Abstract
Variation in traits across species or populations is the outcome of both environmental and historical factors. Trait variation is therefore a function of both the phylogenetic and spatial context of species. Here we introduce a method that, within a single framework, estimates the relative roles of spatial and phylogenetic variations in comparative data. The approach requires traits measured across phylogenetic units, e.g. species, the spatial occurrences of those units and a phylogeny connecting them. The method modifies the expected variance of phylogenetically independent contrasts to include both spatial and phylogenetic effects. We illustrate this approach by analysing cross-species variation in body mass, geographical range size and species-typical environmental temperature in three orders of mammals (carnivores, artiodactyls and primates). These species attributes contain highly disparate levels of phylogenetic and spatial signals, with the strongest phylogenetic autocorrelation in body size and spatial dependence in environmental temperatures and geographical range size showing mixed effects. The proposed method successfully captures these differences and in its simplest form estimates a single parameter that quantifies the relative effects of space and phylogeny. We discuss how the method may be extended to explore a range of models of evolution and spatial dependence.
Keywords: comparative method, spatial analysis, mammals
1. Introduction
Organisms are the dual products of the environment in which they currently live and their evolutionary history. Thus, species that live in similar environments, or have similar ecologies, would be expected to have common adaptations, and their similarity should be correlated with spatial proximity (Cliff & Ord 1981; Ripley 1981; Borcard et al. 1992; Legendre 1993; Legendre et al. 1997; Lennon 2000); similarly, closely related species would be expected to show more similarity than those that are distantly related because they share more common evolutionary history (Ridley 1986; Harvey & Pagel 1991; Harvey & Purvis 1991; Price 1997; Harvey & Rambaut 2000; Freckleton & Harvey 2006). In other words, species' traits may be conserved across both space and phylogeny as a consequence of selection for ecological adaptation and the constraints of past evolutionary history. Potential ecological or environmental determinants of trait variation may similarly contain both phylogenetic (Grafen 1989; Westoby et al. 1995; Diniz-Filho et al. 1998; Desdevises et al. 2003; Wiens & Graham 2005) and spatial (Sokal 1983; Borcard et al. 1992; Legendre 1993; Peres-Neto 2006) signals and both need to be identified.
The comparative approach seeks to disentangle the roles of such processes. In analyses looking at cross-species variation in traits, similarity resulting from shared evolution is regarded as a potentially confounding factor; by incorporating phylogenetic information into comparative analyses, it is possible to address these statistically while analysing correlations between traits and the environment in order to reveal environmentally driven evolutionary patterns (Felsenstein 1985; Grafen 1989; Harvey & Pagel 1991; Lynch 1991). The phylogenetic component of trait variation is typically marginalized, so that such approaches do not explicitly measure what proportion of the variation in trait values in a clade is driven by the environment relative to the proportion that is explained by history. Consequently, it is not generally well understood which of history or environment is more important in determining trait variation across species. Yet, the distinction between phylogenetically structured and more plastic and environmentally driven trait variations has gained new and particular importance in the context of potential range shifts under climate change (Ackerly 2003; Diniz-Filho & Bini 2008).
In analyses with a geographical focus, the importance of spatial non-independence of data has become appreciated, and various techniques have been developed to address it (e.g. reviews by Clifford et al. 1989; Haining 1990; Legendre et al. 2002; Dormann et al. 2007). To date, these approaches have found extensive use for modelling the spatial abundance or richness of species (Borcard et al. 1992; Lennon 2000; Jetz & Rahbek 2002; Lichstein et al. 2002; Diniz et al. 2003; McPherson & Jetz 2007), and also the spatial analysis of genetic variation and diversity (Sokal & Oden 1978; Sokal et al. 1989; Escudero et al. 2003).
Common to both phylogenetic and spatial analyses is the problem of non-independence, and any process that yields non-independence can result in unwelcome correlation structures in data when confronted with statistical methods that assume independence. In the analyses of spatial and time-series data, it has been recognized that diagnosing and measuring such non-independence is an important step in analysing and understanding data (e.g. for overviews see Haining 1990; Chatfield 1996). In those cases, model choice for the spatial pattern is often not easy. By contrast, in comparative analysis, we have the advantage that it is frequently possible to specify models of trait evolution on the phylogeny (e.g. Hansen 1997; Pagel 1997, 1999; Felsenstein 2008) and hence tackle the source of non-independence head-on.
As yet, there have been few attempts to synthesize methods for measuring spatial and phylogenetic signals in comparative datasets. Of course, many studies have looked at how the effects of environmental drivers (e.g. latitude, temperature, altitude) influence species' traits (Ashton et al. 2000; Freckleton et al. 2003; Blackburn & Hawkins 2004; McKechnie et al. 2006). However, spatial non-independence can be pervasive: the influence of unmeasured and hidden variables can dramatically affect the analyses (Lennon 2000). Comparative biologists have developed a suite of statistical techniques for analysing trait data containing phylogenetic signal. Although the issue has been recognized and possible methods discussed (e.g. Legendre et al. 2004), so far none have considered how spatial effects may be also included within a single statistical framework.
In this paper, we provide an illustration of the joint effects of phylogenetic and spatial dependence on trait variation across species, the 891 species of carnivores, even-toed ungulates and primates of the world. We introduce a method allowing the effects of phylogenetic and spatial processes to be measured simultaneously, and show how it may be used to reveal how spatial and phylogenetic factors simultaneously shape the evolution and distribution of traits.
2. Material and methods
(a) Phylogenetic distribution of traits
The model of trait distribution we use is the Brownian model, which forms the basis for many commonly employed phylogenetic methods (Felsenstein 1985; Harvey & Pagel 1991; Martins & Hansen 1997; Pagel 1997, 1999). The Brownian model is essentially a neutral model of trait evolution in which changes in trait values occur continuously and in which increases and decreases in traits are equally as likely and independent of the current state. This is a simple model; however, a range of more complex models can be reduced to a Brownian form or accommodated in the same framework (Hansen 1997; Pagel 1997). We consider a single trait evolving among a set of n species. The state of the trait is denoted by a vector x. Under the Brownian model, if t is the time over which the trait is evolving, then Δx, the change in x, is a multivariate normal (MVN) random deviate,
| (2.1) |
where Σ is a (n×n) matrix proportional to the expected variances and covariances for trait changes among species, which are given by the shared path lengths on the phylogeny (e.g. Martins & Hansen 1997; Pagel 1997), and σ2 is the rate at which variance accumulates per unit time. After T units of time, x(T) is a multivariate normally distributed with mean x(0) and variance–covariance matrix σ2ΣT.
