Abstract
Extensive growth in the field of molecular biology in recent decades has led to the development of new and powerful experimental and computational tools that enable the analysis of complex biological responses to chemical exposure on both a functional and structural genetic level. The ability to profile global responses on a transcriptional level has become a valuable resource in the science of toxicology and attempts are currently being made to further understand toxicity mechanisms by incorporating metabolomics and proteomics approaches. In addition, recent progress in understanding the extent of the genetic diversity within and between species allows us to take a fresh look at research on genetic polymorphisms which may influence an individual’s susceptibility to toxicity. While new technologies have the potential to make a sizeable impact on our understanding of the mechanisms of toxicity, considerable challenges remain to be addressed, especially with regards to the regulatory acceptance and successful integration of omics data. This review highlights recent advancements in the application of functional and structural genomics techniques to chemical hazard identification and characterization, and to the understanding of the inter-individual differences in susceptibility to adverse drug reactions.
Keywords: toxicogenomics, toxicogenetics, susceptibility, systems biology, high-throughput toxicity screening
1. Introduction
Advances in genomics in recent decades have led to exponential growth in the understanding of genome structure and function in both humans and other organisms. Due to the wealth of resources available, including databases of sequence variation and transcriptional changes following chemical exposure, both the industry and the federal government have begun to invest more resources into using genomic tools to improve drug development and drug and chemical safety evaluation. Systems toxicology, or the integration of traditional toxicology approaches with the development and implementation of toxicogenomics, proteomics, and metabolomics, offers the promise of the development of novel reliable toxicity biomarkers and more accurate predictions of adverse health effects in humans.
The need for development of sensitive biomarkers to predict drug toxicity was recently reinforced by the Food and Drug Administration (FDA) Critical Path Initiative which emphasizes the development of diagnostic tests to improve clinical trials and post-market surveillance1. In 2005, the FDA released a final guidance document on genomic data submissions, Guidance for Industry: Pharmacogenomic Data Submissions2, and announced the creation of a new FDA program entitled the Interdisciplinary Pharmacogenomic Review Group3. The importance of genomic information in FDA regulatory decisions is reflected in the increasing voluntary inclusion of these types of data in FDA submissions3. To establish reliable protocols for data analysis and quality control in genomics studies, several consortia have been established which included the academia, industry, FDA, Environmental Protection Agency, the National Institute of Standards and Technology, and the National Institutes of Health4–6. In the near future, a strong understanding of genomic data and its potential uses will be critical to the drug development process and clinical practice. The literature reviewed here focuses on the current progress in the application of genomic and genetic data to drug safety and highlights recent successes and current knowledge gaps in the following areas: 1) the discovery of toxicity biomarkers and identification of compounds that are predicted to cause toxicity, 2) the identification of susceptible individuals, and 3) integration of various omics datasets to improve understanding of toxicity mechanisms within a biological system.
2. Functional Genomics Strategies
2.1 Functional genomics allows for global gene transcript profiling
The major challenge of toxicology today is the ability to extrapolate risk from experimental systems to human populations. Rodents are most frequently used for in vivo toxicity testing, yet there often are major differences in clearance, metabolic activity of enzymes and other key factors between species. While it is difficult to directly translate rodent findings to human population, human testing is most often not an option and the answer to this problem is to develop biomarkers of toxicity that are sub-clinical and that can be assayed from a non-invasive tissue. The promise of toxicogenomics is to identify gene expression patterns or signatures that will indicate an adverse health event at low doses. However, to do this, it needs to be determined which changes and pathways are best associated with impending pathology vs. benign adaptive changes that are responsive to the chemical but are not associated with toxicity.
At the forefront of these emerging technologies is the use of functional genomics or analysis of molecular perturbations as measured by transcriptomics assayed by microarray technology. Gene expression analysis using current microarray platforms that encompass whole genomes allows for a comprehensive picture of cellular responses7, especially when examined in concert with proteomic and metabolomic data. While early microarray experiments were often criticized as “fishing expeditions”, current testing strategies enable the generation of testable hypothesis that aid in understanding the mechanism of toxicity8. The ultimate value of using transcript profiling data within the context of systems toxicology is that responses that are predictive of cellular injury can be sampled across doses, time points, and species, facilitating risk extrapolation to human populations.
2.2 Improving safety assessment through genomic profiling
Microarray profiling has become a major omics tool for the characterization of drug toxicities by allowing for large-scale determination of gene expression changes associated with a defined pathology. A key step in toxicogenomics is the ability to link a chemical-elicited phenotype with gene expression changes, termed “phenotypic anchoring”9. Experimentally determined gene expression signatures can serve as a guide to determining biomarkers that are indicative of toxicological responses that may be as-yet sub-clinical, with no observable morphological changes10. To confirm the utility of the approach, Heinloth et al.11 demonstrated that patterns of gene expression perturbations observed at sub-toxic doses of acetaminophen in rats may indicate subtle cellular injury that was not detectable by histopathology or clinical chemistry methods within the liver. At toxic doses, expression changes in the same subset of genes associated with mitochondrial dysfunction and oxidative stress were more exaggerated and changes were detected in additional genes associated with these processes. These data indicate that gene expression profiling has the potential to identify subtle markers of cellular injury that precipitate overt organ toxicity.
2.3 Identification of biomarkers of toxicity
Identification of sensitive biomarkers that will assist in monitoring drug therapy for evidence of toxicity or therapeutic outcome, and (in acute poisoning cases) to predict exposure levels, is a critical gap where omics technologies hold promise. Genomic biomarkers of toxicity have recently been identified for a wide variety of toxicants including nephrotoxic agents12, testicular toxicants13, and for keratinocyte proliferation in papilloma murine skin model14, to name only a few. The potential of using this technology to identify safety biomarkers is great and may help to create better diagnostic tools for traditionally difficult toxicodynamic monitoring, such as in patients receiving immunosuppressive therapy15.
