Abstract
Matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS) is a powerful tool that has allowed researchers to directly probe tissue molecular structure and drug content with minimal manipulations, while maintaining anatomical integrity. In the present work glycerophospholipids and sphingolipids images were acquired from 16 µm thick coronal rat brain sections using MALDI-MS. Images of phosphatidylinositol 38:4 (PI 38:4), suifatide 24:1 (ST 24:1), and hydroxyl sulfatide 24:1 (ST 24:1 (OH)) were acquired in negative ion mode, while the images of phosphatidylcholine 34:1 (PC 34:1), potassiated phosphatidylcholines 32:0 (PC32:0 + K+) and 36:1 (PC 36:1 +K+) were acquired in positive ion mode. The images of PI 38:4 and PC 36:1+K+ show the preferential distribution of these two lipids in gray matter; and the images of two sulfatides and PC 32:0+K+ show their preferential distribution in white matter. In addition, the gray cortical band and its adjacent anatomical structures were also identified by contrasting their lipid makeup. The resulting images were compared to lipid images acquired by secondary ion mass spectrometry (SIMS). The suitability of TLC sprayers, Collison Nebulizer, and artistic airbrush were also evaluated as means for matrix deposition.
Introduction
Mass spectrometry has opened new vistas into the probing of biological problems. Because of its sensitivity and accuracy it has had a major impact on qualitative and quantitative detection and identification of biological molecules. Direct tissue imaging is a powerful tool, and at the same time a “minimalist approach” as with a modicum of manipulations, it gives a complete and accurate picture that permits the localizaton of drugs and endogenous molecules in tissue while preserving their anatomical structure. This approach also provides a highly efficient tool for the better understanding of physiological processes as well as the pathophysiology of disease while significantly shortening sample processing and turnaround time.
MALDI-MS has certain advantages over other MS techniques. To just mention a few, MALDI instruments have a wide mass range, are easy to use, require little maintenance have a higher tolerance to contaminants, their ion source design allows the adaptation of sample plates for direct analysis of biological molecules and pharmaceutical compounds; and sample preparation is fairly straight forward. The above mentioned advantages result in the gathering of a wealth of data from complex biological matrices, within a short sample turn around time. [1–6] The initial success of these applications has promoted the development of MALDI imaging mass spectrometry (MALDI-IMS) for visualizing endogenous and exogenous molecules in tissue sections that would otherwise require elaborate, time-consuming, and costly (immuno-) histochemistry and autoradiography techniques.
Several investigators have applied MALDI-IMS for direct tissue profiling and imaging of proteins and peptides with various level of success. [2, 3, 7, 8] Others applied this technique for visualizing drug distribution in rat brains, [9] throughout the whole animal body [10] to image agrochemicals accumulated in the soy bean plants, [11] and to detect protein and peptide in the ganglia of invertebrates.[12] All these examples support the notion that MALDI-IMS is a versatile analytical technique that has wide application possibilities. In spite of its promising potential, MALDI-IMS is nonetheless confronted by several technical issues in detection sensitivity and sample preparation. However, to date only a few studies devoted their attention to the optimization of sample preparation and handling for MALDI-IMS. [13, 14]
In addition to MALDI-MS, other mass spectrometric techniques, such as desorption electrospray ionization mass spectrometry (DESI), [15] and secondary ion mass spectrometry (SIMS) [16, 17] have also been adapted for tissue imaging studies. The lateral resolution of SIMS is far superior to what MALDI-IMS can offer. However, the sample preparation for SIMS imaging [16] and the profile depth of the samples [18] are very different from those of MALDI-IMS technique. Hence the differences between SIMS and MALDI-IMS imaging come as no surprise.
Although most MALDI-IMS applications focus on imaging of proteins and small organic molecules, a recent study has shifted the focus to brain lipids.[19] The authors utilized new matrix deposition approaches and obtained fairly interesting results. Nevertheless, some aspect of their matrix application methods deserves further evaluation and investigation for brain lipid imaging studies.
In this study, we further investigated and optimized one of the previously reported MALDI-IMS matrix deposition methods [19] and compared it with other available matrix deposition methods. We acquired ion density maps of such as phosphatidylinositol and phosphatidylcholines (glycerophospholipids) and sulfatides (sphingolipids) in a rat brain section.
Material and Methods
Matrix
2,5-dihydroxy benzoic acid solution (DHB, Aldrich Milwaukee WI) 100 mg/mL in 90% ethanol.
Animals
Male Sprague-Dawley rats (Harlan Industries, IN) between 280g to 420 g . All animal use and handling procedures were approved by the Animal Care and Use Committee (ACUC) of NIDA-IRP, NIH. Rats were euthanized with over dose isoflurane and decapitated upon cessation of breathing. Rat brains were rapidly removed from the skull and frozen in isopentane pre-chilled on dry ice for 10–15 second, then wrapped in aluminum foil, and stored at −80 °C until sectioning.
Tissue Sectioning and Handling
Before starting sectioning, the frozen rat brains were transferred from the −80 °C freezer to the cryostat chamber (CM 3050S; Leica Microsystems Nussloch GmbH, Nussloch, Germany) at −20 °C for 45 minutes to thermally equilibrate the tissue. The brains were trimmed and mounted on the sample stage using water-ice slush, and cut into 16 µm sections.[20] Brain sections were collected onto MALDI stainless steel target plate inserts and briefly thaw-mounted, then stored at −80 °C until matrix deposition. Shortly before matrix deposition, the target inserts were placed in a −20 °C freezer for 20 minutes, then immediately placed in a vacuum desiccators for 20–30 minutes to warm up to room temperature while preventing water condensation over the target inserts and brain sections.