(b) Phylogenetic contrasts
In order to develop the method for simultaneously incorporating spatial and phylogenetic effects, we estimated phylogenetic contrasts. This is a computationally efficient method for fitting a Brownian model to comparative data and for estimating the parameters of the Brownian process. The unstandardized contrasts (u) and their variances (v) are calculated following the detailed algorithm given in Felsenstein (1985).
(c) Incorporating spatial effects
In order to model the spatial effect, we assume an analogous linear variance model of spatial similarity and modify the method of contrasts to account for additional processes (see Garland et al. (1992) for an earlier discussion of the idea of modifying contrasts). If dij is the spatial distance between species i and j, and the phylogenetic distance is pij, then the net variance for the distance between their traits is
| (2.2) |
In this model, ϕ measures the relative contribution of phylogenetic and spatial effects. A value of ϕ equal to 0 is a model in which there are only phylogenetic effects and a value of 1 is the one in which there are only spatial effects. According to this model, traits evolve as a function of both phylogenetic and spatial distances. This model therefore allows for closely related species to be geographically close together, as would be expected in many datasets.
Alternative models are possible and we outline in table 1 a series of simple functions varying in complexity, in terms of nonlinearity and number of parameters that could be used in applications of this approach. Exploratory analysis however indicated that the linear model worked well for the data that we analysed (e.g. see below). In practice, we would recommend that a range of functions are explored.
Table 1.
Possible alternative models of spatial versus phylogenetic effects. These could be used to replace equation (2.2) in the text.
| equation | description |
|---|---|
| variance is a linear function of spatial and phylogenetic distances; ϕ measures the relative contribution of each. | |
| variance is a linear function of phylogenetic distance and an exponential function of spatial distance. ϕ measures the relative contribution of each; α models the change in autocorrelation in space. | |
| variance is a linear function of phylogenetic distance and a power function of spatial distance. ϕ measures the relative contribution of each; α models the change in autocorrelation in space. | |
| variance is a linear function of spatial and phylogenetic distances, plus a term measuring the contribution of other effects (vm); ϕ measures the relative contribution of spatial and phylogenetic effects; a measures the relative contribution of the spatial+phylogenetic versus other effects. |
Equation (2.2) does not incorporate any non-spatial or non-phylogenetic component to trait variance. To address this, we simultaneously also used the λ transformation suggested by Pagel (1997, 1999). This transformation allows for phylogenetically uncorrelated variance, for example, resulting from species-specific adaptation. This transformation is achieved by lengthening the branches of the phylogeny leading to the tips by a fraction relative to the internal branches. This is done by multiplying the internal branches by λ where usually 0<λ<1. A value of λ=0 indicates that there is no phylogenetic signal in the trait and a value equal to 1 indicates that traits vary as predicted by the Brownian model.
In the context of modelling spatial and phylogenetic effects simultaneously, the λ-statistic allows us to include trait variation independent of both phylogeny and space in our analysis. This is akin to including a ‘nugget’ in a spatial model (e.g. Haining 1990).
(d) Estimation
Under the Brownian model, the expected distribution of traits is a MVN distribution. This distribution has two parameters, μ, the weighted mean of the trait at the basal node and, σ2, the variance parameter. The distribution is characterized by the expected variance–covariance matrix, V, which is an n×n matrix. When only phylogenetic effects are modelled, this is identical to Σ in equation (2.1). Because in equation (2.2) the net variance is a linear combination of the phylogenetic and spatial variances, when spatial and phylogenetic effects are combined, V is given by
| (2.3) |
where W is the variance–covariance matrix generated by the spatial distribution of the observations. There are numerous ways to generate W (for alternatives to the evolutionary model of W we use, see Dormann et al. 2007). In the specific model given by equation (2.2), it is assumed that the ancestor originated a single point in space and that the degree of variance between species that results from spatial proximity is given by the accumulated distance between them, so that entries of W are the accumulated spatial distance from the root until the most recent common ancestor of each pair of species.
For data x and variance matrix V(ϕ), the log likelihood of parameters ϕ, μ and σ2 is
| (2.4) |
In equation (2.4) X is the design matrix, in the case of a single trait containing a column of 1 s. The theory of Felsenstein (1973) shows that the multinormal log likelihood given by equation (2.4) is exactly equal to
| (2.5) |
Equation (2.5) is the likelihood of the estimated changes leading to the observed data according to the Brownian process. The summation in equation (2.5) is across all of the internal nodes of the phylogeny and is the sum of the log likelihoods of the individual changes at each of the nodes. Vi is the variance for node i, calculated according to the algorithm in Felsenstein (1985) to account for estimation error in the ancestral traits. Although it is perfectly possible to maximize equation (2.4) directly (and for some choices of W, this would be the only approach), maximization based on equation (2.5) is simpler because it does not require the inversion of the variance matrix, which is potentially numerically inaccurate and computationally inefficient. The parameters μ and σ2 are estimated as the weighted mean trait value at the basal node and the mean value of the squared standardized contrasts, respectively, for a given value of ϕ.
The spatial distances for internal nodes in equation (2.2) were estimated using ancestral state reconstructions on the phylogeny. We used the pic function in the R package analysis of phylogenetics and evolution (APE; Paradis et al. 2004) to do this. When calculating contrasts, the spatial distances and phylogenetic variances were scaled so that the maximum was identical in each case.
In order to obtain the maximum-likelihood estimate of ϕ, we used a one-dimensional parameter search, employing the optimize routine in the statistical package R (R Development Core Team 2007). This is an implementation that uses a combination of golden section and parabolic interpolation algorithms (Brent 1973).
For the simultaneous estimation of λ at the same time as ϕ, the analysis proceeds in the same way: the only extra step is that the phylogeny is first transformed as described above. The maximum likelihood is then found by jointly maximizing over ϕ and λ. To do this, we used the optim function in R employing the L-BRGS-B algorithm (Byrd et al. 1995) and constraining the search to find optimum values of both ϕ and λ between 0 and 1 as these parameters are generally undefined outside this range. Previous simulations have shown that for given V, maximum-likelihood values of the λ-statistic can be accurately tested against null values (0 and 1) using likelihood-ratio tests (Freckleton et al. 2002).