The majority of recent investigations have used microarrays to study toxicity in target organ tissue or in cultured cells. While these experiments often yield important insight into the mechanism of toxicity, they provide limited information for monitoring drug safety in patient populations through non-invasive means. To address this limitation, Bushel et al.16 investigated the utility of using gene expression signatures in peripheral blood as an early indicator of pathological changes in the liver following administration of varying doses of acetaminophen. In this study, a prediction algorithm using classifiers and a pattern-based method that was weighted toward non-injurious exposure levels was used to discriminate sub-toxic and toxic exposure doses. Characterization of acetaminophen-induced liver injury using gene expression profiles derived from blood was shown to better predict acute chemical exposure levels than clinical chemistry, hematology, or histopathology analysis, indicating that transcript profiles derived from blood may be a good marker for specific organ toxicity.
2.4 High-throughput chemical toxicity testing is facilitated by microarrays
Microarrays have also enabled high-throughput screening of chemicals for potential toxicity. Increased sensitivity to detect molecular signatures or “fingerprints” that are associated with a particular class of chemicals or toxic response in both in vitro17, 18 and in vivo19, 20 systems, coupled with data-rich output, offer promise in screening for potential adverse health effects. In addition, approaches have been developed that do not rely upon a priori knowledge of expression signatures using well-characterized reference compounds, instead providing an ab initio estimation of compound-induced liver injury based upon microarray transcript profiling21. Furthermore, gene expression profiling has been suggested to have better predictive power than standard toxicological endpoints in a two-year rodent bioassay to detect carcinogenic potential of compounds22.
To validate the technology as a tool that may be useful in the classification of compound toxicity, an in vivo approach has often been utilized, in which defined organ pathology was present. It is assumed that toxicants that elicit similar pathology or disease will elicit a common pattern of gene expression changes. Recent studies aimed at prediction of toxicity based upon gene expression signatures have shown some success at distinguishing between non-genotoxic and genotoxic carcinogenic hepatotoxicants23, and renal and hepatic toxicants24, and have enabled classification of hepatotoxicants that are macrophage activators, peroxisome proliferators, and oxidative stressors25.
Because an in vivo approach would be impractical for use in high-throughput toxicity screening, in vitro culture systems offer an attractive alternative. For example, it was demonstrated that gene expression signatures can distinguish between toxic and non-toxic chemicals in HepG2 cells26. However, until recently there was little data that suggested that toxicity signatures determined in an in vitro system would predict in vivo responses. The Toxicogenomics Project (TGP), a consortia composed of the Japanese National Institute of Health Sciences, the National Institute of Biomedical Innovation, and 15 pharmaceutical companies, was developed in 2002 to address the problem of connecting model systems with prediction of toxicity in humans27. From the data collected by the TGP on gene expression profiles collected from rat liver and rat and human hepatocytes treated with 150 chemicals, it became clear that a major challenge to overcome was the identification of species-specific and common responses. The possibility of an informational bridge connecting transcript responses between rat and human hepatocytes and rat liver in vivo was investigated after administration of coumarin27. In this experiment, gene expression profiles were compared first between in vivo rat liver samples and coumarin-treated rat hepatocytes to determine those genes that were induced by treatment and that were significant in both models and that served as an in vivo-in vitro bridge. Orthologous bridging genes were then measured in coumarin-treated human hepatocytes and a signature of 14 up-regulated and 11 down-regulated mRNA transcripts was identified. While more data is needed to connect species and model systems with risk assessment to humans, the approach represents an important step in enabling high-throughput chemical toxicity screening.
The ability of traditional cell cultures of primary or cultured cells to predict toxicity can also suffer from an inability to accurately model multi-cellular processes and the myriad effects of cell-cell communication. A precision-cut liver slice model was suggested as a promising model system28. Liver slices offer the advantage of containing all relevant cell types within their original spatial arrangements in which cellular communication is maintained29. The presence of all relevant cell types is particularly important because liver non-parenchymal have been linked to the mediation of liver toxicity for diverse chemicals such as alcohol30, 31, cyclohexamide32, thioacetamide33, and acetaminophen34. Prototype experiments using rat liver slices demonstrated good prediction for toxicity and pathology based on microarray data for liver slices treated with acetaminophen, lipopolysaccharide, and carbon tetrachloride and these data correlated well with in vivo profiles28; however experiments using human liver slices are needed to enhance risk estimates.
2.5 Pathway analysis aids in determination of the mode of action
A major knowledge gap in connecting gene expression profiles to classes of toxicants that elicit similar phenotypes is a lack of complete information on the complexity of cellular molecular pathways. Regulation of responses can be relatively straightforward (i.e., at the transcriptional level), but it can also be quite complex and controlled by multiple genes, proteins, and metabolites. Therefore, accurate identification of gene-gene interactions and regulation is essential for determining well-defined pathways that could serve as potential targets in therapeutic development or intervention. The importance of understanding subtle pathway changes and the need for applying such data to risk assessment have been underlined in the recent National Research Council’s report entitled “Toxicity Testing in the 21st Century”35. Transcriptional pathway-level analysis may prove essential for determining molecular perturbations that are associated with sub-clinical pathologies because alterations in gene expression are likely to be a more sensitive endpoint than the clinically observed toxicity.
There are several public databases available which aid in the annotation and interpretation of gene expression data in terms of cellular process, functions, and pathways. These databases include the Gene Ontology (GO; www.geneontology.org), Gene Map Annotator and Pathway Profiler (www.genmapp.org), the Science Signaling Connections Map (stke.sciencemag.org/cm/), BioCarta (www.biocarta.com/genes/index.asp), Reactome (www.genomeknowledge.org), and KEGG (www.genome.jp/kegg/pathway.html). Most often, these tools are used to associate a set of differentially expressed genes from a microarray experiment with a particular pathway with the goal of identifying key modes of action. Several groups have developed statistical methods for associating cellular pathways and annotations with gene expression changes such as GOMiner36, Significance Analysis of Functional Categories37, and Onto-Tools38. While the methods employed by these statistical techniques vary, each is aimed at assigning biological meaning to gene expression data. A broad understanding of the pathways affected by a variety of xenobiotics is essential for the future of drug safety assessment. A complete understanding of the molecular perturbations that precipitate a toxic pathology in vivo will enable a shift from high dose to low dose testing and may enable a better prediction of pathology from in vitro data when the same molecular pathways are affected.