Matrix deposition
MALDI matrix solution was deposited onto target inserts with a TLC reagent sprayer (Kimble Kontes, product number 422530-0050, Vineland, NJ), a Collison Nebulizer (BGI Inc., Waltham, MA; product number CN24), or an artistic airbrush (Aztek A470/80 Airbrush System, The Testors Corporation, Rockford, IL).[19] Each apparatus was pre-adjusted to deliver the finest possible mist for matrix deposition and spraying. DHB matrix was sprayed onto the target inserts in a well ventilated chemical hood. When sprayed with TLC sprayer and airbrush, the spraying conditions and distance were adjusted to prevent wetting of tissue surface and target insert. The TLC sprayer was driven by a compressed air cylinder with 3–5 psi of pressure according to the manufacturer’s recommendation, while the airbrush was driven by a mini air pump purchased from the air brush manufacturer (Blue Mini Airbrush Compressor, The Testors Corporation, Rockford, IL; maximum pressure: 40 psi). The Collison Neulizer was also driven by a compressed air cylinder at a pressure of less than 20 psi. Each target insert was sprayed with 5–6 spray cycles. A two-minute wait between spray cycles was observed to ensure sufficient evaporation of organic solvents from the target insert surface. One spray cycle is defined as an organized rastering of spraying that results in complete coverage of the target insert surface by the spraying mist. The distance between the nozzle of airbrush and the target plate was kept between 15 and 20 cm. The matrix was perpendicularly sprayed onto the target plate. When spraying with the Collison Nebulizer, the extent of matrix deposition was visually inspected throughout the matrix deposition process.
Mass Spectrometer
An Applied Biosystems 4700 Proteomic Analyzer coupled with 4000 series MSI image acquisition software [21–23] (Norvatis, Basel, Switzerland; available at http://www.maldi-msi.org/) was used for MALDI-IMS data acquisition. The images of phosphatidylinositol and sulfatides were collected in negative reflectron mode, while the images of phosphatidylcholines were acquired in positive reflectron mode. The target plate stepping distance was set to 100 µm for both the X and Y directions by the MSI image acquisition software. In negative ion mode, each spot (spectral pixel) was an average of 160 laser shots across m/z 650 to 1050. For positive ion mode, each spot was an average of 176 laser shots across m/z 700 to 905. A test of laser ablation pattern over a blank target insert sprayed with matrix confirmed that the stepping distance at either axis did not overlap. The two dimensional ion density maps (i.e., images) were generated using BioMap software (Norvatis, Basel, Switzerland; also available free at http://www.maldi-msi.org/) and exported as TIFF image files for subsequent image cropping and resizing using GNU Image Manipulation Program (GIMP; Version 2.2.12; available free at http://www.gimp.org/) running on a PC.
Light microscope photography
The surface pictures of target insert after matrix deposition were taken with a stereo zoom dissecting microscope (Nikon SMZ 1500) coupled with a digital camera (Nikon DXM 1200F) to visualize the surface of the target plates after matrix deposition.
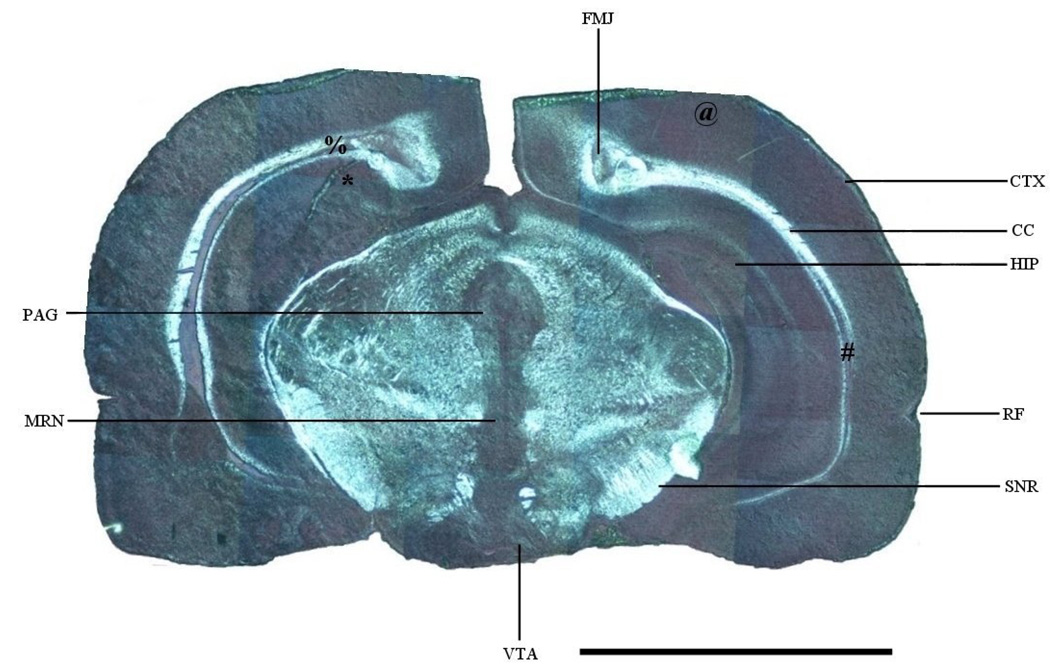
For the light microscope picture of a brain section, digital pictures containing a field of view that covered a partial area of the brain section were sequentially taken directly from a target insert under one setting using a laboratory compound microscope (Leica DMR Microsystems, Wetzlar GmbH, Germany) with a 10× objective lens. The section was illuminated by epiluminescence method under a long wavelength UV light that permits a direct distinction between gray and white matter from an unprocessed brain section. The images were captured with a digital camera (Nikon DXM 1200F) and reassembled offline using the GIMP program to reconstruct the brain section image (Figure 1).
