Abstract
Inhibiting angiogenesis has become a major therapeutic strategy for cancer treatment. To identify common intra-cellular mediators, we previously analyzed gene expression profiles of endothelial cells after treatment with angiogenesis inhibitors. Filamin A interacting protein 1-like (FILIP1L; previously known as down-regulated in ovarian cancer 1) was identified as one of the genes up-regulated in endothelial cells in response to these inhibitors. However, the expression and function of FILIP1L protein is uncharacterized. Here, we provide the first description of the expression and specific subcellular localization of FILIP1L protein in human tissue. Overexpression of FILIP1L resulted in inhibition of cell proliferation and migration and increased apoptosis. In addition, overexpression of FILIP1L truncation mutants showed differential antiproliferative activity. A COOH terminal truncation mutant (FILIP1LΔC103) was more potent than wild-type FILIP1L in mediating this activity. Targeted expression of FILIP1LΔC103 in tumor vasculature inhibited tumor growth in vivo. Overall, these findings suggest that the novel protein FILIP1L may be an important mediator of the effects of angiogenesis inhibitors and that FILIP1L has the potential to be an antivascular reagent for cancer therapy.
Introduction
Angiogenesis, the formation of new blood vessels from pre-existing ones, is required for the sustained growth, invasion, and spread of tumors. Thus, the inhibition of tumor angiogenesis has been considered to be an important component of anticancer therapy. Many angiogenesis inhibitors that prevent endothelial cells from proliferating and migrating or result in the induction of apoptosis have been identified (1, 2). Endostatin, the 20-kDa COOH terminal fragment of collagen XVIII, inhibits endothelial cell proliferation, migration, invasion, and tube formation and induces apoptosis (3–6). Fumagillin, a natural metabolite from Aspergillus fumigatus, induces cell cycle arrest and apoptosis in endothelial cells (7–9). EMAP-II, a 20-kDa fragment of the aminoacyl-tRNA synthetase complex, induces endothelial cell apoptosis, inhibits endothelial cell proliferation, and up-regulates tissue factor expression on the endothelial cell surface (10–14). Previously, our laboratory examined the effects of these different angiogenesis inhibitors, endostatin, fumagillin, and EMAP-II, on the gene expression profiles of human umbilical vascular endothelial cells (HUVEC) to elucidate commonly affected pathways (15, 16). Interestingly, the majority of gene expression changes were observed as early as 1 and 2 hours after treatment. Among these, expression of genes, such as filamin A interacting protein 1-like [FILIP1L; synonym of down-regulated in ovarian cancer 1 (DOC1), National Center for Biotechnology Information (NCBI) accession number NP_055705], KLF4, and TC-1 was shown to be rapidly up-regulated. Using small interfering RNA, we showed that, when FILIP1L was silenced, KLF4 and TC-1 failed to show up-regulation in gene expression in response to endostatin treatment, suggesting that FILIP1L may be upstream of KLF4 and TC-1 in a pathway that mediates the antiangiogenic response to endostatin (15). Likewise, silencing of FILIP1L gene expression resulted in abrogation of the modulatory effect of EMAP II on ADM, KLF4, SOCS3, and TNFAIP3 gene expression (16). Collectively, these data suggest that the FILIP1L gene may play a central role in mediating the common early response pathways seen in endothelial cells after exposure to angiogenesis inhibitors.
In the present study, we define expression and specific subcellular localization of FILIP1L protein in human tissue for the first time. Additionally, we show that overexpression of FILIP1L in endothelial cells results in a similar profile of antiangiogenic activity as the angiogenesis inhibitors. Finally, using a tumor vascular targeted gene therapy vector, we show that targeted expression of FILIP1L in the tumor vasculature results in inhibition of tumor growth in vivo, suggesting that FILIP1L could be developed as a potential cancer therapeutic.
Materials and Methods
Production of mouse monoclonal anti-FILIP1L antibody
The construct encoding a full-length FILIP1L cDNA was generated and expressed in baculovirus. The purified full-length FILIP1L protein (893 amino acids) was used as an antigen to immunize mice. Immunization of mice, production of hybridoma cells, screening by ELISA, and purification of monoclonal antibody were performed by Green Mountain Antibodies, Inc. Antibodies that recognize FILIP1L were further tested by Western blot, and a monoclonal antibody was selected.
Cell culture
HUVECs were cultured in complete EGM-2 medium as recommended by the manufacturer (Lonza). HEK293 cells were grown in DMEM containing 10% fetal bovine serum (FBS). DU145 human prostate carcinoma cells and M21 human melanoma cells were grown in RPMI 1640 containing 10% FBS.
Western blot
HUVECs were cultured, harvested, and fractionated with ProteoExtract Subcellular Proteome Extraction kit according to the manufacturer’s protocol (Calbiochem). For endostatin experiment, HUVECs were starved in EGM-2 basal medium containing 1% FBS for 16 h, treated with 1 µg/mL endostatin for 2, 4, and 8 h, and lysed with radio-immunoprecipitation assay (RIPA) buffer. HEK293 cells were transfected using Lipofectamine 2000 (Invitrogen), with a series of NH2 terminal and COOH terminal truncation mutants of FILIP1L containing a COOH terminal hemagglutinin (HA) tag, harvested at 24 h and lysed with RIPA buffer. Empty lentivirus or lentivirus expressing FILIP1L mutant 1–790 (hereafter called FILIP1LΔC103)–transduced DU145 clones were cultured in the presence or absence of 1 µg/mL doxycycline and lysed with RIPA buffer. Tumors from PBS-treated, null–adeno-associated virus-phage (hereafter called AAVP-null)–treated, and AAVP expressing FILIP1LΔC103 (hereafter called AAVP-ΔC103)–treated mice were removed 4 d after tail vein injection and snap frozen. Whole tumor lysates were prepared from RIPA buffer lysis of 60-µm tumor section. Cellular fractionation (25–50 µg), whole cell lysates, or whole tumor lysates prepared by above methods were separated on SDS-PAGE and transferred to nitrocellulose membrane. The membranes were blotted with antibodies against FILIP1L, HA tag (Covance), and glycerol-dehyde-3-phosphate dehydrogenase (GAPDH; Chemicon) followed by incubation with antimouse antibody conjugated to horseradish peroxidase. The signal was detected using chemiluminescence (Millipore).
Immunohistochemistry
Frozen human colon tumors and their adjacent normal colon samples were obtained under an Institutional Review Board–approved protocol. Tissue sections (10 µm) were fixed with 4% paraformal-dehyde for 20 min and stained with mouse monoclonal antibodies against FILIP1L (7.5 µg/mL) and CD31 (10 µg/mL; DAKO). After visualization of staining by 3,3′-diaminobenzidine tetrahydrochloride, the slides were counterstained with hematoxylin. Images were acquired by Axioplan 2 microscope using a 20×/0.75 objective with Axiovision 4.1 software (Zeiss).