3. Structural Genomics
3.1 Identifying sensitive individuals
Toxicogenetics is a discipline that evaluates genetic sequence variations that may impact individual’s susceptibility to toxicity. In the pharmaceutical arena, these efforts are largely aimed at identification of susceptible individuals within the population that enables personalized drug treatment and improves drug safety. It is well accepted that genetic variants affect responses to drugs39, 40. One of the first studies in this field compared plasma drug half-lives in identical and fraternal twin pairs and showed that greater differences existed between fraternal twins41. Recently, several more monogenic toxicogenetic traits have been reported42. Such research has yielded some success into the identification of genetic alleles that predict drug responses, for example it has been demonstrated that apolipoprotein E (APOE)-4/4 allele carriers are the worst responders to conventional Alzheimer’s disease treatments43.
Successes in this field have led to important regulatory action by the FDA that has allowed a number of drugs to remain on the market due to the availability of genetic testing. Current genetic tests include assigning the dose of 6-mercaptopurine based on the genotype of thiopurine S-methyltransferase (TPMT)44, 45, and the dose of warfarin based on the genotypes of vitamin K epoxide reductase complex (VKORC1)46 and CYP2C947. In the case of the anti-coagulant drug warfarin, the combination of VKORC1 haplotyping and CYP2C9 genotyping explained an estimated 25% and 6–10% of the variance in warfarin dose, respectively48. In fact, warfarin represents one of the first drugs for which toxicogenetic information (by VKORC1 haplotyping) better explained the dose variance than pharmacokinetic pharmacogenomic data (through CYP2C9 genotyping)49. The data on warfarin, therefore, represents a shift from monogenic toxicogenetic testing to a polygenic model, which is expected to be encountered with increasing frequency as research in this field expands to include a greater variety of pharmaceuticals.
The wealth of information on genetic polymorphisms now available through the Human Genome Project has led to a dramatic increase in studies that seek to connect genetic variants with toxicity and pharmacologic phenotypes. Because the base pair sequence variation among individuals averages to be about 1 in 500–1000 base pairs50, it is reasonable to expect that a significant number of genes will contain polymorphisms that contribute to disease and that many will play a role in adverse drug responses. Most of the current research focuses on the association between phenotype/disease and single nucleotide polymorphisms (SNPs). SNPs are an attractive choice for biomarkers of adverse responses because, unlike other factors which contribute to a toxicity phenotype such as age, co-morbidity, and environment, an individual’s genetic code remains stable throughout their lifetime. Genetic testing offers the potential of replacing empirical dose adjustment for many drugs that is based upon therapeutic assessment of pharmacologic or toxic effect after initial dosing. In addition, predictive genetic tests could also be of value in the drug development process by rescuing drugs that failed Phase III clinical trials due to toxicity within a subset of participants51. A key example of this is the genetic testing available to patients with HIV who are prescribed the drug abacavir, in which screening for major histocompatibility complex, class I, B (HLA-B)*5701 reduces the risk of hypersensitivity reactions52.
3.2 Toxicogenetic models and approaches
A number of publications have reported a significant association between SNPs and disease phenotypes in many areas of research; however few have been validated and have often been followed by reports that refute the original conclusion53. Reasons for discrepancies between genotype-and-phenotype are numerous, but are often due to a low sample size in the study, population stratification of alleles, and heterogeneity of phenotypic classification54, 55. To better facilitate a clinical translation of toxicogenetic data, efficient and validated strategies are needed. Clinical data has been used to determine the genetic basis of complex traits such as Parkinson disease56 and susceptibility to HIV infection57.
Classical approaches in this area have focused on mapping quantitative trait loci (QTL) within the genome that influence a specific phenotype. While there are many approaches to identify QTL, all involve a population of individuals with a measurable phenotype, a database of genotypic variation present within that population, and statistical measures that serve to link the magnitude of the phenotype with a specific genotype or polymorphism58. Classical approaches have often sought to utilize the genotypic and phenotypic diversity present in F2 or backcross mouse populations; however, this approach is limited by the necessity to genotype all individuals within a population. Due to the relatively low number of recombination events in F2 and backcross populations, identification of precise QTL locations is often more difficult. Recombinant inbred lines offer the advantage of fixed genomes, but these lines can be expensive to acquire and maintain.
A promising new alternative is a method known as genomic association mapping in classical inbred mouse lines, which takes advantage of the genetic variation that arose naturally across inbred mouse lines over decades of crosses and inbreeding by scientists and fanciers. In this approach, large SNP datasets for several dozen strains and a database of over 8 million SNPs for 15 inbred mouse strains have recently become publicly available59. Computational methods for this approach have been described58, 60.
Applications of genomic association algorithms to a toxicogenetic approach have been performed in vivo within inbred mouse strains to determine additional genetic factors that affect the metabolism of warfarin61. In that study, the metabolism of warfarin, specifically the generation of 7-hydroxywarfarin, was shown to vary across inbred strains and this phenotype was then computationally associated with the mouse genomic region that encodes for Cyp2c enzymes. Experimental validation narrowed the list of potential genomic candidates to show that Cyp2c29 polymorphisms altered hepatic protein expression of this enzyme. In a subsequent publication62, this same group demonstrated the utility of the approach for use in an in vitro drug biotransformation system in which genomic association was performed for downstream metabolism of testosterone and irinotecan across 15 mouse lines. The results of these studies showed that genetic variation within the Cyp2b9 and Ugt1a loci influenced the metabolism of α-hydroxytestosterone61 and irinotecan glucuronidation62, respectively; these results were then confirmed experimentally via recombinant enzymes. These results suggest that genomic association using inbred mouse strains has the potential to aid in identification of toxicogenetic alleles, however further studies that demonstrate a clear translation to human populations is required.