Figure 1.
Surface light microscope pictures of a target plate sprayed with DHB. Left panel: Sprayed with TLC sprayer. Middle panel: Sprayed with Collison Nebulizer over a tissue section. Note the section was not covered by the matrix. Right panel: Sprayed with air brush. Note the homogeneous deposition of matrix over the target surface as compared to the TLC deposition method. Scale bar: 5 mm.
Results
Different techniques were used by various groups to deposit matrix onto tissue sections for MALDI-IMS studies. In this work we evaluated three matrix deposition methods: spraying with a TLC sprayer, a Collision Nebulizer, or an artistic airbrush to compare their suitability for brain lipid imaging.
Although widely accepted as a matrix deposition method for MALDI-IMS application, the TLC sprayer appeared to generate coarse and unevenly sized droplets in spite of careful adjustment of the air flow and pressure of the nebulizing gas. The result was easily observed from the target insert surface’s appearance (Figure 1, left panel) and tissue sprayed with the TLC sprayer gave poorly defined images. The Collison Nebulizer, on the other hand, generated a fine mist that is most suitable for matrix deposition for MALDI-IMS studies. Unfortunately, most of the matrix only deposited onto the tissue-free region of the target plate (Figure 1, middle panel). Further, the spraying process consumes excessive amount of matrix solution, yet deposits disproportionately little amount of matrix onto the target plate. Such inefficiency prolongs the spraying duration for over one hour in order to cover a brain section with enough matrix, and exposes the tissue section to room temperature for too long a time before sufficient matrix can be deposited. Overall, the disadvantages incurred by Collison Nebulizer outweighed its advantage in particle size, thus excluding it as a suitable means of MALDI-IMS matrix deposition, as we were never able to obtain publication quality images.
The airbrush spraying seems to be a compromise of the above two methods and also offers several advantages not shared by the above two. The airbrush appears to generate more homogenous droplet size than the TLC sprayer could (Figure 1, right panel). When spraying onto tissue sections, this method rarely causes uneven matrix deposition. Further, matrix deposition by airbrush can be carried out in a timely manner without consuming excessive amount of matrix solution. When the distance between the target plate and the airbrush nozzle is properly adjusted, airbrush spraying does not cause surface over wetting as was cautioned by other investigators.
Figure 2 shows the light microscope image of a brain section that was also used to generate images in Figure 5. Due to the nature of the light source and illumination method, the gray matter in this figure emits a metallic grayish color, while the white matter shows a bright, white color. The color contrast significantly enhances the distinction between gray and white matter. The major anatomical structures are also labeled for reference.
Figure 2.
Light microscope photograph of a rat brain section. CC: corpus callosum; CTX: cortex; FMJ: forceps major of corpus callosum; HIP: hippocampus; MRN: midline raphe nuclei; PAG: periaquiductal gray; RF: rhinal fissure; SNR: substantia nigra par reticulata; VTA: ventral tegmental area. *: Figure 4A region; #: Figure 4B region; @: Figure 6A region; %: Figure 6B region. Scale bar: 5 mm.
Figure 5.
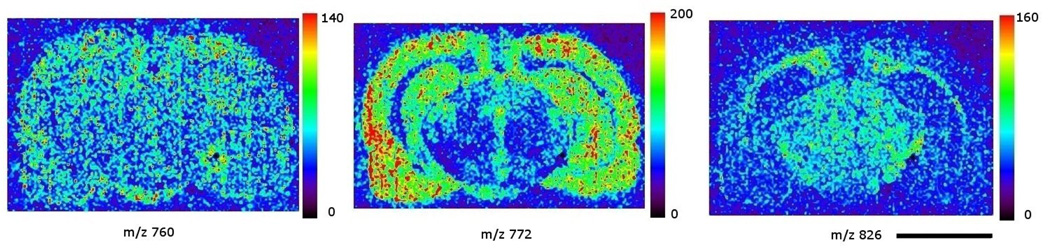
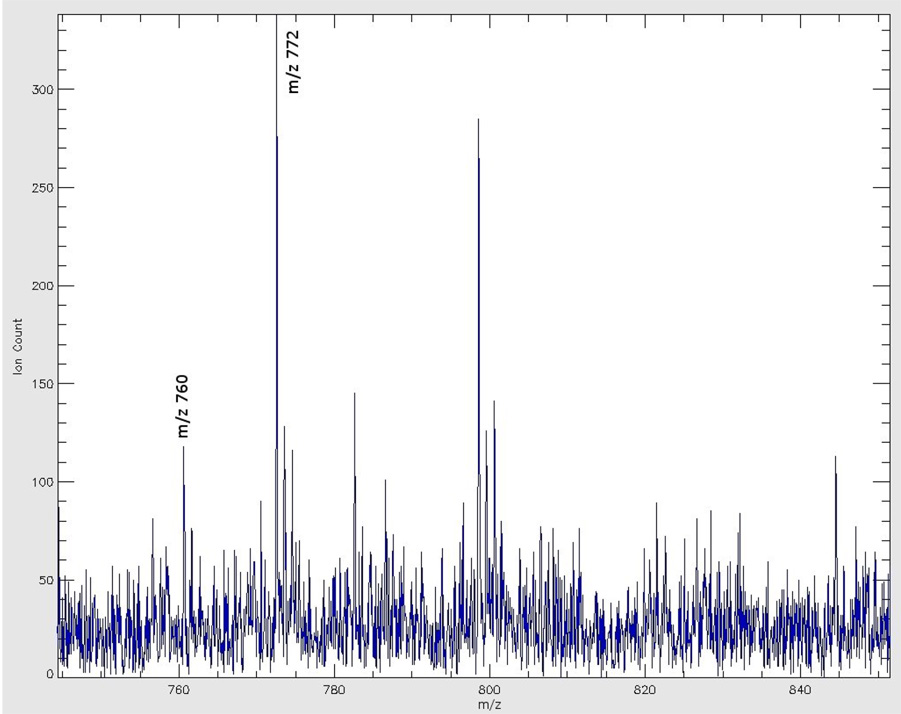
images of three common lipids acquired in positive ion mode: left panel: phosphatidylcholine 34:1, m/z 760; middle panel: potassiated phosphatidylcholine 32:0, m/z 772; right panel: phosphatidylcholine 36:1, m/z 826. The color scale to the right of each panel represents the raw ion count. Scale bar: 5 mm.