Immunofluorescence and vessel density determination
HUVECs were starved in EGM-2 basal medium containing 1% FBS for 16 h and treated with 1 µg/mL endostatin for 4 h. The cells were fixed with 4% paraformaldehyde for 10 min followed by permeabilization with 0.1% Triton X-100 for 5 min. The cells were washed with PBS, blocked with 5% bovine serum albumin (BSA) in PBS, and treated with mouse anti-FILIP1L antibody (4 µg/mL) preincubated with 500-fold molar excess of either BSA or FILIP1L. The cells were then incubated with 2 µg/mL Alexa Fluor 488 antimouse IgG (Invitrogen) and treated with 4′,6-diamidino-2-phenylindole (DAPI) mounting media (Vector Laboratories). Images were acquired on an LSM-510 confocal microscope using a 25×/0.8, 40×/1.3, or 63×/1.4 objective and analyzed by AxioVision LE software (Zeiss). For AAVP-targeting experiment, tumors from PBS-treated, AAVP-ΔC103–treated, and AAVP-ΔC243–treated mice were removed at 30 min and 6 d after tail vein injection (30 min only for PBS-treated tumor), snap frozen, and cut into 10-µm tumor sections. These tumor sections were subjected to immuno-fluorescent staining as described above, except rabbit anti-AAVP antibody (Sigma), followed by Alexa Fluor 594 antirabbit IgG (Invitrogen) and rat anti-CD31 antibody (BD PharMingen), followed by Alexa Fluor 488 antirat IgG (Invitrogen) were used.
CD31-stained tumor sections from PBS-treated, AAVP-null–treated, and AAVP-ΔC103–treated mice were analyzed for vessel density as described previously (17). Three tumors from each treatment group were analyzed. Five random fields per tumor were imaged by Axiovert 200M microscope using a 10×/0.3 objective (Zeiss). Axiovision 4.6 software (Zeiss) was used to quantify CD31-positive vessels. In addition, TUNEL staining was also performed on these AAVP-treated tumors as recommended by the manufacturer (Promega Corporation). Images were taken by Axiovert 200M microscope using a 5×/0.15 objective (Zeiss).
Transfection of HUVECs with FILIP1L plasmids
Plasmids encoding wild-type, as well as truncation mutants of FILIP1L, were purified using Endo-free maxiprep kit (Qiagen). HUVECs were transfected with equimolar amount of each DNA using HUVEC nucleofector solution and Nucleofector II machine as provided by the manufacturer (Amaxa). Transfection efficiency was verified using a plasmid with an enhanced green fluorescent protein (eGFP) marker (2 µg), as was calculated by the GFP expression. The percentage of transfection reached by this method was 50 ± 10%. After transfection, the cells were subjected to proliferation, apoptosis, or migration assays.
Bromodeoxyuridine ELISA cell proliferation assay
The transfected HUVECs were plated with 2 × 104 per well in 96-well culture plates and incubated for 24 h. Cell proliferation was measured by Cell Proliferation Biotrak ELISA (GE Healthcare), as recommended by the manufacturer.
Apoptosis assay
The transfected HUVECs were plated with 2.5 × 104 per well in white-walled 96-well culture plates and incubated for 24 h. Early apoptosis was determined by the measurement of caspase-3/caspase-7 activity using the caspase-Glo 3/7 Assay (Promega Corporation) following manufacturer’s instructions. The transfected HUVECs were plated with 2 × 106/100-mm culture dishes and incubated for 48 h. Late-stage apoptosis was determined by the staining of Annexin V–FITC and 7-amino-actinomycin D (7-AAD) staining using the Annexin V–FITC Apoptosis Detection kit (BD PharMingen) following manufacturer’s instructions. The stained cells were subjected to flow cytometric analysis using a FACSCalibur (BD PharMingen) and analyzed by the CELLQuest program.
Migration assay
The migratory potential of the transfected HUVECs was assessed by Electric Cell-Substrate Impedance Sensing (ECIS Model 9600, Applied Biophysics, Inc.; refs. 18, 19). Cells (1.1 × 105) were inoculated in 8W1E plates in complete EGM-2 medium. The cells were allowed to completely adhere to the electrodes which produced maximum and nonvariable readings of impedance. The monolayers were then wounded (30 s, 4.0 V, 60 kHz) where impedance became a minimum. As cells migrated to heal the wound, the impedance was recorded at 15 kHz every 5 s for 10 h in real time. The differences in migration rate were evaluated by comparison of the slopes of the curves in linear range for early time points.
DU145 clones transduced with either empty lentivirus or lentivirus expressing FILIP1LΔC103 (7.5 × 104 cells per chamber) were plated in the presence or absence of 1 µg/mL doxycycline in upper chamber. Migration toward 10% FBS was measured at 15 h by QCM 24-well colorimetric cell migration assay kit (Chemicon), as recommended by the manufacturer.
Cloning of FILIP1L and its truncation mutants
Genes for FILIP1L and its truncation mutants were cloned into Gateway entry clones using multistep PCR. The subsequent entry clones were sequence verified throughout the entire cloned region. Entry clones were then subcloned by Gateway LR recombination using the manufacturer’s protocols (Invitrogen) into different expression vectors.
Lentivirus generation and development of inducible clones over-expressing FILIP1LΔC103
A lentiviral construct encoding FILIP1LΔC103 was used to generate lentivirus expressing FILIP1LΔC103 by the ViraPower T-REx Lentiviral Expression System (Invitrogen) using the manufacturer’s protocols. DU145 cells were transduced with the Tet repressor-lentivirus and screened for clones that expressed the Tet repressor by Western blot analysis. Tet repressor-expressing DU145 cells were then transduced with either empty lentivirus or lentivirus expressing FILIP1LΔC103, and stable clones were screened by real-time reverse transcription–PCR (RT-PCR) analysis.
Quantitative real-time RT-PCR
DU145 clones transduced with lentivirus expressing FILIP1LΔC103 were cultured in the presence or absence of 1 µg/mL doxycycline for 48 h and harvested. Total RNA was prepared by RNeasy kit (Qiagen), and cDNA was prepared by Superscript II reverse transcriptase (Invitrogen). qPCR was performed using ABI 7500 SDS real-time PCR instrument following manufacturer’s instructions (Applied Biosystems). The expression of the FILIP1L gene was normalized to GAPDH expression. The primers used were 5′-AACGCTGGTATCATGGCTGAA-3′ and 5′-ATCTCTGCACTGCTCCTCCATT-3′ for FILIP1L; 5′-TCACCAGGGCTGCTTTTAACTC-3′ and 5′-GGAATCATATTGGAACATGTAAACCA-3′ for GAPDH.
Construction and generation of targeted AAVP particles
Cloning of both FILIP1LΔC103 (amino acid 1–790) and FILIP1LΔC243 (amino acid 1–650) mutant cDNA into the AAVP vector and the production of AAVP was performed as described previously (20, 21).
Xenograft assay
M21 human melanoma cells were injected s.c. into female athymic nude mice and grown to an average size of 100 mm3. Mice were randomly sorted into four groups (n = 11 for each group), AAVP (1 × 1011 transducing units per dose) was injected i.v. at day 0 and day 7, and tumors were measured in a blinded manner. Tumor volume was calculated as the product of (length × width × height) × 0.52. All animal experiments were conducted according to protocols approved by the NIH Animal Care and Use Committee.
Statistical analysis
Statistical analyses were performed using a two-tailed Student’s t test (GraphPad Prism 3.0), and differences were considered to be statistically significant at a value of P < 0.05. Xenograft and vessel density data were analyzed using one-way ANOVA with Newman-Keuls Multiple Comparison Test. A P value of <0.05 was considered significant.
