Abstract
Comparisons between completely sequenced metazoan genomes have generally emphasized how similar their encoded protein content is, even when the comparison is between phyla. Given the manifest differences between phyla and, in particular, intuitive notions that some animals are more complex than others, this creates something of a paradox. Simplistic explanations have included arguments such as increased numbers of genes; greater numbers of protein products produced through alternative splicing; increased numbers of regulatory non-coding RNAs and increased complexity of the cis-regulatory code. An obvious value of complete genome sequences lies in their ability to provide us with inventories of such components. I examine progress being made in linking genome content to the pattern of animal evolution, and argue that the gap between genomic and phenotypic complexity can only be understood through the totality of interacting components.
Keywords: comparative genomics, evolution, Metazoa, transcription factors, ultraconserved regions
Deus ex machina: A power, event, person, or thing that comes in the nick of time to solve a difficulty; providential interposition…Oxford English Dictionary
1. Introduction
Complete genome sequences provide limits to our imaginations. Even just a few years before the human genome was available in rough draft form, it was widely believed to encode at least 50 000 genes (Fields et al. 1994; Nature Genetics Editorial 2000). In contrast, the initial publications estimated 25–40 000 protein-coding genes (Lander et al. 2001; Venter et al. 2001), and since then estimates have generally carried a downward momentum, most recently approaching 20 000 (Goodstadt & Ponting 2006; Pennisi 2007). Although this number is higher than 16 000 or so found in invertebrate chordates (Dehal et al. 2002), it is roughly the same total as the nematode worm Caenorhabditis elegans (Hillier et al. 2005). Whether or not these low numbers of protein-coding genes for vertebrates stand the test of time, the sense of unease surrounding the lack of correlation between organismal complexity (often measured in numbers of distinct cell types) and protein-coding gene count is evident from the framing of the ‘G-value paradox’ by Hahn & Wray (2002), and the various explanations that have been put forward to ease it, including, for example, miRNAs (Sempere et al. 2006), non-protein-coding DNA (Taft et al. 2007) and alternative splicing (Kim et al. 2007).
Similar gene counts are, of course, a crude measure of biological complexity. There is no reason why two genomes should not encode very different sets of protein-coding genes, but still have similar overall totals. Within the field of animal evolution and the evolution of development (evo–devo), however, the G-value paradox has a particular resonance. Studies in different animal phyla have repeatedly shown the reuse of a core set of developmental genes, the so-called ‘toolkit’ (Carroll et al. 2005), with the HOX genes in particular taking on an iconic significance. Broadly, toolkit genes come from a handful of transcription factor families, defined by the presence of particular structural domains such as the helix-turn-helix (HTH), including the homeobox genes; zinc fingers (ZnFs); leucine zippers and the helix-loop-helix (HLH). As well as transcription factors, there are seven well-conserved pathways responsible for intercellular signalling (Pires-daSilva & Sommer 2003), many of which appear to be present in sponges, the earliest branching clade of animals (Nichols et al. 2006). An extreme interpretation of these data is provided by Davidson (2006): ‘if we focus explicitly on the genes encoding transcription factors, and […] signalling systems required for developmental spatial regulation, there is almost no qualitative variation among the genomes of bilaterians’.
Given all this, where in the genome do the phenotypic differences between animal taxa arise? The undoubted conservation of the protein-coding developmental genes has, particularly in the evo–devo field with its morphological concerns, focused attention on cis-enhancer elements affecting transcription (Carroll et al. 2005; Davidson 2006; Simpson 2007; Wray 2007), although there are alternative views emphasizing the importance of different kinds of regulatory elements (Alonso & Wilkins 2005) and different protein classes, such as structural genes (Hoekstra & Coyne 2007). As well as the presence of particular genes, the role of gene loss, especially with regard to secondarily simplified organisms such as tunicates and nematodes, is also likely to be of major significance. Below I outline some major themes being developed by large-scale genome comparisons, principally of nematodes, insects and vertebrates. My aim is not to present an exhaustive account, but to highlight areas where functionally relevant species-specific differences may arise, within apparently conserved systems. Although I concentrate on the evolution of the systems regulating animal development, this is not to lose sight of the things being regulated: the proteins involved in making nematode cuticles, or asynchronous flight muscles in insects, or the human brain and adaptive immune system, to name but a few, are what make it necessary to evolve those systems.
2. Gene duplication
Usefully summarizing the differences and similarities between more than 10 000 protein-coding genes from several species at once is not necessarily straightforward. Although pairwise similarities between sequences are easy to compute, they suffer from the imposition of arbitrary cut-offs and are less easy to interpret than measures that explicitly reflect phylogeny. Genes in different species are most obviously compared by grouping into sets of orthologues (i.e. genes related by speciation events) and paralogues (genes related by intra-genome duplication events). Closely related species share large numbers of orthologues: 93% of dog (Canis familiaris) and 82% of the marsupial Monodelphis domesticus gene predictions have orthologues in human (Goodstadt et al. 2007). The Linnean hierarchy, however, is not necessarily a good guide of genomic relatedness by this definition of similarity. Within the nematodes, 65% of C. elegans genes share an orthologue with Caenorhabditis briggsae, despite their being from the same genus (Stein et al. 2003). For more distantly related genomes, orthologue counts can drop rapidly. This may be as much a sign of difficulties in reliably assigning gene orthology on a large scale, as a real indication of the extents of the conserved cores.