The genetic variation within panels of inbred mouse strains, while greater than that in RI lines, can be somewhat limited due to the fact that the majority of classical inbred strains are derived from Mus musculus domesticus. To address this concern, the Collaborative Cross mouse strains were recently developed and were designed to incorporate large genetic variation63. To develop this resource, a controlled breeding program was designed to randomize genetic elements among the progeny derived from eight parental strains. A recent study demonstrated that the genetic variation present in the Collaborative Cross represents the optimal polymorphism architecture for the study of systems biology when compared to RI lines or to panels of classical inbred strains, and was demonstrated to be more reflective of the genetic variation expected in natural populations64. Further characterization of this resource may improve the translational ability of toxicogenetic studies in rodents.
It has been suggested that genome-wide association studies that focus on only the top several “most significant” SNPs may have some limitations that could be overcome by a pathway-based approach. These limitations are that genes that contribute in a smaller, but still significant, amount to disease risk may be overlooked, and that variants that confer a large effect may not be included if hundreds of thousands of markers have been tested and the sample size is relatively small. Wang et al. have proposed a method in which the power to detect causal mechanisms of disease may be more robust because multiple contributing factors are considered together, as opposed to focusing on a few SNPs with the highest association score65. This approach, which combines genome-wide association with the gene-set enrichment algorithm66, identified a number of pathways that may be associated with Parkinson disease and age-related macular degeneration.
3.3 Expression QTL – bridging functional and structural genomics
Toxicogenomic and toxicogenetic approaches can be utilized in concert to study the regulation of gene expression that affects the differential toxicity susceptibility between individuals. When combined, these datasets have the potential to provide a mechanistic foundation for determining key pathways where adjuvant therapies could be developed for the prevention of toxicity in genetically sensitive individuals. While this field is still developing, a few proof-of-concept studies were recently published that have integrated QTL analysis with gene expression data; these include a determination of genes that were implicated in insulin resistance67 and in allergic asthma68. Notably, this approach has been adapted to the toxicology field by using recombinant BxD inbred lines derived from C57BL/6J and DBA/2J mice to uncover genes that regulate acute alcohol withdrawal69 and that regulate lipopolysaccharide (LPS)-induced airway disease70.
In a combined expression QTL (eQTL) approach, the gene expression across thousands of genes serves as the phenotype for genome-wide association studies. eQTL data generate a comprehensive picture of the regulation of gene expression on a global scale and of the differences that exist between sensitive and resistant individuals. A recent paper evaluated basal liver gene expression in the genetic context of BxD recombinant inbred and parental mouse strains to identify polymorphic local and distant QTL that control expression of a wide variety of genes in the liver69. When comparing this dataset to similar data generated from nervous system tissue71, tissue-specific differences in the regulation of mRNA transcript expression were identified.
4. Data integration between -omics technologies
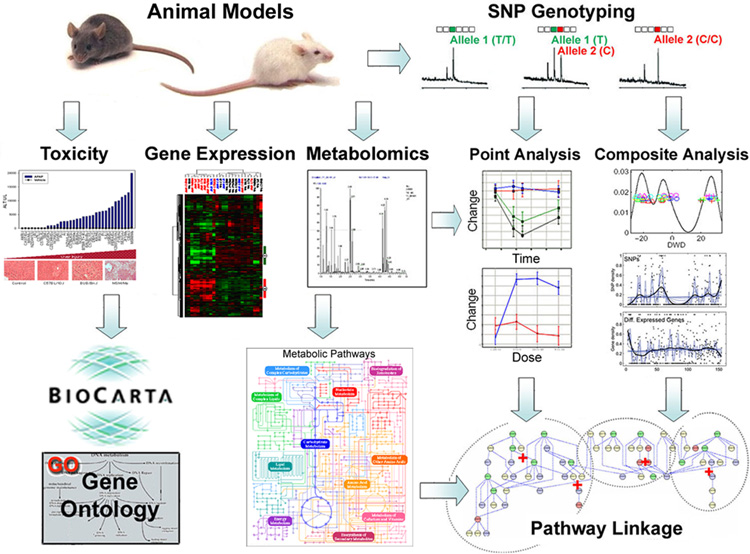
The need to better describe biological systems has led to the study of systems toxicology. Systems toxicology comprises the integration of genetics, toxicogenomics and conventional toxicity endpoints into a systems biology approach. These integrative approaches often use mouse models for an improved characterization of toxicity pathways, the discovery of new molecular and cellular indicators of exposure and outcome, better dose-response assessment and for more accurate inter-individual/cross-species extrapolations (Figure 1).
Figure 1.
Information flow in systems toxicology.
While microarray-based approaches in toxicity studies generate a wealth of data on gene expression and pathways that are affected by treatment or that are associated with a particular phenotype, these data are most often descriptive and may not reflect changes at the protein level. A key limitation of an analysis of toxicogenomic data alone is that these data often do not take into account confounding factors such as the pharmacokinetics of the test chemical72 and environmental factors, such the gut microflora population73. Genomic studies, while more comprehensive than high-throughput metabolite or protein analysis, fail to characterize the full complement of cellular proteins which are subject to post-translational modifications and additional regulation74. To address this concern and to develop additional toxicity biomarkers, metabolomics and proteomic approaches are gaining popularity, however metabolomics data is more frequently utilized due to its relative ease of use and interpretation.