Results
Expression of FILIP1L protein in HUVECs and human tissue
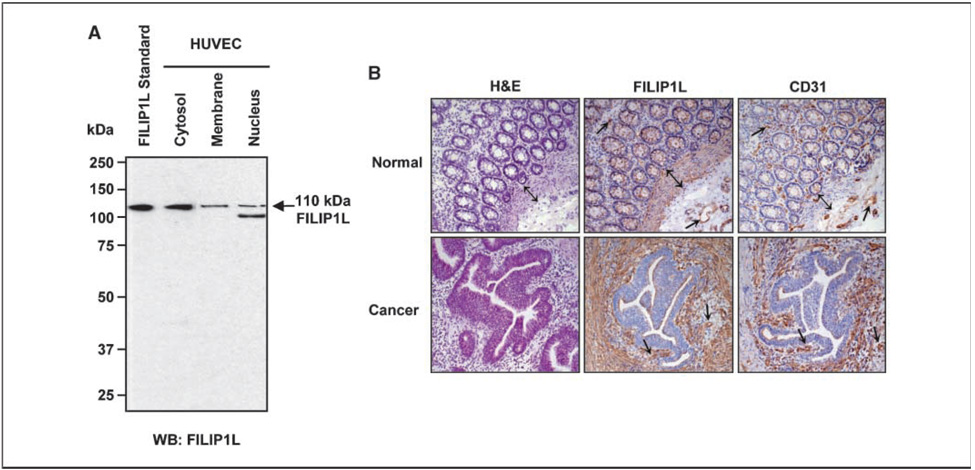
Although FILIP1L mRNA expression has been shown to be up-regulated in human endothelial cells in response to different angiogenesis inhibitors (15, 16), the expression of FILIP1L protein has not been previously investigated. To determine whether FILIP1L protein is endogenously expressed in human tissue, we produced monoclonal antibodies that specifically recognize FILIP1L. To detect FILIP1L protein in endothelial cells and determine its subcellular localization, HUVECs were fractionized and a Western blot was performed using anti-FILIP1L antibody. A specific 110-kDa band, identical size to the purified FILIP1L protein, was detected by anti-FILIP1L antibody, suggesting that HUVECs express a full-length FILIP1L protein (Fig. 1A). In addition, FILIP1L was expressed predominantly in the cytoplasm with less expression in the membrane and nucleus. Having shown the expression of FILIP1L protein in cultured endothelial cells, we then examined the expression of FILIP1L in human tissue. Immunohis-tochemical analysis was performed on 15 frozen human colon cancers and matched normal colon tissues using anti-FILIP1L antibody. In normal colon, FILIP1L was expressed in the vasculature and muscularis mucosa (Fig. 1B, top). In colon cancer, FILIP1L was strongly expressed in tumor stroma and the vasculature (Fig. 1B, bottom). Thus, these data show that FILIP1L is expressed in vasculature and smooth muscle and in desmoplastic stroma in response to tumor invasion.
Figure 1.
Expression of FILIP1L protein in HUVECs and human tissue. A, a 110-kDa FILIP1L protein was detected in the cytoplasm, membrane, and nucleus of HUVECs by Western blot using anti-FILIP1L antibody. A purified FILIP1L protein was used as a standard. B, immunohistochemical staining of FILIP1L and CD31 in human colon cancers and matched normal tissues. H&E staining is also shown. FILIP1L was expressed in the vasculature (→) and muscularis mucosa (↕) of normal human colon tissues. FILIP1L was also expressed in tumor stroma and vasculature (→) of colon cancer tissues. Note that stroma was negative in FILIP1L staining in normal colon, whereas tumor stroma showed a strong positive staining.
Up-regulation of FILIP1L protein by endostatin
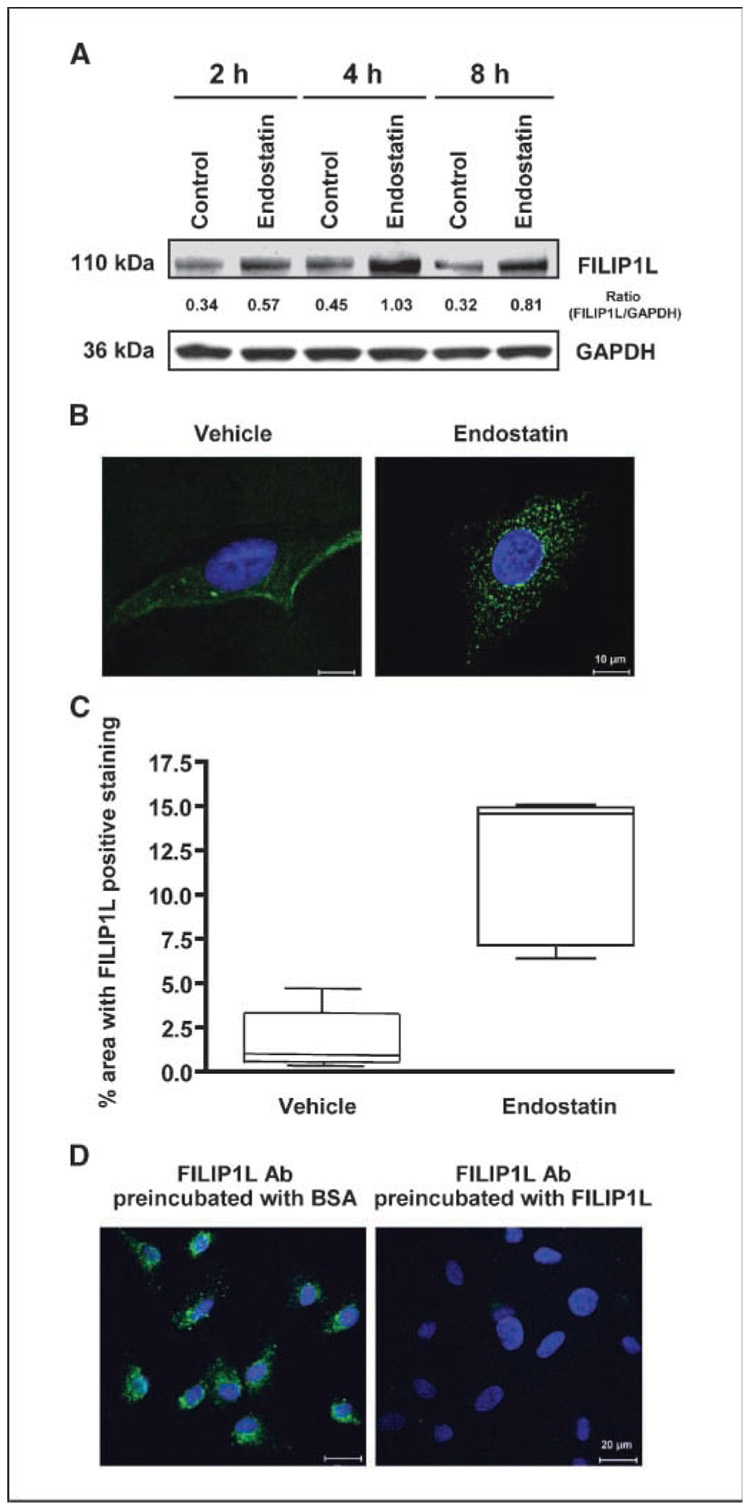
Our previous studies have shown that FILIP1L mRNA expression is up-regulated in HUVECs within 1 hour after the treatment of endothelial cells with the angiogenesis inhibitors endostatin, fumagillin, and EMAP II (15, 16). To further confirm that FILIP1L protein expression is up-regulated in endothelial cells in response to angiogenesis inhibitors, we treated HUVECs with endostatin, harvested cells at 2, 4, and 8 h, and performed Western blot analysis using anti-FILIP1L antibody on whole cell lysates. Compared with vehicle-treated controls, endostatin-treated HUVECs expressed more FILIP1L protein at all the time points tested (densitometric quantitation values are also shown in Fig. 2A). To examine if endostatin treatment affects cellular distribution of FILIP1L protein in endothelial cells, we first serum starved HUVECs to synchronize them. We then treated those HUVECs with endostatin and immunofluorescently stained them with anti-FILIP1L antibody at 4 h. Serum-starved, vehicle-treated control cells showed weak cytoplasmic staining, whereas endo-statin-treated cells showed a stronger punctate distribution of staining in the cytoplasm (Fig. 2B; additional images are shown in Supplementary Fig. S1). FILIP1L expression measured by immuno-fluorescent staining in endostatin-treated HUVECs was significantly more than that in vehicle-treated control cells (P = 0.0012; Fig. 2C). In addition, this staining was FILIP1L-specific, as anti-FILIP1L antibody preincubated with FILIP1L protein, but not with BSA control, failed to show the staining (Fig. 2D). These results suggest that FILIP1L protein expression is increased after endostatin treatment and support our initial observations at the mRNA level.