Paralogues often arise via tandem duplication of genes, giving rise to localized clusters of functionally related genes. As these are the regions where gene content is evolving most rapidly between closely related species, the functions of these genes are of special interest for understanding animal-specific differences. For the most part, for any two closely related vertebrate genomes, the functional classes of genes duplicated in this way are similar—olfaction and chemosensation, reproduction and effectors of the immune response—although the duplications have occurred independently in each lineage (Emes et al. 2003). These large groups of paralogues often show evidence of adaptive evolution in their amino acid sequences, suggesting that new functions have been selected for (Emes et al. 2004a,b).
The recurrent nature of duplications within particular functional classes, coupled with the observed diversifying selection suggests that they are a standard adaptive genomic response to environmental challenges. Does similar rapid duplication occur in the kinds of genes, such as transcription factors, that might be implicated in development? A growing number of examples are known. Perhaps most dramatically, in mice a set of 32 tandemly duplicated homeoboxes have arisen from apparently one or two genes in the common ancestor of humans and rodents; they are believed to play a role in germ cell development and embryonic stem cell differentiation (Maclean et al. 2005; Jackson et al. 2006).
ZnF containing transcription factors have undergone independent rounds of gene duplication in insects and tetrapods. In insects a set of ZnFs is found to co-occur with a ZnF-associated domain (ZAD; Chung et al. 2007); this ZAD class is found in approximately 100 and 150 copies in Drosophila melanogaster and the mosquito Anopheles gambiae, respectively; there is only a single copy in vertebrates (Chung et al. 2007). In D. melanogaster, many are expressed in the female germ line, suggesting a role in oocyte development or embryogenesis (Chung et al. 2007). An analogous story is found with Krüppel-associated box (KRAB) containing Zn fingers in tetrapods. Successive independent tandem duplication events have occurred in different mammalian lineages, leading to over 400 copies in the human genome (Huntley et al. 2006). The KRAB domain itself appears to have been co-opted from a progenitor sequence conserved throughout eukaryotes (Birtle & Ponting 2006), however, it has evolved so much as to make this similarity difficult to detect; clearly identifiable KRAB domains are specific to tetrapods. Their functions are largely unknown, and have not been tied to any general aspects of tetrapod-specific biology. As such, why the family as a whole has expanded is a puzzle.
Nematodes too exhibit lineage-specific expansions of particular transcription factor families, most notably, the nuclear hormone receptors (NHRs). The C. elegans genome encodes 284, far more than the 48 in human and 21 in D. melanogaster. The bulk of these (greater than 200) have arisen from an apparently nematode-specific expansion of a unique gene (Lander et al. 2001; Robinson-Rechavi et al. 2005). Once more, the reasons for such a dramatic lineage-specific expansion of a particular transcription factor family, and any links to taxon-specific biology, are obscure, although it has been speculated that C. elegans relies less on combinatorial reuse of different transcription factors (Antebi 2006). A less dramatic lineage-specific expansion occurs in the case of the T-box-containing transcription factors: there are 21 in C. elegans, with 17 arising from a lineage-specific expansion when compared with D. melanogaster and humans. Ascertaining when and in which taxa these duplications found in C. elegans took place is currently frustrated by a lack of nematode genome sequences—currently, only those of C. elegans and C. briggsae have been published. These T-box genes as a set map to several genomic locations, suggesting that they have arisen over a more protracted time scale than the examples discussed above; some, at least, have known roles in the development of C. elegans (Poole & Hobert 2006).
3. The invention of new genes
A number of gene families appear to be metazoan novelties, with no clear sequence similarity to other genes outside the Metazoa, but present in the more basal animal phyla, such as cnidarians and sponges. These include key families involved in animal development, such as T-box and SMAD transcription factors, and signalling molecules such as WNTs and fibroblast growth factors (FGFs; Putnam et al. 2007). Was the invention of such families a prerequisite for the evolution of the Metazoa, and were analogous protein inventions required for the evolution of particular taxa, such as insects and vertebrates? Analysis of three-dimensional structures (i.e. the protein fold itself) suggests a more subtle transition than large-scale evolution of new protein folds. In many cases, examination of protein three-dimensional structural similarities shows that these genes have distant homologues in non-metazoan genomes. The MH1 (DNA binding) domain of SMADs, for instance, is probably homologous to a family of homing endonucleases found in all kingdoms of life (Grishin 2001); the T-box shares structural similarities indicative of homology with a variety of other transcription factors, such as STAT DNA-binding domains, which are found in other eukaryotes (Murzin et al. 1995; Soler-Lopez et al. 2004); and the signalling domain of metazoan hedgehog proteins shares detailed similarities with members of a family of bacterial peptidases, suggesting that they too are likely to be homologous (Murzin et al. 1995). In these cases, the novel families are likely to be cases of rapid sequence evolution, accompanying functional shifts, within stem lineages leading to the Metazoa. Sparse sequence sampling of non-fungal and metazoan eukaryotic genomes may contribute to the apparent co-origin of these protein domains with the animals.