Methods of data integration for orthogonal omics datasets fall broadly into two categories: 1) biology-driven strategies and 2) data-driven strategies75. Biological interpretations tend to be more hypothesis-driven and take advantage of a contextual understanding of what is currently known about the toxicity mechanism. An example of a biology-driven strategy is a study that integrated gene expression data with metabolomics to identify sensitive biomarkers of human neuroendocrine (NE) cancers76. In this approach, microarray and metabolite data was collected fromNE in vivo mouse tumors, mouse NE tumor cell cultures, and primary human NE tumors and the authors identified from the gene transcript data key metabolism pathways that were altered and validated by mass spectrometry.
Data-driven strategies are comprised of unsupervised methods such as principal components analysis, self-organizing maps, and regression-based partial-least square analysis. While biology-driven interpretations have been more commonly reported, data-driven techniques have been utilized to study diverse areas, such as hepatocellular metabolism77, the unique effects of anti-inflammatory compounds on macrophage cells78, and the identification of regulatory metabolites and gene transcription in glucosinolate and anthocyanin biosynthesis79. Another comprehensive example is a ‘pharmaco-metabolomic’ approach in which pre-dose metabolite profiles of urine and chemometrics were used to predict toxicity responses to acetaminophen in an in vivo rat model80. Genomic, proteomic and metabolomic approaches, when analyzed alone, are limited in their ability to describe the great degree of nonlinearity and complexity that is present within biological systems.
Databases and approaches are currently being developed that can utilize the synergy between these data types with the goal of placing toxicogenomics data into a larger biological perspective. These include the Comparative Toxicology Database (ctd.mdibl.org), the Distributed Structure-Searchable Toxicity database (www.epa.gov/ncct/dsstox/index.html), the Critical Path Institute (www.c-path.org), and systems biology databases developed by Genelogic, Inc. (www.genelogic.com), Iconix Pharmaceuticals (www.iconixbiosciences.com) and Ceetox (www.ceetox.com). As more data is added to publicly- and privately-funded databases, the data can be re-used and integrated to better inform hypothesis-driven research81.
5. Expert Opinion
Without a doubt, toxicogenomics offers great promise for risk assessment and omics technologies have the potential to transform chemical safety testing and to improve clinical intervention. A number of proof-of-principle studies have shown the utility of toxicogenomic data to predict adverse health events and injurious phenotypes. Even though toxicogenomics is unlikely to replace classical toxicological testing paradigms, it enriches the field by allowing researchers to discover biomarkers and subtle changes that occur sub-clinically or that predispose toward an injurious event. The reality is that omics technologies will become increasingly integral to toxicity studies; therefore, it is important that toxicologists have sufficient training and education to be able to design and interpret data from toxicogenomic and toxicogenetic studies.
The realization of the full potential of omics data integration is still several years away due to our currently limited understanding of the complex biology behind many toxicity types in a variety of species. There has been much progress in developing experimental tools to collect data, and some progress in data analysis and interpretation through pathway-based approaches and identification of complex interactions between gene expression, SNPs, other cellular macromolecules, and phenotypes. Focus should be placed on finding the optimal approaches for the integration and interpretation of multiple data types together in order to fully characterize molecular perturbations following exposure to a xenobiotic.
Another arena which requires significant attention from the toxicological community is in improving the use of omic data in high-throughput screening in human and rodent cell lines. Current methods which employ animal-based test methods tend to be expensive, are low-throughput, and often use doses that are much higher than potential human exposures. Toxicogenomic strategies that examine rodent in vivo with in vitro and human data are critical for bridging the uncertainties between species. While the value of in vivo data can not be completely replaced by in vitro screening tools, we anticipate that genomics tools will improve the value of in vitro screening for risk assessment. One current limitation that must be overcome is that very detailed gene annotation is only available for a few mammalian species, such as man and mouse, but is relatively lacking in others, such as in rat. The availability of complete gene information is critical for understanding and extrapolating toxicity risks across model systems and species and, ultimately, to humans.
Furthermore, regulatory agencies, the pharmaceutical industry, and academics must establish clear guidelines for the integration of toxicogenomic and genetic data into drug safety assessment. Evidence of clinical or human in vitro translation of toxicogenomic is essential in order to properly interpret the data and to be able to fully distinguish pathways that are associated with toxic effect vs. a physiological or pharmacological response. In examining datasets of sensitive “omics” data, it is necessary to distinguish between changes that are adaptive in nature and those changes that are within a no observable adverse effect level (NOAEL). Determination of chemical NOAELs can only be accomplished by dose-response modeling of a variety of compounds in which a benchmark dose can be correlated with both transcript changes and in vivo pathology; some progress has been made into developing computational tools that enable genomic benchmark dose analysis82. The question of determining the NOAEL becomes more complex as omics technologies become more refined, with greater throughput and sensitivity, and as data from multiple platforms becomes integrated.
The next wave of toxicogenomic and genetic research is the individualization of toxicity prediction and the ability to tailor drug therapy for specific patients. This area will be greatly facilitated by the development of easy and inexpensive whole-genome genotyping on an individual basis. In order to apply structural genomics data to pharmaceutical development and to clinical safety testing, carefully designed rodent models are needed that can serve as appropriate population-based models. While there is promising data that shows that genome-wide association can predict human toxicity responses to xenobiotics, unequivocal data that demonstrates that this method can identify human “susceptibility” genes is still lacking. Current mouse models used for toxicity testing are not optimized for modeling the genetic diversity present within human populations. Because genome association using inbred or complex recombinant-inbred lines is an emerging technique, we expect that the animal models will be refined to facilitate personalized assessment of efficacy and toxicity. We anticipate that controlled mouse genetic breeding programs, such as the Collaborative Cross63 that enable a superior precision in genetic mapping than current resources83, will revolutionize the ability to detect the genetic control of toxicity in individuals. Translational research bridging rodent and human toxicity will be the key to the success of this field and great emphasis should be given to collaboration across laboratories and disciplines. This effort will require team research within centers and consortia in order to be fully realized.