Figure 2.
Up-regulation of FILIP1L protein by endostatin. A, increased expression of FILIP1L protein was detected in HUVECs treated with endostatin for 2, 4, and 8 h by Western blot using anti-FILIP1L antibody. GAPDH blot is shown as the loading control. The numbers underneath the blot are the densitometric values calculated as FILIP1L-GAPDH ratios using ImageQuant software. The result is representative of two independent experiments. B, the punctate distribution in the cytoplasm was detected in HUVECs treated with endostatin for 4 h by immunofluorescent staining using anti-FILIP1L antibody. Vehicle-treated control cells showed diffused cytoplasmic staining. Nuclear staining with DAPI is shown in blue. Scale bar, 10 µm. The result is a representative image from two independent experiments. Additional images are shown in Supplementary Fig. S1. C, FILIP1L expression in endostatin-treated HUVECs was significantly more than that in vehicle-treated control cells (P = 0.0012). Five images from each treatment group (B) were analyzed. Axiovision 4.6 software (Zeiss) was used to quantify the percentage area with FILIP1L-positive staining. Box and whiskers plot (GraphPad Prism 3.0) is shown. D, preincubation of anti-FILIP1L antibody with FILIP1L protein, but not with BSA, abrogated the punctuate staining seen in HUVECs treated with endostatin. Nuclear staining with DAPI is shown in blue. Scale bar, 20 µm.
Overexpression of FILIP1L in endothelial cells leads to inhibition of cell proliferation and an increase in apoptosis
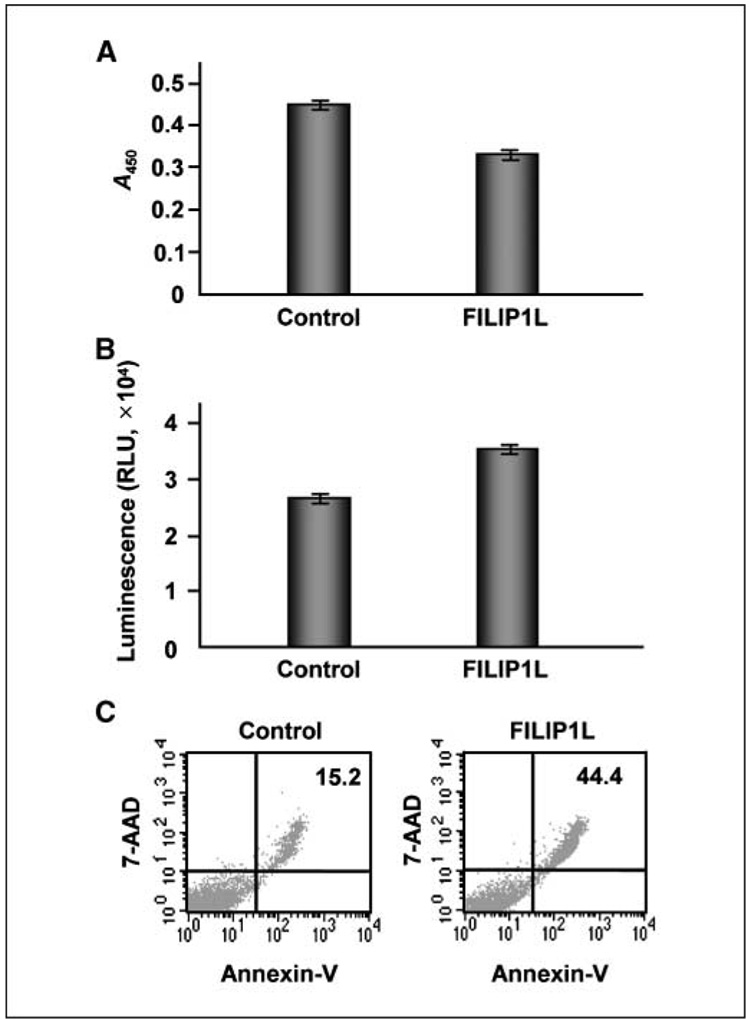
Because FILIP1L protein expression was up-regulated by the angiogenesis inhibitor endostatin, we hypothesized that FILIP1L may mediate antiangiogenic activity in endothelial cells and that overexpression of FILIP1L in endothelial cells may result in antiproliferative and proapoptotic activity. To test this, we transfected HUVECs with a plasmid encoding FILIP1L cDNA and measured cell proliferation by bromodeoxyuridine (BrdUrd) ELISA 24 hours after transfection. Transfection efficiency was 50 ± 10%, as verified by GFP expression after transfection of HUVECs with a control plasmid encoding an eGFP. Compared with control empty vector-transfected cells, FILIP1L-transfected cells showed a de-crease in cell proliferation (P < 0.0001; Fig. 3A). To determine if overexpression of FILIP1L in endothelial cells results in an increase in apoptosis, we measured caspase-3/caspase-7 activity at 24 hours after transfection of HUVECs with FILIP1L cDNA. Although caspase-3/caspase-7 activity in control vector-transfected cells was present due to the cytotoxicity caused by the transfection procedure, FILIP1L-transfected cells showed significantly more activity (P < 0.001; Fig. 3B). To further detect apoptosis in these cells, we stained the transfected cells with Annexin V–FITC and 7-AAD at 48 hours after transfection and measured staining using flow cytometry. As shown in Fig. 3C, FILIP1L overexpression resulted in increased staining of both Annexin V–FITC and 7-AAD (44.4% versus 15.2%), suggesting that late-stage apoptosis is increased in FILIP1L-transfected cells compared with control vector-transfected cells.
Figure 3.