In order to interpret the model including both parameters, we note that when λ is estimated at the same time as ϕ, equation (2.3) becomes (where h is a vector formed from the leading diagonal of Σ representing the heights of the tips)
| (2.6) |
Equation (2.6) can be written as
| (2.7) |
In equation (2.7) γ=(1−ϕ)(1−λ) is the relative contribution of effects independent of both phylogeny and space; λ′=(1−ϕ)λ is the relative contribution of phylogeny and ϕ is the relative spatial effect. These composite parameters allow simple interpretation of the joint estimates of ϕ and λ. In equation (2.7) the sum of γ, λ′ and ϕ is always 1, therefore these parameters can be interpreted as the individual proportional contributions to variance of the different components if Σ and W are appropriately scaled.
(e) Data for analysis
Spatial and environmental data used in the analysis were based on the extent of occurrence range maps provided in geographical information systems vector format by Ceballos (for data sources and mapping methodology, see Ceballos et al. 2005; Ceballos & Ehrlich 2006). We calculated geographical range midpoints from latitudinal and longitudinal extents of range maps and used them to characterize the spatial distance of species data points to each other. For the purpose of this analysis, we consider and analyse the following three attributes as ‘species traits’: geographical range size (range size); the environmental temperature characteristic for a species range (temperature); and a species' typical body mass (body mass). We estimated range size (in km2) directly from range maps in cylindrical equal area projection and log-transformed data for analysis. The same maps formed the basis for characterizing the broad-scale temperature niche of species. Average annual temperature data (in °C) came from the Climatic Research Unit gridded climatology 1961–1990 dataset (New et al. 2002) at native 10 min resolution and was log transformed. We resampled both sets of data to 0.01° spatial resolution and extracted range map occurrences and temperature map across a 55×55 km2 equal area grid (in cylindrical equal area projection). We then calculated for each species temperature as the average environmental temperature across all grid cells it occupies. Finally, we extracted body masses for 183 artiodactyls, 201 carnivores and 207 primates from Smith et al. (2003) and log-transformed data for analysis (see source for details on body size compilation). Phylogenetic relationships among species of the three mammal clades came from select sources (Purvis 1995; Purvis et al. 1995; Bininda-Emonds et al. 1999; Price et al. 2005; Vos & Mooers submitted) and were extracted as subsets from the recently published mammalian supertree (Bininda-Emonds et al. 2007). Tree resolution (internal nodes/tips) was 0.76 for carnivores, 0.62 for artiodactyls and 0.73 for primates.
(f) Simulations
We used simulations to validate the method for the datasets that we analysed. We did this to determine the behaviour of the estimator ϕ in terms of its power and type I error rates. For each node in the tree, we generated the phylogenetic variances and geographical distances to form the net variances for contrasts at nodes using the variances of the reconstructed contrasts under the Brownian model. To generate simulated data, we generated random normal deviates with mean zero and variance given by equation (2.2), and with the given value of ϕ. We first used this simulation method to simulate estimated values of ϕ for known values of ϕ between 0 and 1. We also generated distributions of values of ϕ, repeating 10 000 replicates for each of the three phylogenies.
3. Results
(a) Simulations
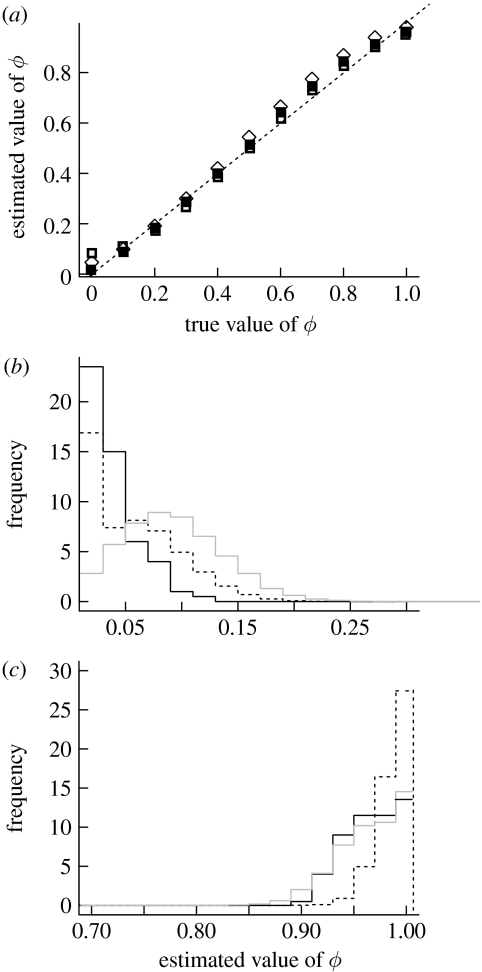
There is generally a close correspondence between the values of ϕ estimated from simulated data with known values (figure 1a). It is important to note, however, that at the extremes there are small but influential biases: this is unsurprising given that the value of ϕ is restricted between 0 and 1. Consequently, any sampling error will yield some degree of bias: overestimation would be expected at the lower bound and underestimation expected at the higher bound. The degree of bias varies between the phylogenies, being the least for the carnivore dataset and slightly greater for the other two.
Figure 1.
Simulations to determine the extent of bias in the estimate of ϕ. A value of ϕ close to 0 indicates that the phylogenetic signal is stronger than the spatial signal and a value close to 1 indicates that the spatial signal is stronger. (a) Correspondence between observed and simulated values. The dashed line is the line of 1 : 1. The points represent data simulated for the different datasets used in the analysis (ν, carnivores; squares, artiodactyls; diamonds, primates). Simulated distributions of ϕ under a true distribution of (b) ϕ=0 and (c) ϕ=1.
As shown in figure 1, this bias leads to small but important consequences for the simulated distributions of ϕ under true values of ϕ=0 (figure 1b) or ϕ=1 (figure 1c). As would be expected given the results in figure 1, the distribution of ϕ varies between datasets, particularly in the case of simulated data under a true value of ϕ=0. The reasons for this difference between the datasets probably are that (i) the sizes of the datasets are different (albeit not vastly) and (ii) there is error in the phylogeny and the extent of this error varies between the datasets. In summary, these results underline the importance of simulation methods to generate sampling intervals for null values of ϕ for different datasets.