As this type of domain evolution is occurring from pre-existing domain types, the process fits within a standard framework of accelerated point mutation and selection for new functions. The invention of the domain type is not a key innovation in itself; rather, it can be seen as the extension of functional diversification of subfamilies of the kind that is apparent when comparing more closely related species. The fact that so many new domain types are found to be coincident within the origin of metazoans suggests that the selective pressures giving rise to this kind of accelerated sequence evolution were greater in the metazoan stem lineage.
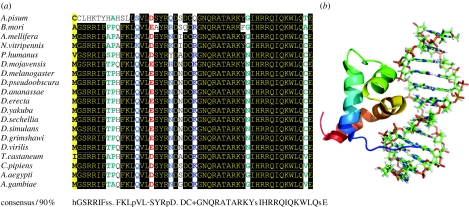
An example of a more recent domain innovation is found in the Drosophila gene brinker, which plays a key role in the establishment of dorsoventral patterning. Although the protein-coding sequence of its DNA-binding domain is well conserved in insects, using current sequence databases it shows no significant sequence similarity to proteins from any other taxa (figure 1), although there is weak (non-significant) similarity to pogo-like transposases, and the structure, which is only folded when complexed with DNA, suggests similarity to various transcription factors (Cordier et al. 2006).
Figure 1.
The DNA-binding domain of brinker is conserved within insects, but has no significantly similar sequences in other taxa. (a) The alignment shows the conserved core from selection of insect species. Drosophila species sequences were taken from the UCSC web browser (http://genome.ucsc.edu), Anopheles and Aedes from ENSEMBL (http://www.ensembl.org), other predictions were made from sequences at the NCBI. GI Accessions: N.vit 146253130, T.cas 73486274, C.pip 145464888, P.hum 145365328, A.mel 63051942, B.mor 91842977 and A.pis 47522326. (b) The three-dimensional structure of the aligned region when binding DNA. The structure was taken from the PDB file 2glo.
4. Evolution of transcription factors: the animal in the orthologue
Lineage-specific duplication followed by sequence divergence provides one route to species-specific biology, but what scope is there for lineage-specific functional shifts within orthologous genes? In the absence of gene duplication, it is hard to imagine how the DNA specificity of a particular factor might be significantly changed in such a way that it targets new genes, without deleterious consequences. The modular structure of proteins, however, suggests that other routes of functional evolution are available. A protein may have pleiotropic effects, but that is not the same as saying that every amino acid in the protein will be directly involved in all those effects. A recent illustrative example from the hox gene Ultrabithorax, is of an insect-specific ‘QA’ protein motif, found outside the homeodomain. The region is involved in limb repression; the effects of deleting the motif are strong in some tissues but close to undetectable in others (Hittinger et al. 2005). Clearly, changes in the protein-coding sequences of transcription factors, apart from their more obvious DNA-binding residues, must be integrated into our understanding of the evolution of developmental regulation.
The majority of residues in metazoan transcription factors do not fall within regions of well-defined globular structure, with many belonging to so-called ‘intrinsically disordered’ regions—regions that may form a structure when complexed with other macromolecules (Liu et al. 2006; Minezaki et al. 2006). The specific sequences of these regions are typically not obviously conserved between paralogues; because they are unique to particular families they are not covered in domain databases such as SMART and Pfam (Finn et al. 2006; Letunic et al. 2006). The lack of extreme conservation between distant species has sometimes masked the fact that within closely related species, these regions are conserved. Comparisons of orthologous sequences from closely related genomes (e.g. vertebrates or drosophilids) often show that substantial proportions of these non-domain sequences are undergoing strong purifying selection—they accumulate many more synonymous nucleotide changes than non-synonymous changes—and are thus functional. For the large part, precisely what these biological functions are is unknown; two possibilities, however, suggest themselves. Firstly, they may have relatively uninteresting non-specific effects, such as facilitating folding of the major domain (e.g. by reducing aggregation) or acting as spacers between globular domains. Secondly, and more interestingly from the point of view of animal evolution, they may include short linear peptide motifs that mediate protein–protein interactions (Dyson & Wright 2005; Neduva & Russell 2005; Neduva et al. 2005).
There are numerous examples of regulatory motifs found outside of transcription factor domains. Many hox proteins include a YPWM-like hexapeptide motif that interacts with other homeodomain-containing proteins (In der Rieden et al. 2004); Drosophila ftz orthologues have lost this motif but acquired an LXXLL motif coupled to a new role in segmentation (Lohr & Pick 2005); and an N-terminal SSYF-like motif believed to be involved in transcriptional activation is conserved across Hox orthologues and paralogues from different phyla (Tour et al. 2005). Interaction motifs can be coupled with signalling pathways to create cell-type specificity. They can, for instance, be regulated by phosphorylation, such that the phosphorylation status governs what interactions can be made (e.g. Sapkota et al. 2007), or alternative splicing can result in protein–protein interaction motifs being included or excluded from particular cell types, providing additional layers of regulatory complexity that are likely to be species specific (Neduva & Russell 2005).