Overexpression of FILIP1L in endothelial cells leads to inhibition of cell proliferation and an increase in apoptosis. A, inhibition of cell proliferation by overexpression of FILIP1L in HUVECs was analyzed by BrdUrd ELISA 24 h after transfection. The amount of BrdUrd incorporated was measured by absorbance at 450 nm. Bars, SE (n = 4, P < 0.0001). The result is representative of three independent experiments. B, increased apoptosis by overexpression of FILIP1L in HUVECs was analyzed by caspase-3/caspase-7 assay 24 h after transfection. Caspase-3/caspase-7 activity was measured by luminescence. Bars, SE (n = 4, P < 0.001). The result is representative of three independent experiments. C, increased apoptosis by overexpression of FILIP1L in HUVECs was analyzed by Annexin V–FITC and 7-AAD staining followed by flow cytometry analysis 48 h after transfection. The numbers 15.2 for control and 44.4 for FILIP1L indicate the percentage of cells in late apoptosis. The result is representative of two independent experiments.
FILIP1L truncation mutants have differential antiprolife-rative activity
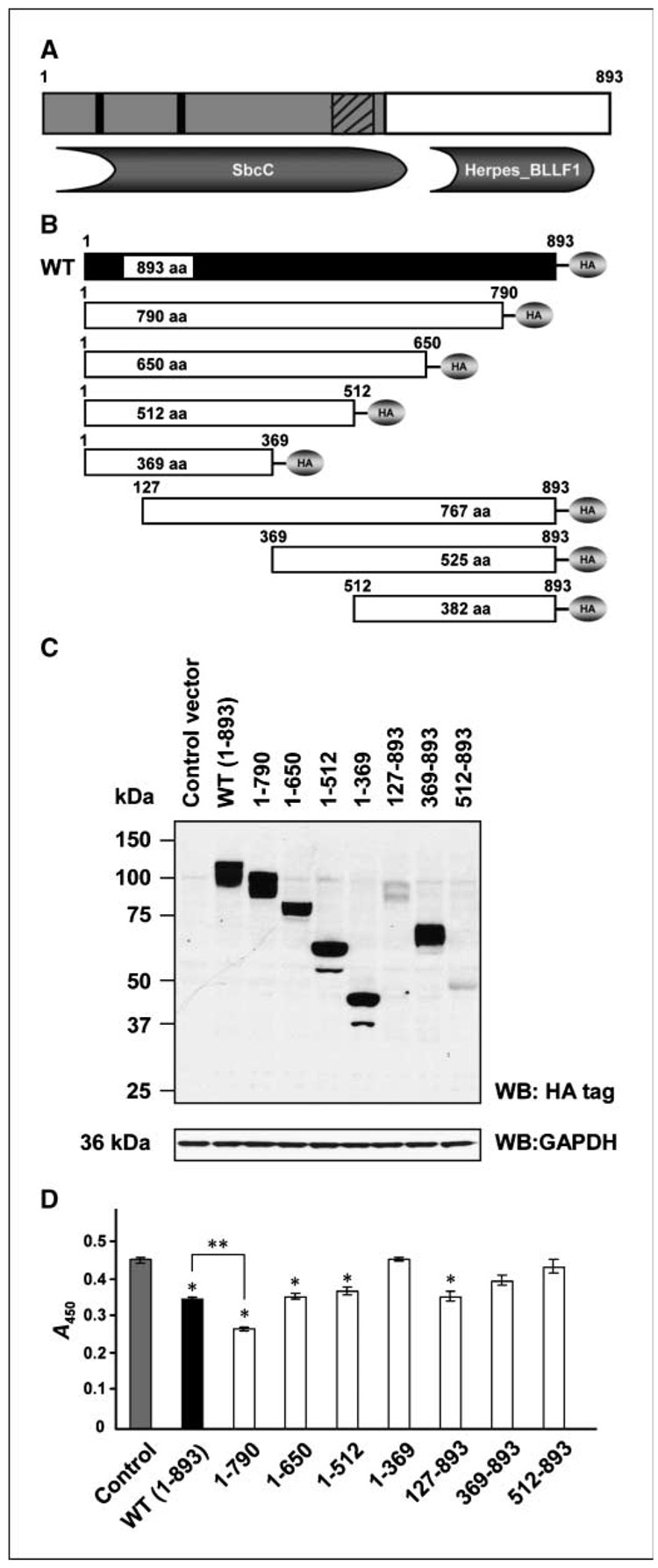
A coiled-coil region (residues 3–542), two leucine zipper motifs (residues 83–111 and 218–253), and a prefoldin domain (residues 465–535) can be recognized in the NH2 terminal half of the FILIP1L protein. In addition, an NCBI conserved domain search5 reveals that FILIP1L has an SbcC (COG0419; ATPase involved in DNA repair; residues 19–576) conserved domain in its NH2 terminal half and a Herpes_BLLF1 (pfam05109; Herpes virus major outer envelope glycoprotein; residues 640–829) conserved domain in its COOH terminal half (Fig. 4A).
Figure 4.
FILIP1L truncation mutants have differential antiproliferative activity. A, a schematic representation of FILIP1L protein (893 amino acids). Two leucine zipper motifs (black rectangles) and a prefoldin domain (striped rectangles) were recognized in the NH2 terminal half of a coiled-coil region (gray area). A SbcC (COG0419; ATPase involved in DNA repair) conserved domain in its NH2 terminal half and a Herpes_BLLF1 (pfam05109; Herpes virus major outer envelope glycoprotein) conserved domain in its COOH terminal half. B, a schematic diagram of FILIP1L truncation mutants. HA indicates COOH terminal HA tag. Amino acid residues are shown on top of each construct. C, expression of each mutant was confirmed in HEK293 cells transfected with each construct by Western blot analysis using anti-HA tag antibody. GAPDH blot is shown as the loading control. The result is representative of two independent experiments. D, differential inhibition of HUVECs proliferation by FILIP1L truncation mutants was analyzed by BrdUrd ELISA 24 h after transfection. The amount of BrdUrd incorporated was measured by absorbance at 450 nm. Bars, SE (n = 4). FILIP1L truncation mutants 1–790 (P = 0.0001), 1–650 (P = 0.004), 1–512 (P = 0.0114), and 127–893 (P = 0.0021) significantly inhibited cell proliferation compared with control (*). COOH terminal mutant 1–790 was more potent than wild-type FILIP1L in mediating antiproliferative activity (**, P = 0.001). The result is representative of two independent experiments.
To examine which part of the FILIP1L protein mediates the antiproliferative activity in endothelial cells, we generated a series of NH2 terminal and COOH terminal truncation mutants of FILIP1L as a fusion protein containing a COOH terminal HA tag (Fig. 4B). To determine if these mutant constructs produce proteins in cells, we transfected HEK293 cells with each construct and performed Western blot analysis using anti-HA tag antibody. All the constructs produced proteins of the predicted size, although NH2 terminal truncation mutants 127–893 and 512–893 showed low levels of expression (Fig. 4C). To determine if these proteins are functional, we transfected HUVECs with the plasmid encoding each FILIP1L mutant and measured cell proliferation by BrdUrd ELISA 24 hours after transfection. FILIP1L truncation mutants 1–790, 1–650, 1–512, and 127–893 significantly inhibited cell proliferation compared with control. COOH terminal truncation mutant 1–790 was more potent in its ability to inhibit cell proliferation than wild type (P = 0.001; Fig. 4D). Thus, we chose to further evaluate the function of the COOH terminal truncation mutant 1–790 (called FILIP1LΔC103). To examine if overexpression of FILIP1LΔC103 in endothelial cells results in an increase in apoptosis, we measured caspase-3/caspase-7 activity at 24 hours after transfection of HUVECs with FILIP1LΔC103 cDNA. As shown in Supplementary Fig. S2, FILIP1LΔC103-transfected cells showed significantly more apoptotic activity than control cells (P < 0.0001).