Imaging of phosphatidylinositol and sulfatides in rat brain section
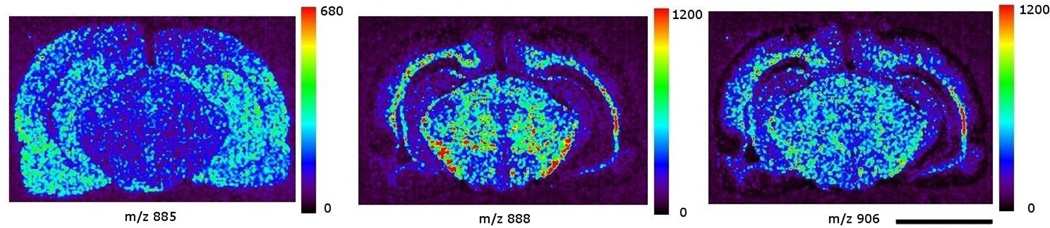
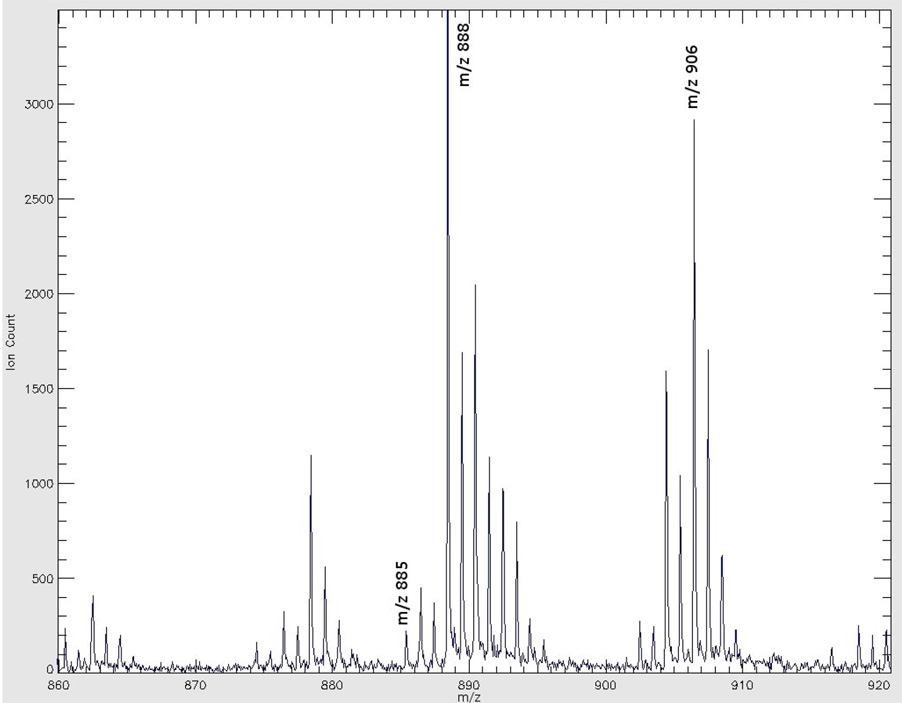
Images in Figure 3 show the distribution of three most commonly encountered brain lipid species in negative ion mode MS. The tissue section used for this group of images was 16 µm caudal to (deeper than) the section in Figure 2. There was no apparent anatomical structure difference between these two sections. The images were extracted from a single data file that covered spectral data from m/z 650 to 1050. The left panel of Figure 3 shows the image of m/z 885, the molecular ion of phosphatidylinositol 38:4 (PI 38:4), which is present in large amounts in gray matter regions such as cortex and hippocampus. The middle and right panels show the image of m/z 888, the molecular ion of sulfatide 24:1 (ST 24:1), and m/z 906, the molecular ion of hydroxy sulfatide 24:1 (ST 24:1 (OH)), respectively. Both sulfatide species were detected in high abundance in white matter. The moderate abundance of both ST species in midbrain also reflects the anatomical nature where gray matter regions such as nuclei are intertwined with white matter structures like tracts, bundles, and fasciculi.