(b) Analysis of data
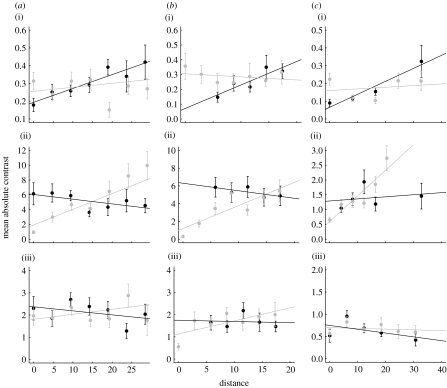
We conducted initial analyses using diagnostic plots showing the value of the unstandardized contrasts as a function of phylogenetic or spatial distance (figure 3). These plots are comparable to variograms, with data grouped into distance classes (e.g. Haining 1990). They effectively visualize the apparently strong relationships between phylogenetic distance and contrasts in body mass, and between geographical distance and temperature contrasts, with somewhat shallower slopes for range size.
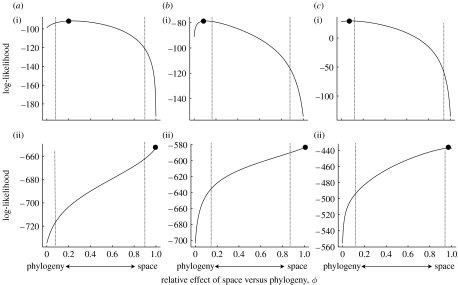
Figure 3.
Log-likelihood profiles for estimates of ϕ for the three datasets described in the text ((a) carnivores, (b) artiodactyls and (c) primates: (i) body size and (ii) environmental temperature). The lines show the log likelihood as ϕ is varied and the filled point indicates the maximum. Simulations were used to determine sampling intervals. The vertical dashed line on the right-hand side in each case shows the lower 2.5 per cent for the distribution when the true value is 1 and that on the left-hand side shows the upper 2.5 per cent for the sampling distribution when the true value is 0. Note that calculated this way, the type I error rate for each analysis is equal to 5 per cent when testing the composite hypothesis that the observed value is different from 0 or 1.
The parameter ϕ jointly quantifies the relative contribution of phylogenetic and spatial effects, ranging from 0 (phylogeny only) to 1 (space only). The estimated likelihood profiles of ϕ illustrate the combined assessment provided by this approach (figure 3). Table 2 summarizes the maximum-likelihood values of estimated parameters. Body mass yields a low maximum-likelihood value of ϕ confirming a dominant phylogenetic effect (figure 4a–c). In all three cases, the maximum-likelihood value of ϕ is not significantly different from 0. In turn, the maximum-likelihood value of ϕ for temperature is not significantly different from 1 in any clade (figure 3d–f), indicating that the spatial dependence of species' environmental temperature niche is considerably stronger than the phylogenetic effect. This reflects the clear relationship between the phylogenetic distance and the mean unstandardized contrast values found for temperature in figure 2.
Table 2.
Estimates of parameters describing the relative effects of spatial and phylogenetic effects on traits in three orders of mammals. As described in the text, these effects are estimated by fitting either a single parameter ϕ that measures the relative contributions of phylogenetic and spatial processes or a further parameter λ that allows for a separate phylogenetic and non-phylogenetic component to trait variation in addition to the spatial effect. The table shows the maximum-likelihood value of ϕ when estimated singly, as well as jointly estimated with λ. Note that if the maximum-likelihood value of ϕ is 1, then the value of λ cannot be estimated as ϕ=1 indicates that there is no non-spatial component to trait variation. The right-hand side of the table lists the joint model where the composite parameters summarize the net relative effects of phylogeny λ′, spatial effects and variation independent of both (see text for details).
| single parameter | both parameters | composite parameters | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ϕ | L | ϕ | λ | L | independent (γ) | phylogenetic (λ′) | spatial (ϕ) | ||
| carnivores | mass | 0.237 | −91.51 | 0.237 | 1.000 | −91.51 | 0.000 | 0.763 | 0.237 |
| temp | 1.000 | −652.70 | 1.000 | n.a. | −652.70 | 0.000 | 0.000 | 1.000 | |
| range | 0.505 | −512.94 | 0.000 | 0.000 | −477.83 | 1.000 | 0.000 | 0.000 | |
| artiodactyls | mass | 0.109 | −78.59 | 0.109 | 1.000 | −78.59 | 0.000 | 0.891 | 0.109 |
| temp | 1.000 | −583.79 | 1.000 | n.a. | −583.79 | 0.000 | 0.000 | 1.000 | |
| range | 0.908 | −424.64 | 0.512 | 0.000 | −399.73 | 0.488 | 0.000 | 0.512 | |
| primates | mass | 0.086 | −29.50 | 0.071 | 0.999 | −29.54 | 0.001 | 0.928 | 0.071 |
| temp | 0.983 | −436.66 | 0.943 | 0.000 | −421.17 | 0.057 | 0.000 | 0.943 | |
| range | 0.844 | −927.45 | 0.000 | 0.216 | −887.52 | 0.784 | 0.216 | 0.000 | |
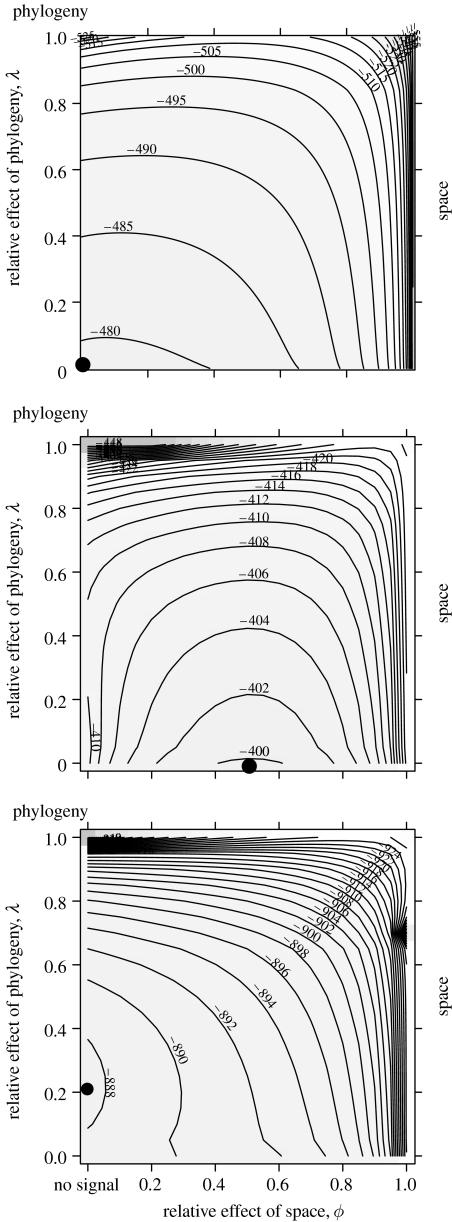
Figure 4.