Overexpression of FILIP1LΔC103 in HUVECs and DU145 prostate cancer cells leads to inhibition of cell migration
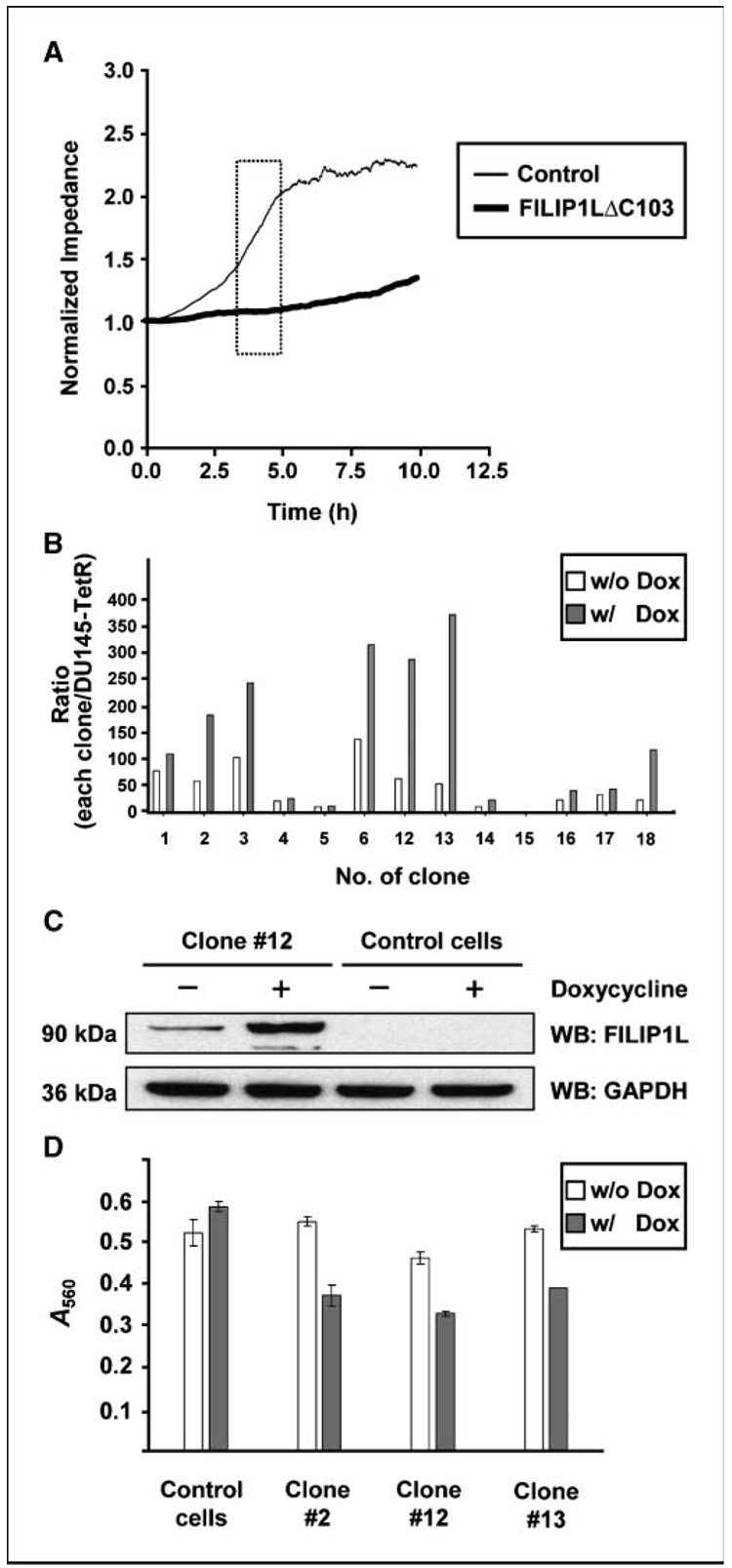
Because inhibition of cell migration is one of the important characteristics of angiogenesis inhibitors, we tested whether overexpression of FILIP1LΔC103 results in inhibition of cell migration. To do this, we transfected HUVECs with a plasmid encoding FILIP1LΔC103 cDNA and measured cell migration by Electric Cell-Substrate Impedance Sensing system (Applied Biophysics, Inc.; refs. 18, 19). Compared with control empty vector-transfected cells, FILIP1LΔC103-transfected cells showed a significantly slower migration rate (P < 0.0001; Fig. 5A), suggesting that overexpression of FILIP1LΔC103 in HUVECs results in inhibition of cell migration.
Figure 5.
Overexpression of FILIP1LΔC103 in HUVECs, as well as DU145 prostate cancer cells, leads to inhibition of cell migration. A, FILIP1LΔC103-transfected HUVECs showed a significantly slower migration rate than control vector-transfected HUVECs as measured by Electric Cell-Substrate Impedance Sensing System in real time (P < 0.0001). Square box, linear range in the curve that was used for analysis. The result is representative of three independent experiments. B, real-time RT-PCR analysis for FILIP1L on cDNA from DU145 clones transduced with FILIP1LΔC103-lentivirus. Each clone was treated with either PBS (w/o Dox) or 1 µg/mL doxycycline (Dox) for 48 h before harvest. The y axis represents a ratio between each clone and the parental Tet repressor-expressing DU145 cells where each value was standardized with housekeeping gene GAPDH. Columns, average of three experiments. C, a 90-kDa FILIP1LΔC103 protein was detected in Tet repressor-expressing DU145 cells transduced with FILIP1LΔC103-lentivirus, but not control lentivirus, by Western blot using anti-FILIP1L antibody. The result shown is a representative (clone 12) from several clones. GAPDH blot is shown as the loading control. D, all three FILIP1LΔC103 clones, but not mixed population of control cells, showed a significantly slower migration in the presence of doxycycline. Error bars indicate SE (n = 3). P value comparisons between in the presence and in the absence of doxycycline were as follows: control cells, P = 0.141; clone 2, P = 0.0014; clone 12, P = 0.0005; and clone 13, P < 0.0001. The result is representative of two independent experiments.