The challenge of identifying small regulatory motifs means that their species distributions, and how their presence might produce taxon-specific differences in protein functions, have not been well studied. Examples that tie cleanly to one taxonomic group are less common, but an interesting case has been proposed in bilaterian orthologues of the Brachyury gene. These possess an N-terminal motif that is not found in non-bilaterian Metazoa (Marcellini 2006), which instead have a well-defined EH1-like motif (Copley 2005). The bilaterian motif is believed to be responsible for an interaction with Smad1, and hence to link gastrulation to bilateral pattern formation (Marcellini 2006).
5. Enhancers: transcription factor-binding sites and ultraconserved regions
Theoretical considerations have led to an intense focus on transcription factor-binding sites (TFBSs) as a major molecular source of morphological novelty (Wray et al. 2003; Carroll et al. 2005; Davidson 2006; Wray 2007), although see (Hoekstra & Coyne 2007) for a critique. Individual TFBSs show rapid turnover in comparisons of closely related genomes, with many being lineage specific (Dermitzakis & Clark 2002; Moses et al. 2006). This dynamic nature may not be revealed in the phenotype—patterns of gene expression may be conserved even though regulatory sequences change at the molecular level (Ludwig et al. 2000; Romano & Wray 2003; Fisher et al. 2006). On the other hand, the gain and the loss of individual TFBSs have been implicated in several recent cases of morphological evolution, in both vertebrates and invertebrates (reviewed in Simpson (2007) and Wray (2007)). The relationship between individual TFBSs and enhancer function is clearly not straightforward, beyond the fact that clustering of individual binding sites can identify some enhancer regions (Markstein et al. 2002). Cases of functional linkages between particular transcription factors have been proposed, for example, between dorsal, twist, Su(H) and an unidentified motif in neurogenic ectoderm formation in Diptera (Markstein et al. 2004), and even a coupling originating prior to the origin of Bilateria, of hairy and E(spl) promoting neural cell fate (Rebeiz et al. 2005).
Comparisons of vertebrate genomes have revealed large regions (more than 100 nucleotides) of extreme conservation of non-coding sequences (conserved non-coding elements (CNEs); Bejerano et al. 2004). These regions are often found near transcription factors and other developmental genes (Sandelin et al. 2004). Outside of the vertebrates, there is evidence for similar regions occurring near developmental genes in flies (Glazov et al. 2005) and nematodes (Vavouri et al. 2007). Although in many cases the conserved regions are even found near orthologous genes, there is no evidence that they are homologous; they appear to have evolved independently in each of the phyla (Vavouri et al. 2007). Experimental evidence from vertebrates shows that many instances have roles as tissue-specific enhancer elements (Woolfe et al. 2005; Pennacchio et al. 2006).
The length and lack of inter-phylum conservation of CNEs is in contrast to individual TFBSs. The DNA specificity of orthologous transcription factors is usually well conserved over large phylogenetic distances, but typical TFBSs are short, of the order of 6–10 nucleotides. An obvious possibility is that longer CNEs are composed of overlapping or adjacent TFBSs. This would suggest a tight packing of transcription factor proteins on the genomic DNA of these CNEs. There is direct evidence for this: some fragments of highly conserved non-coding sequences are present in crystal structures of transcription factor complexes. An atomic model based on known crystal structures of the interferon-β enhancer, for example, shows 50 consecutive nucleotides in contact with eight different proteins; these nucleotides are well conserved in mammalian species (Panne et al. 2007; see figure 2 for another example). Given that such structures exist, it is not such a leap to imagine 16 proteins binding to 100 nucleotides, or even bigger complexes. This suggests a model where CNE enhancer regions controlling orthologous genes in different phyla are controlled by multiple TFBSs, although not necessarily the same transcription factors or in the same orientation. Moreover, the tight packing of transcription factors on the genomic DNA suggests that the proteins themselves may be co-adapted to interact with each other and aid the cooperative formation of enhancer complexes. Previously, Ruvinsky & Ruvkun (2003) have presented experimental evidence that enhancers and transcription factors co-evolve in this way, with neuronal and muscle-specific enhancer elements from D. melanogaster failing to drive expression in homologous tissue types in C. elegans, and Dover and co-workers have argued for coevolution of bicoid protein and hunchback regulatory regions (McGregor et al. 2001; Shaw et al. 2002).
Figure 2.
Adjacent TFBSs cause extended regions of DNA sequence conservation. Structure of CEBPβ homodimer and Runx-1 (Tahirov et al. 2001). Three transcription factors (2xCEBPB and RUNX1) bind in a region of 25 nucleotides conserved throughout placental mammals. The DNA-binding domains represented as three-dimensional structures are boxed and colour coded in the schematic of the proteins. In each case, the majority of the protein is not represented in the structure; these regions could interact with other transcription factors, activators and repressors. The human sequence coordinates are chromosome 5, bases 149 446 373–149 446 396 of the NCBI build 36. The alignment is taken from the UCSC web browser http://genome.ucsc.edu.