Contour plots for joint likelihoods of ϕ and λ for the data on range sizes in three orders of mammals ((a) carnivores, (b) artiodactyls and (c) primates). The contours show parameter combinations of equal likelihood and the filled circles indicate the joint maximum-likelihood values (see table 2 for details).
Figure 2.
Variograms of the relationship between geographical or phylogenetic distance and trait variance. ((a) carnivores, (b) artiodactyls and (c) primates: (i) body size, (ii) environmental temperature and (iii) range size.) The plots are constructed in the manner of a conventional variogram (e.g. Haining 1990), with data organized into distance intervals and then the mean variance in unstandardized contrasts calculated for data at each distance interval. Only density classes with more than 10 observations are included. The grey and black circles are variances calculated for groups based on geographical (spatial effect) and phylogenetic distances, respectively. The lines are regression lines. As with variograms, these plots are intended to be illustrative only. The formal tests of the parameter values are shown in figures 3 and 4. Table 2 summarizes the analyses.
We additionally fitted λ to the data that measure the relative phylogenetic and non-phylogenetic components of variance. For body mass and temperature, a model including both ϕ and λ does not offer an improved fit. The simple model given by equation (2.2) is thus sufficient to describe the combined spatial and phylogenetic dependence in these data. This means that the variation in traits is explained best by either phylogenetic effects (body mass) or spatial effects (temperature). In the case of environmental temperature in artiodactyls, there was an increase in log likelihood resulting from including λ in the model (increase in log likelihood of 15.5 units); however, the resultant net effect was small (γ=0.057).
For range size we found very different patterns. For this variable, a model including λ in addition to ϕ yielded considerably better models (increase in log likelihood of 35.1 units in carnivores, 24.9 units in artiodactyls and 39.9 units in primates). This suggests that in this variable a strong component of the trait variation was independent of phylogeny or spatial effects. In carnivores, there was no phylogenetic or spatial signal in the data (figure 4a). Range size in artiodactyls showed no phylogenetic signal, with a moderate amount of spatial signal (γ=0.488 and ϕ=0.512; figure 4b). Finally, in the case of the primates, there was a weak phylogenetic signal in the range sizes (λ′=0.216), but no spatial effect (figure 4c), and most variation was independent of either (γ=0.784).
4. Discussion
We have shown that across 591 mammal species, key traits may often be significantly affected by both spatial and phylogenetic factors, indicating the need for a joint rather than separate assessment of these effects. We introduce a method for such a joint measurement of phylogenetic and spatial signals that are a simple and logical extension of existing approaches. This integrative approach allows questions about the role of environment and history to be asked, which could not be addressed using separate diagnostics provided by the existing techniques. Furthermore, this approach can be readily adapted to form the basis for conducting comparative tests (e.g. regression, analysis of variance and other linear models) that allow both sources of non-independence to be controlled for simultaneously.
(a) Autocorrelation versus direct effects
Whether we are examining phylogenetic or spatial effects, an obvious issue is whether non-independence in data should be modelled as the direct effect of predictors (e.g. by regressing trait values onto a set of predictors) or indirectly through expected covariance or autocorrelation (as in the generalized least squares (GLS) model above). The method we describe is readily expanded to consider predictors of the trait of interest.
The distinction between models of autocorrelation and those of direct effects of predictors is important as frequently non-independence in data can be generated by ‘hidden’ variables, so that as successive predictors are added to an analysis, the degree of non-independence of data may decrease (e.g. McKechnie et al. 2006). In the model we employ, the non-independence is contained in the residual portion of the variation in traits, so that if predictors are used to directly explain residual variation, the degree of independence of the residuals should increase. Therefore, if the predictors are phylogenetically or spatially non-independent, and these play a significant role in determining trait values, the spatial or phylogenetic components in traits can be directly explained.
In the analyses we report above, the aim was not to explain the variation in traits by direct effects: although this may be possible, at this point we simply sought to measure the broad contribution of both space and phylogeny to trait variation without asking what drives this variation. This is informative in itself, for instance in asking how the data are structured (e.g. as in figure 3), as well as being a useful first step in developing more complex analyses.
Neither space nor phylogeny can in themselves generate a direct effect on trait values: the effects of either have to be mediated through other variables, for example through changes in the environment or species' niches. One issue that is likely to be important when considering spatial effects is that the degree of overlap in species' ranges will be an important factor in determining the degree of similarity resulting from spatial proximity. Including this is a more complex issue. The approach that has been recently suggested by Felsenstein (2002, 2008) could be possibly adapted for such an analysis: in this approach intraspecific variation is also modelled and could be adapted to consider the within-species spatial distribution of populations. In general, it would be expected that the more species' ranges overlap with each other, the less important spatial proximity would be.
As with other comparable methods (e.g. the λ-statistic of Pagel 1999), the GLS methods we employ may be adapted for considering the effects of predictor variables, and ϕ estimated simultaneously with fitting statistical models (e.g. Freckleton et al. (2002) for an illustration of how such a method would be developed). Such an approach would be very useful in constructing analyses in which the combined effects of spatial and temporal non-independence are controlled for. The estimation of parameters such as ϕ to optimize a variance–covariance matrix in GLS analysis is known as estimated generalized least squares, and is known to be a very flexible technique (e.g. Ives et al. 2007).
The only issue with developing multi-predictor analyses is that the value of ϕ would have to be interpreted with caution: as outlined above, if the predictors in an analysis were environmental predictors, and hence likely to be spatially correlated, it would be no surprise if the spatial signal in the residuals of the analysis was weakened leaving only phylogenetic signal. Consequently, ϕ would not be interpretable in an evolutionary sense in such an analysis.