Footnotes
Conflict of Interest
The authors declare that they have no competing interests.
Reference List
- 1.Gutman S, Hackett J. Search for shortcuts on the critical path to market: US FDA perspectives from the diagnostic side. Pharmacogenomics. 2006 Dec;7(8):1223–1227. doi: 10.2217/14622416.7.8.1223. [DOI] [PubMed] [Google Scholar]
- 2.U.S. Food and Drug Administration. Guidance for Industry: Pharmacogenomic Data Submissions. 2005. [Google Scholar]
- 3.Frueh FW. Impact of microarray data quality on genomic data submissions to the FDA. Nat Biotechnol. 2006 Sep;24(9):1105–1107. doi: 10.1038/nbt0906-1105. [DOI] [PubMed] [Google Scholar]
- 4.Bammler T, Beyer RP, Bhattacharya S, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2(5):351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 5.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006 Sep;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyer RP, Fry RC, Lasarev MR, et al. Multicenter study of acetaminophen hepatotoxicity reveals the importance of biological endpoints in genomic analyses. Toxicol Sci. 2007 Sep;99(1):326–337. doi: 10.1093/toxsci/kfm150. [DOI] [PubMed] [Google Scholar]
- 7.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004 Mar;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 8.Arcellana-Panlilio M, Robbins SM. Cutting-edge technology. I. Global gene expression profiling using DNA microarrays. Am J Physiol Gastrointest Liver Physiol. 2002 Mar;282(3):G397–G402. doi: 10.1152/ajpgi.00519.2001. [DOI] [PubMed] [Google Scholar]
- 9.Paules R. Phenotypic anchoring: Linking cause and effect. Environ Health Perspect. 2003;111(6):A338–A339. doi: 10.1289/ehp.111-a338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powell CL, Kosyk O, Ross PK, et al. Phenotypic anchoring of acetaminophen-induced oxidative stress with gene expression profiles in rat liver. Toxicol Sci. 2006 Sep;93(1):213–222. doi: 10.1093/toxsci/kfl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinloth AN, Irwin RD, Boorman GA, et al. Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci. 2004 Jul;80(1):193–202. doi: 10.1093/toxsci/kfh145.*One of the first toxicogenomics papers that demonstrated the sensitivity of microarray data analysis for detecting organ toxicity.
- 12.Wang EJ, Snyder RD, Fielden MR, et al. Validation of putative genomic biomarkers of nephrotoxicity in rats. Toxicology. 2008 Apr 18;246(2–3):91–100. doi: 10.1016/j.tox.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Khor TO, Ibrahim S, Kong AN. Toxicogenomics in drug discovery and drug development: potential applications and future challenges. Pharm Res. 2006 Aug;23(8):1659–1664. doi: 10.1007/s11095-006-9003-8. [DOI] [PubMed] [Google Scholar]
- 14.Ridd K, Zhang SD, Edwards RE, et al. Association of gene expression with sequential proliferation, differentiation and tumor formation in murine skin. Carcinogenesis. 2006 Aug;27(8):1556–1566. doi: 10.1093/carcin/bgl007. [DOI] [PubMed] [Google Scholar]
- 15.Christians U, Schmitz V, Schoning W, et al. Toxicodynamic therapeutic drug monitoring of immunosuppressants: promises, reality, and challenges. Ther Drug Monit. 2008 Apr;30(2):151–158. doi: 10.1097/FTD.0b013e31816b9063. [DOI] [PubMed] [Google Scholar]
- 16.Bushel PR, Heinloth AN, Li J, et al. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):18211–18216. doi: 10.1073/pnas.0706987104.**This study is an example of translational toxicogenomics research which shows that blood mRNA is a sensitive predictor of chemical exposure levels that cause liver toxicity.
- 17.Burczynski ME, McMillian M, Ciervo J, et al. Toxicogenomics-based discrimination of toxic mechanism in HepG2 human hepatoma cells. Toxicol Sci. 2000 Dec;58(2):399–415. doi: 10.1093/toxsci/58.2.399. [DOI] [PubMed] [Google Scholar]
- 18.Waring JF, Ulrich RG, Flint N, et al. Interlaboratory evaluation of rat hepatic gene expression changes induced by methapyrilene. Environ Health Perspect. 2004 Mar;112(4):439–448. doi: 10.1289/ehp.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamadeh HK, Bushel PR, Jayadev S, et al. Gene expression analysis reveals chemical-specific profiles. Toxicol Sci. 2002 Jun;67(2):219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- 20.Inadera H, Tachibana S, Takasaki I, et al. Expression profile of liver genes in response to hepatotoxicants identified using a SAGE-based customized DNA microarray system. Toxicol Lett. 2008 Feb 28;177(1):20–30. doi: 10.1016/j.toxlet.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Dai X, He YD, Dai H, et al. Development of an approach for ab initio estimation of compound-induced liver injury based on global gene transcriptional profiles. Genome Inform. 2006;17(2):77–88. [PubMed] [Google Scholar]
- 22.Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci. 2007 Sep;99(1):90–100. doi: 10.1093/toxsci/kfm156.*Study suggesting that gene expression profiling is a sensitive predictor of carcinogenesis in rodent two-year bioassay.