We also tested the effects of overexpression of FILIP1LΔC103 on migration of neoplastic cell lines. We selected DU145 prostate cancer cells as a model system because FILIP1L mRNA expression was shown to be repressed in immortalized prostate epithelial cells (22, 23), and FILIP1L mRNA expression is relatively low in this cell line compared with other cancer cell lines.6 Overexpression of FILIP1LΔC103 in DU145 cells also resulted in inhibition of cell proliferation (Supplementary Fig. S3). Thus, we chose to develop inducible FILIP1LΔC103-overexpressing clones. The system we used was the ViraPower T-REx Lentiviral Expression System (Invitrogen). In this system, the expression of a gene of interest is repressed by a Tet repressor in the absence of tetracycline (or doxycycline), whereas it is derepressed in the presence of tetracycline (or doxycycline). Clones were screened by real-time RT-PCR analysis. Several clones showed a 2.5-fold to 7-fold increase in FILIP1LΔC103 mRNA expression after doxycycline induction compared with the uninduced condition (Fig. 5B). Unexpectedly, however, we observed that, at the uninduced basal level, most of these clones expressed 20-fold to 60-fold more FILIP1LΔC103 mRNA than parental Tet repressor-expressing DU145 cells (Fig. 5B). FILIP1LΔC103 protein levels were shown to be increased considerably by doxycycline, but the basal level expression was also detectable in these clones (Fig. 5C). Control cells that were a mixed population from empty lentivirus-transduced Tet repressor-expressing DU145, however, did not produce any FILIP1LΔC103 protein (Fig. 5C). To measure cell migration for these cells, we used the Boyden chamber assay. Using this system, we measured cell migration for the FILIP1LΔC103 clones 2, 12, and 13, as well as control cells. All the FILIP1LΔC103 clones, but not control cells, showed a significantly slower migration in the presence of doxycycline (P < 0.005; Fig. 5D). Therefore, these data suggest that overexpression of FILIP1LΔC103 in DU145 cells also results in inhibition of cell migration.
Targeted expression of FILIP1LΔC103 in tumor vasculature results in inhibition of tumor growth in vivo
We have shown that overexpression of FILIP1L results in inhibition of cell proliferation and migration and increased apoptosis in vitro. We then set out to evaluate the effects of targeted FILIP1L expression in vivo. In particular, we chose to selectively deliver FILIP1L to tumor vasculature to determine if overexpression of FILIP1L in tumor vasculature leads to an antitumor effect. To achieve this, we used a hybrid AAVP vector, which has been shown to specifically target tumor vasculature in an RGD peptide–restricted manner (20, 21, 24). These hybrid vectors rely on the specific binding relationship between an RGD peptide and αv integrin expressed on the surface of tumor vasculature. These vectors have been shown to specifically traffic to and specifically transfect tumor associated endothelial cells without evidence of transfection of normal endothelial cells. We tested two FILIP1L mutants for this purpose: FILIP1LΔC103 (amino acid 1–790) and FILIP1LΔC243 (amino acid 1–650). We cloned each FILIP1L mutant cDNA into the AAVP vector, produced RGD-targeted AAVP, and screened for the AAVP, which showed the highest expression of each mutant by real-time RT-PCR analysis.
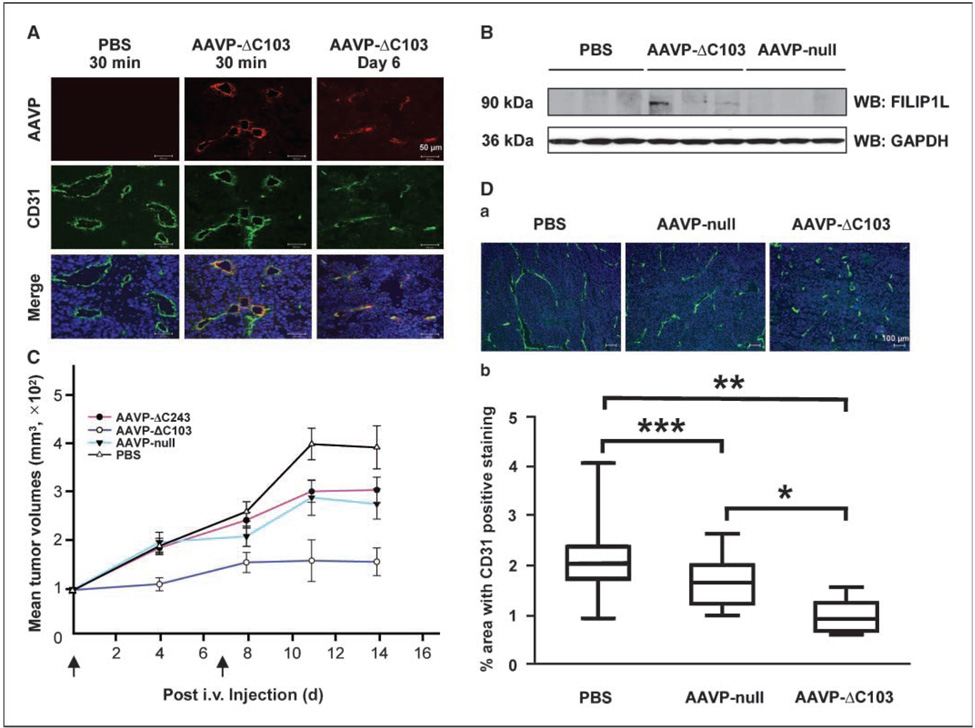
To examine whether the AAVP specifically targets tumor vasculature in our M21 human melanoma model, the RGD-targeted AAVP was injected into the tail vein of female athymic nude mice harboring a 100 to 150 mm3 subcutaneous M21 tumor. After injection, tumors were harvested by time course and analyzed by immunofluorescent staining with anti-CD31 and anti-AAVP antibodies. Both RGD-4C-FILIP1LΔC103 AAVP (AAVP expressing FILIP1L mutant 1–790; called AAVP-ΔC103) and RGD-4C-FILIP1LΔC243 AAVP (AAVP expressing FILIP1L mutant 1–650; hereafter called AAVP-ΔC243) were shown to specifically target tumor vasculature (Fig. 6A; data for AAVP-ΔC243 shown in Supplementary Fig. S4), but not tumor cells (Fig. 6A) or the vasculature of normal organs (Supplementary Fig. S5). We also measured FILIP1L protein expression in these tumors by Western blot analysis using anti-FILIP1L antibody on whole tumor lysates. Tumors from AAVP-ΔC103–treated mice, but not PBS and RGD-4C AAVP (control null AAVP; called AAVP-null)–treated mice, showed FILIP1L protein expression (Fig. 6B). Therefore, these data support the observation that the FILIP1L mutant is expressed in the tumor vasculature of mice treated with AAVP-ΔC103.
Figure 6.