(b) Evolutionary interpretations
According to the Brownian model of trait evolution, the differences between species traits grow linearly with evolutionary time. Thus, closely related species are more similar than distantly related ones. This model does not preclude an adaptive component to trait evolution (Blomberg et al. 2003): on the contrary, the model implies that species traits are continually changing and, unless the traits examined are neutral for the fitness of organisms, this change presumably must have an adaptive basis and be driven by changes to exogenous drivers.
If the value of ϕ is close to 1, we can be sure that trait evolution is being driven by geographical or environmental factors unrelated to phylogeny. If ϕ>0 then if two sister species live close together they will be more similar than a pair of species that split at the same time but live farther apart. By contrast, if ϕ=0, geographical distance is unimportant and only phylogenetic distance matters, presumably with environment playing little role.
According to this interpretation, the value of ϕ can be informative about the degree to which species' niches are conserved or changed relative to geography or phylogeny. A value of ϕ>0 indicates that niches are changing faster with respect to spatial variation than with evolutionary distance and that this is changing traits faster than the null Brownian model. Under such circumstances, the conclusion would be that traits and species niches are more labile than is expected based on phylogenetic distance alone.
Clearly, as indicated by the results in figures 3 and 4, not all traits are under the same kind of selective pressure, even for the same set of species. For the groups we have analysed, body mass is closely associated with evolutionary distance; however, environmental temperature and range size are much more closely allied to spatial factors. The variance in responses indicated in figures 3 and 4 is a clear evidence of mosaic evolution of species traits in response to a suite of selective pressures: range size and temperature are clearly a great deal more labile with respect to environmental variation than is body size.
In the specific dataset we have analysed, there is a question of whether variables such as range size and temperature can be considered as traits (see also the discussion of this in Freckleton et al. 2002), although the literature clearly does. For example, in climate envelope modelling (e.g. Thomas et al. 2004), the mean temperature at which a species exists is regarded as a fundamental measure of a species' niche and this seems a reasonable measure of climate tolerance. Our finding is that the mean temperatures at which species exist are more variable (with respect to phylogeny) than predicted by the null model. Although the sample on which we have based our conclusion is small (three groups), if repeated more widely, this finding would have important implications for our understanding of how species evolve to respond to changing environments.
There has been a lively debate about how range size evolves and whether species' ranges are in some sense ‘heritable’ (e.g. Webb & Gaston 2003). Our analysis suggests how such debates can be addressed empirically by measuring phylogenetic (i.e. heritable) and spatial (i.e. non-heritable) components. In our data, it is clear that the phylogenetic drivers of range size are weak and that in at least two cases spatial effects are more dominant (table 2; figure 4). In these data, the evidence is that range size variation is driven by where species live rather than by evolutionary history (and presumably therefore life history). Indeed, the evidence that range size shows any phylogenetic signal at all is weak (table 2) with the conclusion being that range size is an extremely labile trait with low heritability in these groups.
(c) Developing the method
As mentioned above, a suite of other methods exist, particularly for conducting spatial analyses. Those most commonly used in ecology and evolutionary biology are summarized by Dormann et al. (2007). There exists a range of very similar methods based on autocorrelation models and GLS, typically employing a single parameter to measure spatial dependence, with different approaches varying in how the expected variance among species is calculated. The spatial approach that we have taken is essentially the same as the spatial GLS approach (e.g. Dormann et al. 2007) and the single trait GLS model including λs is essentially the same as this. Our approach is therefore set firmly among conventional approaches to measuring spatial dependence. Moreover, the method is economical in that it involves the estimation of only one parameter, although the approach is flexible enough that more complex models can be constructed (table 1). Moreover, the method is readily implemented using freely available software (e.g. using the APE package; Paradis et al. 2004).
Changes to the spatial model to account for different assumptions and modes of trait variation are readily incorporated. In order to distinguish between models, likelihood ratio statistics, Akaike information criteria or Bayesian information criteria (e.g. Burnham & Anderson 2002; Link & Barker 2006) or simulation approaches (as above) could be used. For instance, highly parametrized models of spatial variation could be incorporated. The important consideration in doing so, however, is to maintain a balance between the models of the two processes. For example, if the model of spatial variation is more complex than that of phylogenetic signal, it would be sensible to compare the variation attributable to the two directly, unless there were a priori reasons for employing the more complex model.
5. Conclusions
We have shown that species trait shows a variety of phylogenetic and spatial structures. Across 891 species of mammals, body size is mainly correlated with phylogeny, environmental temperature mainly a function of spatial processes and range size shows intermediate responses. Such a range of behaviour in a relatively small sample is evidence that a combined phylogenetic and spatial approach will be frequently warranted and that useful information on the evolution of traits will be gained by looking at both aspects of trait variation simultaneously. We illustrate how widely used phylogenetic methods can be adapted to also measure the strength of spatial dependence in comparative data. Such integrative assessment of both phylogenetic and spatial effects on biological variation is likely to facilitate more rigorous inference in ecology, behaviour, conservation and global change biology.
Acknowledgments
The authors are indebted to Gerardo Ceballos, UNAM, for allowing the use of mammal geographical range maps compiled in his laboratory for this analysis. R.P.F. is funded by a Royal Society University Research Fellowship. W.J. gratefully acknowledges support by NSF (BCS—0648733). R code for implementing the methods described is available from R.P.F.