- 23.Eun JW, Ryu SY, Noh JH, et al. Discriminating the molecular basis of hepatotoxicity using the large-scale characteristic molecular signatures of toxicants by expression profiling analysis. Toxicology. 2008 Jul 30;249(2–3):176–183. doi: 10.1016/j.tox.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Bartosiewicz MJ, Jenkins D, Penn S, et al. Unique gene expression patterns in liver and kidney associated with exposure to chemical toxicants. J Pharmacol Exp Ther. 2001 Jun;297(3):895–905. [PubMed] [Google Scholar]
- 25.McMillian M, Nie AY, Parker JB, et al. Inverse gene expression patterns for macrophage activating hepatotoxicants and peroxisome proliferators in rat liver. Biochem Pharmacol. 2004 Jun 1;67(11):2141–2165. doi: 10.1016/j.bcp.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Harries HM, Fletcher ST, Duggan CM, Baker VA. The use of genomics technology to investigate gene expression changes in cultured human liver cells. Toxicol In Vitro. 2001 Aug;15(4–5):399–405. doi: 10.1016/s0887-2333(01)00043-1. [DOI] [PubMed] [Google Scholar]
- 27.Uehara T, Kiyosawa N, Shimizu T, et al. Species-specific differences in coumarin-induced hepatotoxicity as an example toxicogenomics-based approach to assessing risk of toxicity to humans. Hum Exp Toxicol. 2008 Jan;27(1):23–35. doi: 10.1177/0960327107087910.**One of the first explorations into creating an in vivo-in vitro bridge for the validation of toxicogenomic biomarkers.
- 28.Elferink MG, Olinga P, Draaisma AL, et al. Microarray analysis in rat liver slices correctly predicts in vivo hepatotoxicity. Toxicol Appl Pharmacol. 2008 Jun 15;229(3):300–309. doi: 10.1016/j.taap.2008.01.037.*Study that suggests that a rat liver slice model can be used successfully to model in vivo toxicity responses.
- 29.Lerche-Langrand C, Toutain HJ. Precision-cut liver slices: characteristics and use for in vitro pharmaco-toxicology. Toxicology. 2000 Nov 16;153(1–3):221–253. doi: 10.1016/s0300-483x(00)00316-4. [DOI] [PubMed] [Google Scholar]
- 30.Adachi Y, Bradford BU, Gao W, et al. Inactivation of Kupffer cells prevents early alcohol-induced liver injury. Hepatology. 1994 Aug;20(2):453–460. [PubMed] [Google Scholar]
- 31.Wheeler MD, Kono H, Yin M, et al. The role of Kupffer cell oxidant production in early ethanol-induced liver disease. Free Radic Biol Med. 2001 Dec 15;31(12):1544–1549. doi: 10.1016/s0891-5849(01)00748-1. [DOI] [PubMed] [Google Scholar]
- 32.Kumagai K, Kiyosawa N, Ito K, et al. Influence of Kupffer cell inactivation on cycloheximide-induced hepatic injury. Toxicology. 2007 Nov 30;241(3):106–118. doi: 10.1016/j.tox.2007.08.090. [DOI] [PubMed] [Google Scholar]
- 33.Andres D, Sanchez-Reus I, Bautista M, Cascales M. Depletion of Kupffer cell function by gadolinium chloride attenuates thioacetamide-induced hepatotoxicity. Expression of metallothionein and HSP70. Biochem Pharmacol. 2003 Sep 15;66(6):917–926. doi: 10.1016/s0006-2952(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 34.Ito Y, Bethea NW, Abril ER, McCuskey RS. Early hepatic microvascular injury in response to acetaminophen toxicity. Microcirculation. 2003 Oct;10(5):391–400. doi: 10.1038/sj.mn.7800204. [DOI] [PubMed] [Google Scholar]
- 35.National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press; 2007. **A comprehensive volume with broad recommendations for improving toxicity testing to better inform risk assessment.
- 36.Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4(4):R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005 May 1;21(9):1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 38.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns JJ., IV Pharmacogenetics and drug toxicity. Variation of drug metabolism in animals and the prediction of drug action in man. Ann N Y Acad Sci. 1968 Jul 31;151(2):959–967. doi: 10.1111/j.1749-6632.1968.tb48282.x. [DOI] [PubMed] [Google Scholar]
- 40.Kalow W. Contribution of hereditary factors to the response to drugs. Fed Proc. 1965 Nov;24(6):1259–1265.*One of the original papers that demonstrated that genetics can affect drug responses.
- 41.Vesell ES, Page JG. Genetic control of dicumarol levels in man. J Clin Invest. 1968 Dec;47(12):2657–2663. doi: 10.1172/JCI105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev. 2008;40(2):187–224. doi: 10.1080/03602530801952864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cacabelos R. Pharmacogenetic basis for therapeutic optimization in Alzheimer's disease. Mol Diagn Ther. 2007;11(6):385–405. doi: 10.1007/BF03256262. [DOI] [PubMed] [Google Scholar]
- 44.Yates CR, Krynetski EY, Loennechen T, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997 Apr 15;126(8):608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 45.Relling MV, Hancock ML, Rivera GK, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999 Dec 1;91(23):2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 46.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004 Feb 5;427(6974):537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 47.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. J Am Med Assoc. 2002 Apr 3;287(13):1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 48.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005 Jun 2;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 49.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genomics Hum Genet. 2006;7:223–245. doi: 10.1146/annurev.genom.6.080604.162315.**An comprehensive review of pharmacogenetics and pharmacogenomics research.