If protein–protein interactions between transcription factors are often required for the formation of enhancer complexes, close analysis of transcription factor sequence and structure may reveal evidence for co-adaptation of proteins, such as the Hox hexapeptide motif, through which homeotic proteins form complexes with TALE class homeodomains (LaRonde-LeBlanc & Wolberger 2003). We might expect instances of co-adapted transcription factor combinations to be taxon specific, to match the taxon specificity of enhancer sequences.
6. Alternative splicing
Not all CNEs are associated with enhancer regions. There is good evidence that many are involved in regulating alternative splicing events, including the alternative splicing of mRNAs of proteins which themselves regulate alternative splicing (Lareau et al. 2007; Ni et al. 2007). The presence of highly conserved control elements to regulate alternative splicing indicates that the functional consequences are of importance. Although large very conserved elements may be the exception rather than the rule, detailed comparative analyses have identified smaller conserved motifs regulating alternative splicing, for instance in nematodes (Kabat et al. 2006) and vertebrates (Sorek & Ast 2003; Yeo et al. 2005).
Alternative splicing is often touted as a mechanism by which proteomic complexity is increased. Although early reports suggested that levels of alternative splicing were comparable in vertebrates and invertebrates (Brett et al. 2002), more recent studies suggest that there is indeed more alternative splicing of transcripts in vertebrates (Kim et al. 2007), suggesting a link with increased phenotypic complexity. How relevant is alternative splicing for species-specific biology and morphological differences? Quantitatively, the gene products that appear to be most affected by alternative splicing are typically involved in nervous and immune system function (Modrek et al. 2001). There are, however, ample examples of alternatively spliced transcription factors—as many as 63% of mouse transcription factors have variant exons (Taneri et al. 2004). Although the differences in molecular roles of the alternatively spliced products are often unknown, the genes themselves include developmental classics such as members of Hox, SMAD and T-box families (Fan et al. 2004; Dunn et al. 2005; Noro et al. 2006), although they do not necessarily present obvious morphological correlates (Yoder & Carroll 2006). Alternative splicing of modular proteins is an obvious route through which functions can be changed, by including or excluding particular combinations of domains. In this regard, it is interesting that alternative splicing often affects intrinsically disordered regions outside known protein domains (Romero et al. 2006)—this again points to a critical role for finely tuned protein–protein interactions among transcriptional regulators.
There are few known cases of distant conservation of alternative splice variants of transcription factors; typically, examples are conserved within phyla at best. Widening the search to other classes of gene again suggests that splice variants are not conserved over long periods, although it should be remembered that transcript coverage of most species from which evidence of alternative splicing is obtained is very restricted. Perhaps the best counter-example is currently that of fibroblast growth factor receptor 2 (FGFR2), where an exon configuration diagnostic of mutually exclusive alternative splicing is found in both vertebrates and the sea urchin Strongylocentrotus purpuratus (Mistry et al. 2003). Examples of orthologous ion channel encoding genes showing similar alternative splicing patterns in D. melanogaster, C. elegans and humans are likely to be cases of parallel evolution (Copley 2004). The shared ability of vertebrates and at least insects and C. elegans to produce alternative transcripts in a regulated manner, but the absence of large numbers of conserved alternative splicing between protostomes and deuterostomes suggests that gene products have become alternatively spliced in parallel between different lineages, while at the same time hinting that the functions performed by alternative splice variants may, over time, be replaced by different genomic solutions.
7. Summary
Although most major classes of protein involved in animal development may be conserved throughout the Metazoa, detailed comparative analysis of these gene types reveals a more dynamic picture, with frequent gene duplication, gene loss, couplings with new motifs and other processes such as alternative splicing and regulation by micro-RNAs, all of which are likely to be important for a full understanding of function. Cis-regulatory variation may well be revealed to be quantitatively the most common form of variation between species, but it seems probable that the cumulative effects of multiple cis-regulatory changes will have required that protein networks evolve to accommodate and correctly regulate changed enhancer structures.
Our knowledge of animal evolution and the picture presented here is currently based on a very small sampling of almost exclusively nematode, insect and vertebrate genomes. Although this situation is beginning to change, the fact that many important functional regions, especially those that do not encode proteins, are only revealed by having sets of closely related genome sequences, and that there are 35 or so animal phyla gives some idea of the enormity of the challenges ahead. The rapidly falling costs of genome sequencing do, however, give grounds for optimism.
Acknowledgments
I thank the organizers and participants of the Novartis Foundation symposium on Animal Evolution, 20 June 2007, for helpful discussions, two anonymous referees for perceptive comments, and the Wellcome Trust for support.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Evolution of the animals: a Linnean tercentenary celebration’.