References
- Ackerly D.D. Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 2003;164:S165–S184. doi:10.1086/368401 [Google Scholar]
- Ashton K.G, Tracy M.C, de Queiroz A. Is Bergmann's rule valid for mammals? Am. Nat. 2000;156:390–415. doi: 10.1086/303400. doi:10.1086/303400 [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O.R.P, Gittleman J.L, Purvis A. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia) Biol. Rev. Camb. Philos. Soc. 1999;74:143–175. doi: 10.1017/s0006323199005307. doi:10.1017/S0006323199005307 [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O.R.P, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. doi:10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Blackburn T.M, Hawkins B.A. Bergmann's rule and the mammal fauna of northern North America. Ecography. 2004;27:715–724. doi:10.1111/j.0906-7590.2004.03999.x [Google Scholar]
- Blomberg S.P, Garland T, Jr, Ives A.R. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. doi:10.1554/0014-3820(2003)057[0717:TFPSIC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Borcard D, Legendre P, Drapeau P. Partialling out the spatial component of ecological variation. Ecology. 1992;73:1045–1055. doi:10.2307/1940179 [Google Scholar]
- Brent R.P. Prentice Hall; Englewood Cliffs, NJ: 1973. Algorithms for minimization without derivatives. [Google Scholar]
- Burnham K.P, Anderson D.R. 2nd edn. Springer; London, UK: 2002. Model selection and multimodel inference: a practical information-theoretic approach. [Google Scholar]
- Byrd R.H, Lu P, Nocedal J, Zhu C. A limited memory algorithm for bound constrained optimization. SIAM J. Sci. Comput. 1995;16:1190–1208. doi:10.1137/0916069 [Google Scholar]
- Ceballos G, Ehrlich P.R. Global mammal distributions, biodiversity hotspots, and conservation. Proc. Natl Acad. Sci. USA. 2006;103:19 374–19 379. doi: 10.1073/pnas.0609334103. doi:10.1073/pnas.0609334103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos G, Ehrlich P.R, Soberon J, Salazar I, Fay J.P. Global mammal conservation: what must we manage? Science. 2005;309:603–607. doi: 10.1126/science.1114015. doi:10.1126/science.1114015 [DOI] [PubMed] [Google Scholar]
- Chatfield C. Chapman & Hall; London, UK: 1996. The analysis of time series. [Google Scholar]
- Cliff A.D, Ord J.K.Spatial processes: models and applications1981Pion; London, UK [Google Scholar]
- Clifford P, Richardson S, Hemon D. Assessing the significance of the correlation between two spatial processes. Biometrics. 1989;45:123–134. doi:10.2307/2532039 [PubMed] [Google Scholar]
- Desdevises Y, Legendre P, Azouzi L, Morand S. Quantifying phylogenetically structured environmental variation. Evolution. 2003;57:2647–2652. doi: 10.1111/j.0014-3820.2003.tb01508.x. doi:10.1554/02-695 [DOI] [PubMed] [Google Scholar]
- Diniz-Filho J.A.F, Bini L.M. Macroecology, global change and the shadow of forgotten ancestors. Global Ecol. Biogeogr. 2008;17:11–17. doi:10.1111/j.1466-8238.2008.00395.x [Google Scholar]
- Diniz-Filho J.A.F, de Sant'Ana C.E.R, Bini L.M. An eigenvector method for estimating phylogenetic inertia. Evolution. 1998;52:1247–1262. doi: 10.1111/j.1558-5646.1998.tb02006.x. doi:10.2307/2411294 [DOI] [PubMed] [Google Scholar]
- Diniz-Filho J.A.F, Bini L.M, Hawkins B.A. Spatial autocorrelation and red herrings in geographical ecology. Global Ecol. Biogeogr. 2003;12:53–64. doi:10.1046/j.1466-822X.2003.00322.x [Google Scholar]
- Dormann C.F, et al. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography. 2007;30:609–628. doi:10.1111/j.2007.0906-7590.05171.x [Google Scholar]
- Escudero A, Iriondo J.M, Torres M.E. Spatial analysis of genetic diversity as a tool for plant conservation. Biol. Conserv. 2003;113:351–365. doi:10.1016/S0006-3207(03)00122-8 [Google Scholar]
- Felsenstein J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am. J. Hum. Genet. 1973;25:471–492. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;126:1–25. doi: 10.1086/703055. doi:10.1086/284325 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Contrasts for a within-species comparative method. In: Slatkin M, Veuille M, editors. Modern developments in theoretical population genetics. Oxford University Press; Oxford, UK: 2002. pp. 130–164. [Google Scholar]
- Felsenstein J. Comprative methods with sampling error and within-species variation: contrasts revisited and revised. Am. Nat. 2008;171:713–725. doi: 10.1086/587525. doi:10.1086/587525 [DOI] [PubMed] [Google Scholar]
- Freckleton R.P, Harvey P.H. Detecting non-Brownian trait evolution in adaptive radiations. PLoS Biol. 2006;4:e373. doi: 10.1371/journal.pbio.0040373. doi:10.1371/journal.pbio.0040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freckleton R.P, Harvey P.H, Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 2002;160:712–726. doi: 10.1086/343873. doi:10.1086/343873 [DOI] [PubMed] [Google Scholar]
- Freckleton R.P, Harvey P.H, Pagel M. Bergmann's rule and body size in mammals. Am. Nat. 2003;161:821–825. doi: 10.1086/374346. doi:10.1086/374346 [DOI] [PubMed] [Google Scholar]
- Garland T, Jr, Harvey P.H, Ives A.R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 1992;41:18–32. doi:10.2307/2992503 [Google Scholar]
- Grafen A. The phylogenetic regression. Phil. Trans. R. Soc. B. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. doi:10.1098/rstb.1989.0106 [DOI] [PubMed] [Google Scholar]
- Haining R. Cambridge University Press; Cambridge, UK: 1990. Spatial data analysis in the social and environmental sciences. [Google Scholar]
- Hansen T.F. Stabilizing selection and the comparative analysis of adaptation. Evolution. 1997;51:1341–1351. doi: 10.1111/j.1558-5646.1997.tb01457.x. doi:10.2307/2411186 [DOI] [PubMed] [Google Scholar]
- Harvey P.H, Pagel M.D. Oxford University Press; Oxford, UK: 1991. The comparative method in evolutionary biology. [Google Scholar]
- Harvey P.H, Purvis A. Comparative methods for explaining adaptations. Nature. 1991;351:619–624. doi: 10.1038/351619a0. doi:10.1038/351619a0 [DOI] [PubMed] [Google Scholar]