Figure 3.
images of three phospholipids acquired in negative ion mode: left panel: phosphatidylinositol 38:4, m/z 885; middle panel: sulfatide 24:1, m/z 888; right panel: hydroxy sulfatide 24:1, m/z 906. The color scale to the right of each panel represents the raw ion count. Scale bar: 5 mm.
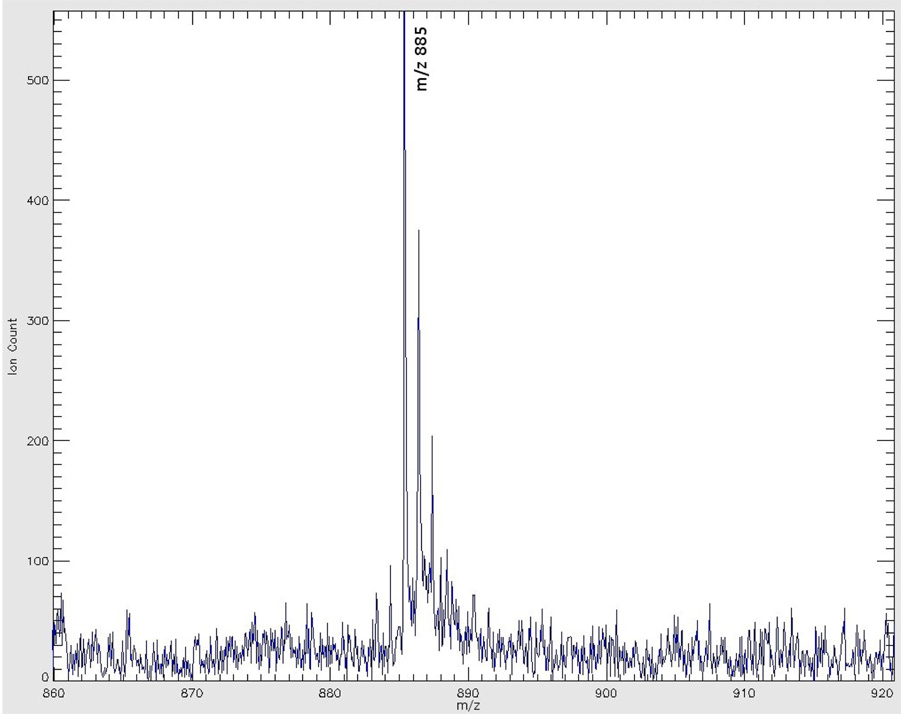
Figure 4A shows the mass spectrum acquired from the left hippocampus (a gray matter region) right underneath the forceps major of the corpus callosum seen in Figure 2. Figure 4B shows a mass spectrum of the right corpus callosum, a white matter region, noted in Figure 2. The mass range for both figures m/z 860 to 920, covers the range of major glycolipid species detected under negative ion mode. Both spectra show the regionally specific spectral profiles pertinent to gray and white matters. Table 1 lists the 11 lipid species identified in this imaging experiment.
Figure 4.
Figure 4A: Negative ion mode MALDI TOF spectrum acquired from gray matter region indicated in Figure 1.
Figure 4B: Negative ion mode MALDI TOF spectrum acquired from white matter region indicated in Figure 1.
Table 1.
Lipid species identified in negative ion mode MALDI-IMS.
| Species | Theoretical m/z | Actual m/z |
|---|---|---|
| ST 18:0 | 806.55 | 806.62 |
| PS 40:6/ ST 20:0 | 834.55 | 834.49 |
| PI 36:4 | 857.52 | 857.38 |
| ST 22:0 | 862.61 | 862.48 |
| ST 22:0 (OH) | 878.60 | 878.47 |
| PI 38:4 | 885.55 | 885.51 |
| ST 24:1 | 888.62 | 888.50 |
| ST 24:0 | 890.64 | 890.55 |
| ST 24:1 (OH) | 904.62 | 904.49 |
| ST 24:0 (OH) | 906.63 | 906.56 |
Imaging of phosphatidylcholines in rat brain section
Most of the lipids identified in positive ion mode were phosphatidylcholines (PCs). Images in Figure 5 show the distributions of the three most commonly encountered lipid species in positive ion mode. The images were extracted from a single data file containing mass spectral information from m/z 700 to m/z 905. The left panel shows an image of m/z 760, molecular ion of PC 34:1 that is omnipresent in the brain. The middle panel shows the image of m/z 772, potassiated PC 32:0 species, which is present in high abundance in gray matter. The right panel shows the image of m/z 826, potassiated PC 36:1, a PC species predominantly present in white matter. Figure 6A shows the mass spectrum acquired from the right parietal cortex, a gray matter region, labeled in Figure 2. Figure 6B shows the mass spectrum from the tail of forceps major from the left corpus callosum, a white matter region, as labeled in Figure 2. Table 2 lists 6 lipid species identified by this imaging result.
Figure 6.
Figure 6A: Positive ion mode MALDI-TOF spectrum acquired from the gray matter region indicated in Figure 1.
Figure 6B: Positive ion mode MALDI-TOF spectrum acquired from the white matter region indicated in Figure 1.
Table 2.