References
- Alonso C.R, Wilkins A.S. The molecular elements that underlie developmental evolution. Nat. Rev. Genet. 2005;6:709–715. doi: 10.1038/nrg1676. doi:10.1038/nrg1676 [DOI] [PubMed] [Google Scholar]
- Antebi, A. 2006 Nuclear hormone receptors in C. elegans In WormBook (ed. The C. elegans Research Community). See www.wormbook.org/chapters/www_nuclearhormonerecep/nuclearhormonerecep.htm. [DOI] [PMC free article] [PubMed]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent W.J, Mattick J.S, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. doi:10.1126/science.1098119 [DOI] [PubMed] [Google Scholar]
- Birtle Z, Ponting C.P. Meisetz and the birth of the KRAB motif. Bioinformatics. 2006;22:2841–2845. doi: 10.1093/bioinformatics/btl498. doi:10.1093/bioinformatics/btl498 [DOI] [PubMed] [Google Scholar]
- Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. doi:10.1038/ng803 [DOI] [PubMed] [Google Scholar]
- Carroll S.B, Grenier J.K, Weatherbee S.D. Blackwell Publishing; Oxford, UK: 2005. From DNA to diversity: molecular genetics and the evolution of animal design. [Google Scholar]
- Chung H.R, Lohr U, Jackle H. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol. Biol. Evol. 2007;24:1934–1943. doi: 10.1093/molbev/msm121. doi:10.1093/molbev/msm121 [DOI] [PubMed] [Google Scholar]
- Copley R.R. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 2004;20:171–176. doi: 10.1016/j.tig.2004.02.001. doi:10.1016/j.tig.2004.02.001 [DOI] [PubMed] [Google Scholar]
- Copley R.R. The EH1 motif in metazoan transcription factors. BMC Genom. 2005;6:169. doi: 10.1186/1471-2164-6-169. doi:10.1186/1471-2164-6-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier F, Hartmann B, Rogowski M, Affolter M, Grzesiek S. DNA recognition by the brinker repressor—an extreme case of coupling between binding and folding. J. Mol. Biol. 2006;361:659–672. doi: 10.1016/j.jmb.2006.06.045. doi:10.1016/j.jmb.2006.06.045 [DOI] [PubMed] [Google Scholar]
- Davidson E.H. Academic Press; London, UK: 2006. The regulatory genome. [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. doi:10.1126/science.1080049 [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T, Clark A.G. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Dunn N.R, Koonce C.H, Anderson D.C, Islam A, Bikoff E.K, Robertson E.J. Mice exclusively expressing the short isoform of Smad2 develop normally and are viable and fertile. Genes Dev. 2005;19:152–163. doi: 10.1101/gad.1243205. doi:10.1101/gad.1243205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H.J, Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. doi:10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- Emes R.D, Goodstadt L, Winter E.E, Ponting C.P. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum. Mol. Genet. 2003;12:701–709. doi: 10.1093/hmg/ddg078. doi:10.1093/hmg/ddg078 [DOI] [PubMed] [Google Scholar]
- Emes R.D, Beatson S.A, Ponting C.P, Goodstadt L. Evolution and comparative genomics of odorant- and pheromone-associated genes in rodents. Genome Res. 2004a;14:591–602. doi: 10.1101/gr.1940604. doi:10.1101/gr.1940604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes R.D, Riley M.C, Laukaitis C.M, Goodstadt L, Karn R.C, Ponting C.P. Comparative evolutionary genomics of androgen-binding protein genes. Genome Res. 2004b;14:1516–1529. doi: 10.1101/gr.2540304. doi:10.1101/gr.2540304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004;64:5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. doi:10.1158/0008-5472.CAN-04-0615 [DOI] [PubMed] [Google Scholar]
- Fields C, Adams M.D, White O, Venter J.C. How many genes in the human genome? Nat. Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. doi:10.1038/ng0794-345 [DOI] [PubMed] [Google Scholar]
- Finn R.D, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. doi:10.1093/nar/gkj149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice E.A, Vinton R.M, Bessling S.L, McCallion A.S. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. doi:10.1126/science.1124070 [DOI] [PubMed] [Google Scholar]
- Glazov E.A, Pheasant M, McGraw E.A, Bejerano G, Mattick J.S. Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. doi:10.1101/gr.3545105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstadt L, Ponting C.P. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. doi:10.1371/journal.pcbi.0020133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstadt L, Heger A, Webber C, Ponting C.P. An analysis of the gene complement of a marsupial, Monodelphis domestica: evolution of lineage-specific genes and giant chromosomes. Genome Res. 2007;17:969–981. doi: 10.1101/gr.6093907. doi:10.1101/gr.6093907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin N.V. Mh1 domain of SMAD is a degraded homing endonuclease. J. Mol. Biol. 2001;307:31–37. doi: 10.1006/jmbi.2000.4486. doi:10.1006/jmbi.2000.4486 [DOI] [PubMed] [Google Scholar]
- Hahn M.W, Wray G.A. The G-value paradox. Evol. Dev. 2002;4:73–75. doi: 10.1046/j.1525-142x.2002.01069.x. doi:10.1046/j.1525-142X.2002.01069.x [DOI] [PubMed] [Google Scholar]
- Hillier L.W, Coulson A, Murray J.I, Bao Z, Sulston J.E, Waterston R.H. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 2005;15:1651–1660. doi: 10.1101/gr.3729105. doi:10.1101/gr.3729105 [DOI] [PubMed] [Google Scholar]