- Harvey P.H, Rambaut A. Comparative analyses for adaptive radiations. Phil. Trans. R. Soc. B. 2000;355:1599–1606. doi: 10.1098/rstb.2000.0721. doi:10.1098/rstb.2000.0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives A.R, Midford P.E, Garland T. Within-species variation and measurement error in phylogenetic comparative methods. Syst. Biol. 2007;56:252–270. doi: 10.1080/10635150701313830. doi:10.1080/10635150701313830 [DOI] [PubMed] [Google Scholar]
- Jetz W, Rahbek C. Geographic range size and determinants of avian species richness. Science. 2002;297:1548–1551. doi: 10.1126/science.1072779. doi:10.1126/science.1072779 [DOI] [PubMed] [Google Scholar]
- Legendre P. Spatial autocorrelation: trouble or new paradigm. Ecology. 1993;74:1659–1673. doi:10.2307/1939924 [Google Scholar]
- Legendre P, Galzin R, HarmelinVivien M.L. Relating behavior to habitat: solutions to the fourth-corner problem. Ecology. 1997;78:547–562. [Google Scholar]
- Legendre P, Dale M.R.T, Fortin M.J, Gurevitch J, Hohn M, Myers D. The consequences of spatial structure for the design and analysis of ecological field surveys. Ecography. 2002;25:601–615. doi:10.1034/j.1600-0587.2002.250508.x [Google Scholar]
- Legendre P, Dale M.R.T, Fortin M.J, Casgrain P, Gurevitch J. Effects of spatial structures on the results of field experiments. Ecology. 2004;85:3202. doi:10.1890/03-0677 [Google Scholar]
- Lennon J.J. Red-shifts and red herrings in geographical ecology. Ecography. 2000;23:101–113. doi:10.1034/j.1600-0587.2000.230111.x [Google Scholar]
- Lichstein J.W, Simons T.R, Shriner S.A, Franzreb K.E. Spatial autocorrelation and autoregressive models in ecology. Ecol. Monogr. 2002;72:445–463. [Google Scholar]
- Link W.A, Barker R.J. Model weights and the foundations of multimodel inference. Ecology. 2006;87:2626–2635. doi: 10.1890/0012-9658(2006)87[2626:mwatfo]2.0.co;2. doi:10.1890/0012-9658(2006)87[2626:MWATFO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lynch M. Methods for the analysis of comparative data in evolutionary biology. Evolution. 1991;45:1065–1080. doi: 10.1111/j.1558-5646.1991.tb04375.x. doi:10.2307/2409716 [DOI] [PubMed] [Google Scholar]
- Martins E.P, Hansesn T.F. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of inter-specific data. Am. Nat. 1997;149:646–667. doi:10.1086/286013 [Google Scholar]
- McKechnie A.E, Freckleton R.P, Jetz W. Phenotypic plasticity in the scaling of avian basal metabolic rate. Proc. R. Soc. B. 2006;273:931–937. doi: 10.1098/rspb.2005.3415. doi:10.1098/rspb.2005.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson J.M, Jetz W. Type and spatial structure of distribution data and the perceived determinants of geographical gradients in ecology: the species richness of African birds. Global Ecol. Biogeogr. 2007;16:657–667. doi:10.1111/j.1466-8238.2007.00318.x [Google Scholar]
- New M, Lister D, Hulme M, Makin I. A high-resolution data set of surface climate over global land areas. Clim. Res. 2002;21:1–25. doi:10.3354/cr021001 [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zool. Scr. 1997;26:331–348. doi:10.1111/j.1463-6409.1997.tb00423.x [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. doi:10.1038/44766 [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analysis of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. doi:10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Peres-Neto P.R. A unified strategy for estimating and controlling for spatial, temporal and phylogenetic autocorrelation in ecological models. Oecologia Brasiliensis. 2006;10:105–119. [Google Scholar]
- Price T. Correlated evolution and independent contrasts. Phil. Trans. R. Soc. B. 1997;352:519–529. doi: 10.1098/rstb.1997.0036. doi:10.1098/rstb.1997.0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price S.A, Bininda-Emonds O.R.P, Gittleman A.L. A complete phylogeny of the whales, dolphins and even-toed hoofed mammals (Cetartiodactyla) Biol. Rev. 2005;80:445–473. doi: 10.1017/s1464793105006743. doi:10.1017/S1464793105006743 [DOI] [PubMed] [Google Scholar]
- Purvis A. A composite estimate of primate phylogeny. Phil. Trans. R. Soc. B. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. doi:10.1098/rstb.1995.0078 [DOI] [PubMed] [Google Scholar]
- Purvis A, Nee S, Harvey P.H. Macroevolutionary inferences from primate phylogeny. Proc. R. Soc. B. 1995;260:329–333. doi: 10.1098/rspb.1995.0100. doi:10.1098/rspb.1995.0100 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2007 R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. See http://www.R-project.org
- Ridley M. Longman; Harlow, UK: 1986. Evolution and classification. [Google Scholar]
- Ripley B.D.Spatial statistics1981Wiley; New York, NY; Chichester, UK [Google Scholar]
- Smith F.A, Lyons S.K, Ernest S.K.M, Jones K.E, Kaufman D.M, Dayan T, Marquet P.A, Brown J.H, Haskell J.P. Body mass of late quaternary mammals. Ecology. 2003;84:3403–3403. doi:10.1890/02-9003 [Google Scholar]
- Sokal R.R. Analyzing character variation in geographic space. In: Felsenstein J, editor. Numerical taxonomy. Springer; New York, NY: 1983. pp. 383–403. [Google Scholar]
- Sokal R.R, Oden N.L. Spatial autocorrelation in biology. 2. Some biological implications and four applications of evolutionary and ecological interest. Biol. J. Linn. Soc. 1978;10:229–249. doi:10.1111/j.1095-8312.1978.tb00014.x [Google Scholar]
- Sokal R.R, Jacquez G.M, Wooten M.C. Spatial autocorrelation analysis of migration and selection. Genetics. 1989;121:845–855. doi: 10.1093/genetics/121.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.D, et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. doi:10.1038/nature02121 [DOI] [PubMed] [Google Scholar]
- Vos, R. A. & Mooers, A. H. Submitted. A dated MRP supertree for the order primates.
- Webb T.J, Gaston K.J. On the heritability of geographic range sizes. Am. Nat. 2003;161:553–566. doi: 10.1086/368296. doi:10.1086/368296 [DOI] [PubMed] [Google Scholar]
- Westoby M, Leishman M.R, Lord J.M. On misinterpreting the ‘phylogenetic correction’. J. Ecol. 1995;83:531–534. doi:10.2307/2261605 [Google Scholar]
- Wiens J.J, Graham C.H. Niche conservatism: integrating evolution, ecology, and conservation biology. Ann. Rev. Ecol. Evol. Syst. 2005;36:519–539. doi:10.1146/annurev.ecolsys.36.102803.095431 [Google Scholar]