks. Paleobiology. 2005;31:36–55. doi:10.1666/0094-8373(2005)031[0036:TAMOEA]2.0.CO;2 [Google Scholar]
- Peterson K.J, Summons R.E, Donoghue P.C.J. Molecular paleobiology. Palaeontology. 2007;50:775–809. doi:10.1111/j.1475-4983.2007.00692.x [Google Scholar]
- Pisani D. Identifying and removing fast-evolving sites using compatibility analysis: an example from the Arthropoda. Syst. Biol. 2004;53:978–989. doi: 10.1080/10635150490888877. doi:10.1080/10635150490888877 [DOI] [PubMed] [Google Scholar]
- Pisani D, Poling L.L, Lyons-Weiler M, Hedges S.B. The colonization of land by animals: molecular phylogeny and divergence times among arthropods. BMC Evol. Biol. 2004;2:1–10. doi: 10.1186/1741-7007-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani D, Benton M.J, Wilkinson M. The congruence of molecular and morphological phylogenies. Acta Biotheor. 2007;55:269–281. doi: 10.1007/s10441-007-9015-8. doi:10.1007/s10441-007-9015-8 [DOI] [PubMed] [Google Scholar]
- Porter S.M. Seawater chemistry and early carbonate biomineralization. Science. 2007;316:1302. doi: 10.1126/science.1137284. doi:10.1126/science.1137284 [DOI] [PubMed] [Google Scholar]
- Regier J.C, Shultz J.W, Kambic R.E. Pancrustacean phylogeny: hexapods are terrestrial crustaceans and maxillopods are not monophyletic. Proc. R. Soc. B. 2005;272:395–401. doi: 10.1098/rspb.2004.2917. doi:10.1098/rspb.2004.2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger A.J, Hug L.A. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Phil. Trans. R. Soc. B. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. doi:10.1098/rstb.2006.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Krüger D, Carroll S.B. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. doi:10.1126/science.1116759 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Runnegar B. A molecular-clock date for the origin of the animal phyla. Lethaia. 1982a;15:199–205. doi:10.1111/j.1502-3931.1982.tb00645.x [Google Scholar]
- Runnegar B. The Cambrian explosion: animals or fossils? J. Geol. Soc. Austr. 1982b;29:395–411. [Google Scholar]
- Runnegar B. Molecular palaeontology. Palaeontology. 1986;29:1–24. [Google Scholar]
- Ruppert E.E. Introduction to the aschelminth phyla: a consideration of mesoderm, body cavities, and cuticle. In: Harrison F.W, Ruppert E.E, editors. Microscopic anatomy of invertebrates. Aschelminthes. vol. 4. Wiley-Liss, Inc; New York, NY: 1991. pp. 1–17. [Google Scholar]
- Sanderson M.J. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 1997;14:1218–1231. [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson, M. J. 2004 r8s, version 1.70. Davis, CA: Section of Evolution and Ecology, University of California, Davis.
- Seilacher A. Biomat-related lifestyles in the Precambrian. Palaios. 1999;14:86–93. doi:10.2307/3515363 [Google Scholar]
- Sempere L.F, Martinez P, Cole C, Baguñà J, Peterson K.J. Phylogenetic distribution of microRNAs supports the basal position of acoel flatworms and the polyphyly of Platyhelminthes. Evol. Dev. 2007;9:409–415. doi: 10.1111/j.1525-142X.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- Smith A.B. The pre-radial history of echinoderms. Geol. J. 2005;40:255–280. doi:10.1002/gj.1018 [Google Scholar]
- Smith A.B, Peterson K.J, Wray G, Littlewood D.T.J. From bilateral symmetry to pentaradiality: the phylogeny of hemichordates and echinoderms. In: Cracraft J, Donoghue M.J, editors. Assembling the tree of life. Oxford University Press; Oxford, UK: 2004. pp. 365–383. [Google Scholar]
- Sperling E.A, Pisani D, Peterson K.J. Poriferan paraphyly and its implications for Precambrian paleobiology. Geol. Soc. Lond. Spec. Publ. 2007;286:355–367. [Google Scholar]
- Thorne J.L, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. doi:10.1080/10635150290102456 [DOI] [PubMed] [Google Scholar]
- Thorne J.L, Kishino H, Painter I.S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Vannier J, Chen J.-Y. The Early Cambrian colonization of pelagic niches exemplified by Isoxys (Arthropoda) Lethaia. 2000;33:295–311. doi:10.1080/002411600750053862 [Google Scholar]
- Vannier J, Chen J. Early Cambrian food chain: new evidence from fossil aggregates in the Maotianshan Shale biota, SW China. Palaios. 2005;20:3–26. doi:10.2110/palo.2003.p03-40 [Google Scholar]
- Vannier J, Steiner M, Renvoisé E, Hu S.-X, Casanova J.-P. Early Cambrian origin of modern food webs: evidence from predator arrow worms. Proc. R. Soc. B. 2007;274:627–633. doi: 10.1098/rspb.2006.3761. doi:10.1098/rspb.2006.3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G.A, Levinton J.S, Shapiro L.H. Molecular evidence for deep Precambrian divergences among metazoan phyla. Science. 1996;274:568–573. doi:10.1126/science.274.5287.568 [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. doi:10.1093/molbev/msj024 [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. doi:10.1126/science.1077061 [DOI] [PubMed] [Google Scholar]