- Hittinger C.T, Stern D.L, Carroll S.B. Pleiotropic functions of a conserved insect-specific Hox peptide motif. Development. 2005;132:5261–5270. doi: 10.1242/dev.02146. doi:10.1242/dev.02146 [DOI] [PubMed] [Google Scholar]
- Hoekstra H.E, Coyne J.A. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. doi:10.1111/j.1558-5646.2007.00105.x [DOI] [PubMed] [Google Scholar]
- Huntley S, Baggott D.M, Hamilton A.T, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. doi:10.1101/gr.4842106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- In der Rieden P.M.J, Mainguy G, Woltering J.M, Durston A.J. Homeodomain to hexapeptide or PBC-interaction-domain distance: size apparently matters. Trends Genet. 2004;20:76–79. doi: 10.1016/j.tig.2003.12.001. doi:10.1016/j.tig.2003.12.001 [DOI] [PubMed] [Google Scholar]
- Jackson M, et al. A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genom. 2006;7:212. doi: 10.1186/1471-2164-7-212. doi:10.1186/1471-2164-7-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat J.L, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler A.M. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput. Biol. 2006;2:e86. doi: 10.1371/journal.pcbi.0020086. doi:10.1371/journal.pcbi.0020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. doi:10.1093/nar/gkl924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lareau L.F, Inada M, Green R.E, Wengrod J.C, Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. doi:10.1038/nature05676 [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N.A, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. doi:10.1101/gad.1103303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley R.R, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. doi:10.1093/nar/gkj079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Perumal N.B, Oldfield C.J, Su E.W, Uversky V.N, Dunker A.K. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. doi:10.1021/bi0602718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr U, Pick L. Cofactor-interaction motifs and the cooption of a homeotic Hox protein into the segmentation pathway of Drosophila melanogaster. Curr. Biol. 2005;15:643–649. doi: 10.1016/j.cub.2005.02.048. doi:10.1016/j.cub.2005.02.048 [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z, Bergman C, Patel N.H, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. doi:10.1038/35000615 [DOI] [PubMed] [Google Scholar]
- Maclean J.A, II, Chen M.A, Wayne C.M, Bruce S.R, Rao M, Meistrich M.L, Macleod C, Wilkinson M.F. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. doi:10.1016/j.cell.2004.12.022 [DOI] [PubMed] [Google Scholar]
- Marcellini S. When Brachyury meets Smad1: the evolution of bilateral symmetry during gastrulation. Bioessays. 2006;28:413–420. doi: 10.1002/bies.20387. doi:10.1002/bies.20387 [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine M.S. Genome-wide analysis of clustered dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. doi:10.1073/pnas.012591199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee K.P, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. doi:10.1242/dev.01124 [DOI] [PubMed] [Google Scholar]
- McGregor A.P, Shaw P.J, Hancock J.M, Bopp D, Hediger M, Wratten N.S, Dover G.A. Rapid restructuring of bicoid-dependent hunchback promoters within and between dipteran species: implications for molecular coevolution. Evol. Dev. 2001;3:397–407. doi: 10.1046/j.1525-142x.2001.01043.x. doi:10.1046/j.1525-142X.2001.01043.x [DOI] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Kinjo A.R, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. doi:10.1016/j.jmb.2006.04.016 [DOI] [PubMed] [Google Scholar]
- Mistry N, Harrington W, Lasda E, Wagner E.J, Garcia-Blanco M.A. Of urchins and men: evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA. 2003;9:209–217. doi: 10.1261/rna.2470903. doi:10.1261/rna.2470903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. doi:10.1093/nar/29.13.2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M, Pollard D.A, Nix D.A, Iyer V.N, Li X.Y, Biggin M.D, Eisen M.B. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. doi:10.1371/journal.pcbi.0020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G, Brenner S.E, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. doi:10.1006/jmbi.1995.0159 [DOI] [PubMed] [Google Scholar]
- Nature Genetics Editorial. The nature of the number. Nat. Genet. 2000;25:127–128. doi: 10.1038/75946. doi:10.1038/75946 [DOI] [PubMed] [Google Scholar]
- Neduva V, Russell R.B. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. doi:10.1016/j.febslet.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson T.J, Lewis J, Serrano L, Russell R.B. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. doi:10.1371/journal.pbio.0030405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Z, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. doi:10.1101/gad.1525507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols S.A, Dirks W, Pearse J.S, King N. Early evolution of animal cell signaling and adhesion genes. Proc. Natl Acad. Sci. USA. 2006;103:12 451–12 456. doi: 10.1073/pnas.0604065103. doi:10.1073/pnas.0604065103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay D.J, Zhang W, Mann R.S. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 2006;20:1636–1650. doi: 10.1101/gad.1412606. doi:10.1101/gad.1412606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison S.C. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. doi:10.1016/j.cell.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. doi:10.1038/nature05295 [DOI] [PubMed] [Google Scholar]