Lipid species identified under positive ion mode MALDI IMS.
| Species | Presentation | Theoretical m/z | Actual m/z |
|---|---|---|---|
| SM 18:0 | [M+K]+ | 769.56 | 769.71 |
| PC 32:0 | [M+Na]+ | 756.55 | 756.64 |
| [M+K]+ | 772.53 | 772.62 | |
| PC 34:1 | [M+H]+ | 760.59 | 760.71 |
| [M+K]+ | 798.54 | 798.66 | |
| PC 36:1 | [M+K]+ | 826.57 | 826.72 |
| PC 36:4 | [M+H]+ | 782.57 | 782.66 |
| [M+K]+ | 820.53 | 820.65 | |
| PC 38:6 | [M+K]+ | 844.53 | 844.63 |
It is interesting to note that the image of m/z 772 in the midbrain region (Figure 5, middle panel) roughly reflects the location of gray matter areas such as the midbrain periaquiductal gray (PAG), the midline raphe nuclei (MRN), and the ventral tegmental area (VTA). The white matter regions, such as corpus callosum and substantia nigra par reticulata (SNR) are seen in the same image as low intensity regions. The white matter structures, on the other hand, are well represented by the m/z 826 image (right panel, Figure 5). Compared to the light microscope picture in Figure 2, the contours of these midbrain regions outlined by ion intensity signals seem to correspond relatively well with the regions in the light microscope picture.
Discussion
One of the most critical steps in tissue MALDI-IMS studies is a well-executed matrix deposition on the tissue section, which permits homogeneous mixing and re-crystallization between analytes and matrix, without causing any noticeable displacement or redistribution of biomolecules. Several investigators have demonstrated various approaches addressing this issue. Airbrush deposition, among the commonly accessible matrix deposition methods, appears to carry significant advantages over others that deserve further evaluation in spite of the preference of other investigators.[19] The two key elements in successful matrix deposition by airbrush are sufficient matrix delivery to the tissue surface while preventing section over wetting which leads to delocalization of analytes. Spraying matrix prepared in high concentration of organic solvent in a well-ventilated chemical hood will speed up the matrix droplet evaporation, thus limiting the extent of analyte delocalization, if any. Properly adjusted distance between the airbrush nozzle and target plate will control the density of the droplets deposited onto the target, and enhances matrix droplet drying. Spraying with high concentration of matrix solution also reduces the number of spraying cycle. Together, these measures could prevent excessive extraction and redistribution of endogenous molecules in tissue sections.
The lipid images in this study revealed the distribution of a phosphatidylinositol, two sulfatides, and three phosphatidlycholines commonly encountered in brain. Their distribution images follow the contours of white and gray matters. The results correspond to our earlier profiling studies, [20, 25, 26] and agree with the conclusion from several early quantitative studies.[27–29] The images of two sulfatide species and the potassiated PC 36:1 clearly outline the white matter regions from the surrounding gray matter structures. The gray matter images of PI 38:4 and potassiated PC 32:0, on the other hand, show a complementary pattern to the sulfatide and potassiated PC 36:1 images.
In the images of ST 24:1 and ST 24:1 (OH), one could follow the extreme dim band immediately medial to the cortical surface to identify the rhinal fissure (RF, Figure 2) and the surrounding convolutions that fold into this fissure. The location of this dim band appears to be in agreement with the description of the cerebrum gray cortical band.[30] The sub-cortical regions medial to this dim band contain projection, association, and commissural fibers whose aggregation results in white matter regions. A good example of such aggregation in the white matter is the corpus callosum, the most prominent commissural fiber structures in the brain. This compacted neural fibers structure generated high intensity signals of STs and postassiated PC 36:1. The region sandwiched between the cortical dim band and corpus callosum shows a moderate intensity of ST signals which reflect the underlying moderate aggregation of neural fibers, and further contrasts the lack of white matter material in the grey cortical band.
The two ST images in our study were significantly different from the images obtained in an earlier TOF-SIMS study on lipid distribution, where the authors barely detected ST signals in the corpus callosum.[17] However, the same study showed a similar pattern of ST signals in the sub-gray cortical band regions. The discrepancy could be attributed to the following: First, the section preparation and handling are quite different between these two studies. In our study, the sections were not rinsed or washed with any reagents. The brain sections in the mentioned TOF-SIMS study were rinsed in NH3HCOO before refreezing. This additional rinsing and refreezing may potentially redistribute, or wash away molecules on the section surface, or change the tissue surface characteristics. Second, the use of MALDI matrix and the resulting solvent extracting effect, even if trivial, permits a deeper penetration into tissue sections.[31] The SIMS technique, on the other hand, usually profiles no deeper than a few hundred nanometers beneath the sample surface.[18] Since the cellular and interstitial content in a brain section is three-dimensionally heterogeneous, different mass spectrometry profiling depth over the same surface may lead to different outcome.
It is interesting to observe the apparent overlapping distribution of ST 24:1 and ST 24:1 (OH) in the brain section. The relative abundance of these two sulfatides in Figure 3 and Figure 4B both indicate that ST 24:1 is slightly more abundant than ST 24:1 (OH) in white matter. This observation agrees with earlier studies where the abundance of ST 24:1 in white matter was quantitatively determined to be slightly higher than that of ST 24:1 (OH).[28, 32] In positive ion mode the preferential distribution of a common phospholipid species like PC is demonstratrd. PC differential distribution in these images also agree with an early study on the distribution of PC in gray and white matter,[33] and further confirmed our previous lipid profiling results.[24] Structurally, these three PC species only differ in their fatty acid chain length and the number of double bonds. When subjected to collision-induced dissociation (CID), all three species produce a m/z 184 fragment ion (phosphocholine). However, the same phosphocholine fragment ion can also come from CID fragmentation of sphingomyelins [34, 35] that are present in higher abundance in white matter and myelin.[28, 29] Therefore, it may not be suitable to associate the distribution of PCs with the distribution of this fragment ion as was proposed by other investigators.[16, 17, 36] Perhaps a more appropriate imaging representation for PC and sphingomyelin distribution should be based on signals f intact molecules, or species-specific fragments.