- 50.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001 Feb 16;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 51.Weiss ST, McLeod HL, Flockhart DA, et al. Creating and evaluating genetic tests predictive of drug response. Nat Rev Drug Discov. 2008 Jul;7(7):568–574. doi: 10.1038/nrd2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008 Feb 7;358(6):568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 53.Nebert DW, Vesell ES. Advances in pharmacogenomics and individualized drug therapy: exciting challenges that lie ahead. Eur J Pharmacol. 2004 Oct 1;500(1–3):267–280. doi: 10.1016/j.ejphar.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 54.Ioannidis JP, Trikalinos TA, Ntzani EE. Contopoulos-Ioannidis DG. Genetic associations in large versus small studies: an empirical assessment. Lancet. 2003 Feb 15;361(9357):567–571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- 55.Ntzani EE, Rizos EC, Ioannidis JP. Genetic effects versus bias for candidate polymorphisms in myocardial infarction: case study and overview of large-scale evidence. Am J Epidemiol. 2007 May 1;165(9):973–984. doi: 10.1093/aje/kwk085. [DOI] [PubMed] [Google Scholar]
- 56.Lesnick TG, Papapetropoulos S, Mash DC, et al. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 2007 Jun;3(6):e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donfack J, Buchinsky FJ, Post JC, Ehrlich GD. Human susceptibility to viral infection: the search for HIV-protective alleles among Africans by means of genome-wide studies. AIDS Res Hum Retroviruses. 2006 Oct;22(10):925–930. doi: 10.1089/aid.2006.22.925. [DOI] [PubMed] [Google Scholar]
- 58.McClurg P, Pletcher MT, Wiltshire T, Su AI. Comparative analysis of haplotype association mapping algorithms. BMC Bioinformatics. 2006;7:61. doi: 10.1186/1471-2105-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frazer KA, Eskin E, Kang HM, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007 Aug 30;448(7157):1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 60.Pletcher MT, McClurg P, Batalov S, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004 Dec;2(12):e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Y, Weller P, Farrell E, et al. In silico pharmacogenetics of warfarin metabolism. Nat Biotechnol. 2006 May;24(5):531–536. doi: 10.1038/nbt1195.*Study that used genetic association among inbred mouse strains to identify a functional polymorphism that affects warfarin metabolism.
- 62.Guo Y, Lu P, Farrell E, et al. In silico and in vitro pharmacogenetic analysis in mice. Proc Natl Acad Sci U S A. 2007 Nov 6;104(45):17735–17740. doi: 10.1073/pnas.0700724104.*Study demonstrating the utility of using panels of inbred strains for pharmacogenetic analysis.
- 63.Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004 Nov;36(11):1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 64.Roberts A, Pardo-Manuel dV, Wang W, et al. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007 Jul;18(6–7):473–481. doi: 10.1007/s00335-007-9045-1.*Study demonstrating that Collaborative Cross mouse strains are an improvement for use in systems biology research over RI lines and inbred mouse panels.
- 65.Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007 Oct 26;81(6) doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collison M, Glazier AM, Graham D, et al. Cd36 and molecular mechanisms of insulin resistance in the stroke-prone spontaneously hypertensive rat. Diabetes. 2000 Dec;49(12):2222–2226. doi: 10.2337/diabetes.49.12.2222. [DOI] [PubMed] [Google Scholar]
- 68.Karp CL, Grupe A, Schadt E, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000 Sep;1(3):221–226. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- 69.Hitzemann R, Reed C, Malmanger B, et al. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol Clin Exp Res. 2004 Oct;28(10):1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- 70.Cook DN, Wang S, Wang Y, et al. Genetic regulation of endotoxin-induced airway disease. Genomics. 2004 Jun;83(6):961–969. doi: 10.1016/j.ygeno.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 71.Chesler EJ, Lu L, Shou S, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005 Mar;37(3):233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 72.Nebert DW, Jorge-Nebert L, Vesell ES. Pharmacogenomics and "individualized drug therapy": high expectations and disappointing achievements. Am J Pharmacogenomics. 2003;3(6):361–370. doi: 10.2165/00129785-200303060-00002. [DOI] [PubMed] [Google Scholar]
- 73.Phipps AN, Stewart J, Wright B, Wilson ID. Effect of diet on the urinary excretion of hippuric acid and other dietary-derived aromatics in rat. A complex interaction between diet, gut microflora and substrate specificity. Xenobiotica. 1998 May;28(5):527–537. doi: 10.1080/004982598239443. [DOI] [PubMed] [Google Scholar]
- 74.Schrattenholz A, Ss Kic V. What does systems biology mean for drug development? Curr Med Chem. 2008;15(15):1520–1528. doi: 10.2174/092986708784638843. [DOI] [PubMed] [Google Scholar]
- 75.Thomas CE, Ganji G. Integration of genomic and metabonomic data in systems biology--are we 'there' yet? Curr Opin Drug Discov Devel. 2006 Jan;9(1):92–100.**An comprehensive review of omics data integration strategies and current research.
- 76.Ippolito JE, Xu J, Jain S, et al. An integrated functional genomics and metabolomics approach for defining poor prognosis in human neuroendocrine cancers. Proc Natl Acad Sci U S A. 2005 Jul 12;102(28):9901–9906. doi: 10.1073/pnas.0500756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Z, Srivastava S, Mittal S, et al. A Three Stage Integrative Pathway Search (TIPS) framework to identify toxicity relevant genes and pathways. BMC Bioinformatics. 2007;8:202. doi: 10.1186/1471-2105-8-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verhoeckx KC, Bijlsma S, de Groene EM, et al. A combination of proteomics, principal component analysis and transcriptomics is a powerful tool for the identification of biomarkers for macrophage maturation in the U937 cell line. Proteomics. 2004 Apr;4(4):1014–1028. doi: 10.1002/pmic.200300669. [DOI] [PubMed] [Google Scholar]
- 79.Hirai MY, Klein M, Fujikawa Y, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem. 2005 Jul 8;280(27):25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- 80.Clayton TA, Lindon JC, Cloarec O, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006 Apr 20;440(7087):1073–1077. doi: 10.1038/nature04648.*Manuscript suggesting that pre-dose metabolomic profiling can predict acetaminophen toxicity responses in a rat model.
- 81.Fostel JM. Future of toxicogenomics and safety signatures: balancing public access to data with proprietary drug discovery. Pharmacogenomics. 2007 May;8(5):425–430. doi: 10.2217/14622416.8.5.425. [DOI] [PubMed] [Google Scholar]
- 82.Yang L, Allen BC, Thomas RS. BMDExpress: a software tool for the benchmark dose analyses of genomic data. BMC Genomics. 2007;8:387. doi: 10.1186/1471-2164-8-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chesler EJ, Miller DR, Branstetter LR, et al. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. doi: 10.1007/s00335-008-9135-8. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]