- Pennisi E. Genetics. Working the (gene count) numbers: finally, a firm answer? Science. 2007;316:1113. doi: 10.1126/science.316.5828.1113a. doi:10.1126/science.316.5828.1113a [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer R.J. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 2003;4:39–49. doi: 10.1038/nrg977. doi:10.1038/nrg977 [DOI] [PubMed] [Google Scholar]
- Poole R.J, Hobert O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr. Biol. 2006;16:2279–2292. doi: 10.1016/j.cub.2006.09.041. doi:10.1016/j.cub.2006.09.041 [DOI] [PubMed] [Google Scholar]
- Putnam N.H, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. doi:10.1126/science.1139158 [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Stone T, Posakony J.W. An ancient transcriptional regulatory linkage. Dev. Biol. 2005;281:299–308. doi: 10.1016/j.ydbio.2005.03.004. doi:10.1016/j.ydbio.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Maina C.V, Gissendanner C.R, Laudet V, Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J. Mol. Evol. 2005;60:577–586. doi: 10.1007/s00239-004-0175-8. doi:10.1007/s00239-004-0175-8 [DOI] [PubMed] [Google Scholar]
- Romano L.A, Wray G.A. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–4199. doi: 10.1242/dev.00611. doi:10.1242/dev.00611 [DOI] [PubMed] [Google Scholar]
- Romero P.R, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. doi:10.1073/pnas.0507916103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Ruvkun G. Functional tests of enhancer conservation between distantly related species. Development. 2003;130:5133–5142. doi: 10.1242/dev.00711. doi:10.1242/dev.00711 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Bailey P, Bruce S, Engstrom P.G, Klos J.M, Wasserman W.W, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genom. 2004;5:99. doi: 10.1186/1471-2164-5-99. doi:10.1186/1471-2164-5-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli F.M, Brivanlou A.H, Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. doi:10.1016/j.molcel.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Sempere L.F, Cole C.N, McPeek M.A, Peterson K.J. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. B: Mol. Dev. Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. doi:10.1002/jez.b.21118 [DOI] [PubMed] [Google Scholar]
- Shaw P.J, Wratten N.S, McGregor A.P, Dover G.A. Coevolution in bicoid-dependent promoters and the inception of regulatory incompatibilities among species of higher Diptera. Evol. Dev. 2002;4:265–277. doi: 10.1046/j.1525-142x.2002.02016.x. doi:10.1046/j.1525-142X.2002.02016.x [DOI] [PubMed] [Google Scholar]
- Simpson P. The stars and stripes of animal bodies: evolution of regulatory elements mediating pigment and bristle patterns in Drosophila. Trends Genet. 2007;23:350–358. doi: 10.1016/j.tig.2007.04.006. doi:10.1016/j.tig.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Soler-Lopez M, Petosa C, Fukuzawa M, Ravelli R, Williams J.G, Muller C.W. Structure of an activated Dictyostelium STAT in its DNA-unbound form. Mol. Cell. 2004;13:791–804. doi: 10.1016/s1097-2765(04)00130-3. doi:10.1016/S1097-2765(04)00130-3 [DOI] [PubMed] [Google Scholar]
- Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. doi:10.1101/gr.1208803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.D, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. doi:10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft R.J, Pheasant M, Mattick J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. doi:10.1002/bies.20544 [DOI] [PubMed] [Google Scholar]
- Tahirov T.H, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. doi:10.1016/S0092-8674(01)00271-9 [DOI] [PubMed] [Google Scholar]
- Taneri B, Snyder B, Novoradovsky A, Gaasterland T. Alternative splicing of mouse transcription factors affects their DNA-binding domain architecture and is tissue specific. Genome Biol. 2004;5:R75. doi: 10.1186/gb-2004-5-10-r75. doi:10.1186/gb-2004-5-10-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tour E, Hittinger C.T, McGinnis W. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development. 2005;132:5271–5281. doi: 10.1242/dev.02138. doi:10.1242/dev.02138 [DOI] [PubMed] [Google Scholar]
- Vavouri T, Walter K, Gilks W.R, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. doi:10.1186/gb-2007-8-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. doi:10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. doi:10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. doi:10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]
- Wray G.A, Hahn M.W, Abouheif E, Balhoff J.P, Pizer M, Rockman M.V, Romano L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. doi:10.1093/molbev/msg140 [DOI] [PubMed] [Google Scholar]
- Yeo G.W, Van Nostrand E, Holste D, Poggio T, Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. doi:10.1073/pnas.0409742102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.H, Carroll S.B. The evolution of abdominal reduction and the recent origin of distinct abdominal-B transcript classes in Diptera. Evol. Dev. 2006;8:241–251. doi: 10.1111/j.1525-142X.2006.00095.x. doi:10.1111/j.1525-142X.2006.00095.x [DOI] [PubMed] [Google Scholar]