Generally speaking, 2,6-dihydroxyacetophenone (DHA) is a more suitable matrix than DHB for MALDI-MS study of lipids. However, DHA usually sublimes in high vacuum within 30 minutes [26], and a MALDI-IMS experiment usually takes more than 5 hours to collect quality data from a 12mm×10 mm area that covers the brain section shown in this study. Such a prolonged data collecting process precludes DHA as a suitable matrix for this imaging study. Therefore, choosing DHB as the matrix is a compromise between adapting optimal conditions for MALDI-MS lipid study and the matrix vacuum stability. Although not as optimal, DHB still allowed us to identify a total of 18 lipid species.
All five phosphatidylcholines were identified as potassiated adducts in positive ion mode. This phenomenon could possibly be due to unavoidable introduction of exogenous potassium during the sample preparation process (matrix, or airbrush, or other sources). Alternatively, those prominent potassium adducts may simply reflect a high abundance of endogenous potassium in brain parenchyma, [37] since 82% of the brain water is confined intracellularly where potassium is the major cation electrolyte.
In this, and other MALDI-IMS studies, the diameter of the laser spot on the target plate is between 100 and 200 µm, and the lateral displacement by the sample stage motor is generally around 100–200 µm. Hence the “spectral pixel” size, i.e., the lateral resolution, is limited by these parameters. The size of spectral pixel is at least twice as large as the diameter of an average cell. Such lateral resolution should be sufficient for imaging across an organ sections prepared for distribution studies. The diameter of primary ion beam in SIMS, on the other hand, could be as narrow as a few micronmeters. Such small impact area could offer a much higher lateral resolution [16] that is more suitable for molecular imaging at the cellular level.
One of the intuitive approaches to enhance the lateral resolution in MALDI IMS is to refocus the laser beam down to a narrower diameter. Although theoretically possible, this approach may ablate more than ionize the target, resulting in a reduction of detection sensitivity.[38] Adjusting the optical modulator on the Nd:YAG laser path could drastically change the energy profiles across the laser beam and significantly improve the above-mentioned shortcomings. However, due to its complexity in optical modulation and instrument liability issue, this approach may not be readily available to most of the bioanalytical laboratories.
In conclusion, by increasing ionization efficiency and selecting matrices of sufficient vacuum stability, MALDI-IMS will broaden its application into lipid distribution studies in normal and pathophysiological conditions, and further extends its advantages for generation of image profiling studies with much higher specificity than what can be had with conventional approaches.
Acknowledgment
This research was supported by the Intramural Research Program of National Institute on Drug Abuse, NIH. The authors thank the Office of National Drug Control Policy (ONDCP) for instrumentation funding, without which this and other projects could not have been accomplished. We also would like to thank Dr. Patricia Tagliaferro and Dr. Marisela Morales for their assistance in light microscope photography.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edmonson RD, Russell DH. High-Resolution Mass Spectrometry and Accurate Mass Measurement of Biopolymers Using MALDI-TOF. In: Larsen BS, McEwen CN, editors. Mass Spectrometry of Biological Materials. 2nd ed. New York: Marcel Dekker; 1998. pp. 29–52. [Google Scholar]
- 2.Chaurand P, Schwartz SA, Caprioli RM. Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr. Opin. Chem. Biol. 2002;6(5):676–681. doi: 10.1016/s1367-5931(02)00370-8. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell RL, Caprioloi RM. Tissue profiling by mass spectrometry: a review of methodology and applications. Mol. Cel. Proteomics. 2005;4(4):394–401. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioloi RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 2003;38(10):1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 5.Troendle FJ, Reddick CD, Yost RA. Detection of pharmaceutical compounds in tissue by matrix-assisted laser desorption/ionization and laser desorption/chemical ionization tandem mass spectrometry with a quadrupole ion trap. J. Am. Soc. Mass Spectrom. 1999;10(12):1315–1321. [Google Scholar]
- 6.Wang HYJ, Jackson SN, McEuen J, Woods AS. Localization and analyses of small drug molecules in rat brain tissue sections. Anal. Chem. 2005;77(20):6682–6686. doi: 10.1021/ac050868d. [DOI] [PubMed] [Google Scholar]
- 7.Han J, Schey KL. MALDI tissue imaging of ocular lens alpha-crystallin. Invest. Ophthal. Vis. Sci. 2006;47(7):2990–2996. doi: 10.1167/iovs.05-1529. [DOI] [PubMed] [Google Scholar]
- 8.Groseclose MR, Andersson M, Caprioli RM. Toward identification of proteins directly from tissue: on-tissue tryptic digestions coupled with imaging mass spectrometry; 54th ASMS Conference on Mass Spectrometry and Allied Topics; Seattle, WA: 2006. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh Y, Casale R, Fukada E, Chen J, Knemeyer I, Wingate J, Morrison R, Korfmacher WA. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun. Mass Spectrom. 2006;20(6):965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 10.Khatib-Shahidi S, Andresson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 2006;78(18):6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 11.Mullen AK, Clench MR, Crosland S, Sharples KR. Determination of agrochemical compounds in soya plants by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19(18):2507–2516. doi: 10.1002/rcm.2078. [DOI] [PubMed] [Google Scholar]
- 12.Krause R, Sweedler JV. Spatial profiling invertebrate ganglia using MALDI MS. J. Am. Soc. Mass Spectrom. 2003;14(7):752–759. doi: 10.1016/S1044-0305(03)00288-5. [DOI] [PubMed] [Google Scholar]
- 13.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 2006;78(3):827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 14.Stoeckli M, Staab D, Capretta S. Automated mtrix deposition for MALDI MSI; 54th ASMS Conference on Mass Spectrometry and Allied Topics; Seattle, WA: 2006. [Google Scholar]
- 15.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Detection Technologies. Ambient mass spectrometry. Science. 2006;311(5767):1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 16.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J. Mass Spectrom. 2001;36(4):355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 17.Sjövall P, Lausmaa J, Johansson B. Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 2004;76(15):4271–4278. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 18.Cheng J, Wucher A, Winograd N. Molecular depth profiling with cluster ion beams. J. Phys. Chem. B: Condens. Matter Mater. Surf. Interfaces Biophys. 2006;110(16):8329–8336. doi: 10.1021/jp0573341. [DOI] [PubMed] [Google Scholar]
- 19.Garrett TJ, Yost RA. Imaging phospholipids in brain tissue by intermediate-pressure MALDI/MSn on a linear ion trap; 54th ASMS Conference on Mass Spectrometry and Allied Topics; Seattle, WA: 2006. [Google Scholar]
- 20.Jackson SN, Wang HYJ, Woods AS, Ugarov M, Egan T, Schultz JA. Direct tissue analysis of phospholipids in rat brain using MALDI-TOFMS and MALDI-ion mobility-TOFMS. J. Am. Soc. Mass Spectrom. 2005;16(2):133–138. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 22.Stoeckli M, Farmer TB, Caprioli RM. Automated mass spectrometry imaging with a matrix-assisted laser desorption ionization time-of-flight instrument. J. Am. Soc. Mass Spectrom. 1999;10(1):67–71. doi: 10.1016/S1044-0305(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 23.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001;7(4):493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 24.Jackson SN, Wang HYJ, Woods AS. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal. Chem. 2005;77(14):4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- 25.Jackson SN, Wang HYJ, Woods AS. In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 2005;16(12):2052–2056. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Jackson SN, Wang HYJ, Woods AS. In Situ Structural Characterization of Glycerophospholipids and Sulfatides in Brain Tissue Using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 2006 doi: 10.1016/j.jasms.2006.08.015. in press. [DOI] [PubMed] [Google Scholar]
- 27.O’Brien JS, Fillerup DL, Mead JF. Brain lipids: I. Quantification and fatty acid composition of cerebroside sulfate in human cerebral gray and white matter. J. Lipid Res. 1964;15:109–116. [PubMed] [Google Scholar]
- 28.O’Brien JS, Sampson EL. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J. Lipid Res. 1965;6(4):537–544. [PubMed] [Google Scholar]
- 29.Suzuki K. Chemistry and metabolism of Brain Lipids. In: Albers RW, editor. Basic Neurochemistry. 1st ed. Boston: Little, Brown, and Co.; 1972. pp. 207–227. [Google Scholar]
- 30.Duus P. Topical Diagnosis in Neurology. 2nd. rev. ed. Stuttgart: Georg Thieme Verlag; 1989. pp. 251–256. [Google Scholar]
- 31.Crossman L, McHugh NA, Hsieh Y, Korfmacher WA, Chen J. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. 2006;20(2):284–290. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien JS, Rouser G. The fatty acid composition of brain sphingolipids: sphingomyelin, ceramide, cerebroside, and cerebroside sulfate. J. Lipid Res. 1964;5(3):339–342. [PubMed] [Google Scholar]
- 33.Svennerholm L. Distribution and fatty acid composition of phosphoglycerides in normal human brain. J. Lipid Res. 1968;9(5):570–579. [PubMed] [Google Scholar]
- 34.Hsu FF, Turk J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2000;11(5):437–449. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- 35.McDonnell LA, Heeren RM, de Lange RP, Fletcher IW. Higher sensitivity secondary ion mass spectrometry of biological molecules for high resolution, chemically specific imaging. J. Am. Soc. Mass Spectrom. 2006;17(9):1195–1202. doi: 10.1016/j.jasms.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Luxembourg SL, McDonnell LA, Duursma MC, Gou X, Heeren RM. Effect of local matrix crystal variations in matrix-assisted ionization techniques for mass spectrometry. Anal. Chem. 2003;75(10):2333–2341. doi: 10.1021/ac026434p. [DOI] [PubMed] [Google Scholar]
- 37.Cserr HF, DePasquale M, Nicholson C, Patlak CS, Pettigrew KD, Rice ME. Extracellular volume decreases while cell volume is maintained by ion uptake in rat brain during acute acute hypernatremia. J Physiol. 1991;442:277–295. doi: 10.1113/jphysiol.1991.sp018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holle A, Haase A, Kayser M, Hohndorf J. Optimizing UV laser focus profiles for improved MALDI performance. J. Mass Spectrom. 2006;41(6):705–716. doi: 10.1002/jms.1041. [DOI] [PubMed] [Google Scholar]