Targeted expression of FILIP1LΔC103 in tumor vasculature results in inhibition of tumor growth in vivo. A, AAVP-ΔC103 injected i.v. specifically targeted tumor vasculature. Tumors from PBS-treated and AAVP-ΔC103–treated mice were immunofluorescently stained with anti-AAVP antibody followed by Alexa Fluor 594 antirabbit IgG (red, AAVP staining) and anti-CD31 antibody followed by Alexa Fluor 488 antirat IgG (green, blood vessel staining). Blue, nuclear staining with DAPI. Scale bar, 50 µm. The result is a representative image from two independent experiments. Data for AAVP-ΔC243 are shown in Supplementary Fig. S4. B, a 90-kDa FILIP1LΔC103 protein was detected in whole tumor lysates from AAVP-ΔC103–treated mice, but not PBS-treated and AAVP-null–treated mice, by Western blot using anti-FILIP1L antibody. Three different tumors per group were analyzed. GAPDH blot is shown as the loading control. C, tumors from AAVP-ΔC103–treated mice (empty circles) were significantly smaller than those from PBS-treated mice (empty triangles ) in M21 xenograft model at day 14 (P < 0.01). Tumors from AAVP-ΔC103–treated mice were also significantly smaller than those from AAVP-null–treated mice (filled triangles; P < 0.05) and AAVP-ΔC243–treated mice (filled circles; P < 0.05) by day 14. AAVP was injected i.v. at day 0 and day 7 (arrow). Bars, SE (n = 11). The result is representative of two independent experiments. D-a, representative images of CD31-stained tumors from PBS-treated, AAVP-null–treated, and AAVP-ΔC103–treated mice. Tumors treated with each AAVP for 4 d were immunofluorescently stained with anti-CD31 antibody followed by Alexa Fluor 488 anti-rat IgG (green, vessel staining). Blue, nuclear staining with DAPI. Scale bar, 100 µm. D-b, vessel density was significantly decreased in tumors from AAVP-ΔC103–treated mice compared with those from AAVP-null–treated mice (P < 0.01) and PBS-treated mice (P < 0.001) at day 4. The percentage of cells in the tumors stained positive for CD31 was quantified using Axiovision 4.6 software (Zeiss). Box and whiskers plot (GraphPad Prism 3.0) of vessel density. Middle lines indicate median values. *, P < 0.01; **, P < 0.001; ***, P < 0.05; n = 3 mice per treatment group.
To evaluate the efficacy of the FILIP1L mutant-AAVP treatment on tumor growth inhibition in vivo, M21 melanoma cells were injected s.c. into female athymic nude mice and grown to an average size of 100 mm3. Mice were randomly sorted into four groups (n = 11 for each group), AAVP (1 × 1011 transducing units per dose) was injected i.v. at day 0 and day 7, and tumors were measured in a blinded manner. The following groups were tested: PBS, AAVP-null, AAVP-ΔC243, and AAVP-ΔC103. PBS control tumors grew aggressively and started to show central necrosis by day 14. Thus, the experiments were terminated at day 14. Only tumors from AAVP-ΔC103–treated mice were significantly smaller than those from PBS-treated mice by day 14 (P < 0.01; Fig. 6C). Although tumors from AAVP-null–treated mice and AAVP-ΔC243– treated mice were smaller than those from PBS-treated mice, the differences were not statistically significant. In addition, tumors from AAVP-ΔC103–treated mice were significantly smaller than those from AAVP-null–treated mice (P < 0.05) and AAVP-ΔC243– treated mice (P < 0.05) by day 14 (Fig. 6C). These results suggest that targeted expression of FILIP1LΔC103 in tumor vasculature results in inhibition of M21 melanoma growth in vivo.
To confirm that the inhibition of tumor growth is dependent on the antivascular effects of FILIP1LΔC103, vessel density was analyzed for these AAVP-treated tumors. The percentage area of CD31-positive cells was used as a measure of vessel density (17). Vessel density from AAVP-ΔC103–treated tumors was significantly less than those from PBS-treated tumors (P < 0.001) and AAVPnull–treated tumors (P < 0.01) at day 4 (Fig. 6D), suggesting that the inhibition of tumor vasculature by AAVP-ΔC103 leads to the inhibition of M21 tumor growth. In addition, AAVP-ΔC103–treated tumors showed extensive apoptosis as measured by TUNEL staining compared with PBS-treated or AAVP-null–treated tumors (Supplementary Fig. S6), further suggesting that the inhibition of tumor vasculature by AAVP-ΔC103 results in induction of apoptosis and necrosis in these M21 tumors.
Discussion
The development and maintenance of a blood supply is critical for the growth, invasion, and metastatic spread of tumors. Inhibiting the process of new blood vessel formation, as well as attacking established tumor vasculature, has been shown to be a viable strategy for treating cancer (25). A number of antiangio-genic agents have been identified, and several have entered clinical trials.7 To better understand the mechanisms of action of these agents on endothelial cells and determine if common pathways are shared between agents, we previously analyzed the gene expression responses of endothelial cells to a variety of angiogenesis inhibitors (15, 16). Based on these studies, we identified FILIP1L (previously termed DOC1) as a potentially important common regulator of the antiangiogenic activity on endothelial cells.
In the present study, we characterized the patterns of FILIP1L expression and elucidated important functions of FILIP1L in endothelial cells and cancer cells. We have shown that FILIP1L protein is expressed in the vasculature and muscularis mucosa of the colon. The same pattern of expression was observed in human prostate (data not shown). In endothelial cells, endogenous FILIP1L protein expression is up-regulated by the angiogenesis inhibitor endostatin, suggesting that FILIP1L may be a downstream mediator of some of endostatin’s functions, including the inhibition of cell proliferation and migration and the induction of apoptosis. Overexpression of FILIP1L in endothelial cells results in inhibition of cell proliferation and increased apoptosis. The COOH terminal truncation mutant 1–790 (FILIP1LΔC103) is more potent than wild-type FILIP1L in mediating this antiproliferative activity. In addition, overexpression of FILIP1LΔC103 in HUVECs, as well as DU145 prostate cancer cells, leads to inhibition of cell migration. Finally, targeted expression of FILIP1LΔC103 in tumor-associated vasculature results in the inhibition of M21 melanoma growth in vivo, demonstrating a similar effect as that seen by the delivery of angiogenesis inhibitors. Therefore, these data suggest that the novel protein FILIP1L could be a potential candidate for cancer therapy by targeted delivery.
Overexpression of FILIP1L in vitro resulted in inhibition of cell proliferation in other cell types, in addition to endothelial cells. Normal fibroblasts, as well as some cancer cells, including DU145 prostate carcinoma cells showed inhibition of cell proliferation after FILIP1L overexpression (Supplementary Fig. S3 and data not shown). To avoid possible toxicity, we chose to selectively deliver FILIP1L to tumor vasculature. To achieve this, we used a hybrid AAVP vector, which has been shown to specifically target tumor vasculature (21). Systemic delivery of our targeted FILIP1LΔC103-expressing AAVP had the greatest effect on tumor growth inhibition (Fig. 6C).
Limited studies of FILIP1L from other laboratories have suggested that gene expression of FILIP1L is also implicated in neoplasia and senescence. FILIP1L mRNA was originally characterized as present in human ovarian epithelial cells, but consistently absent in ovarian carcinoma cells (26). Using cDNA microarray analysis, FILIP1L was identified as one of the genes whose transcription is induced in senescent human prostate epithelial cells, but significantly repressed in immortalized prostate epithelial cells (22, 23). In addition, FILIP1L mRNA expression was shown to be down-regulated in microvascular endothelial cells infected with Kaposi’s sarcoma-associated herpesvirus compared with uninfected microvascular endothelial cells (27). Furthermore, FILIP1L mRNA expression was also shown to be down-regulated in B cells transformed with an oncogene TaxBLV (bovine leukemia virus Tax) compared with untransformed B cells (28).
Although we have shown that overexpression of FILIP1L inhibits cell proliferation and migration, and induces apoptosis, the physiologic function of FILIP1L remains unclear. FILIP1L has a striking similarity to FILIP1 (29). There is 46% amino acid identity between FILIP1L and FILIP1. Both have coiled-coil in NH2 terminal halves, and FILIP1L has two leucine zipper motifs whereas FILIP1 has four leucine zipper motifs in its NH2 terminal halves. Both have two isoforms: isoform 2 of FILIP1L (893 amino acids) lacks the 240 amino-terminal residues of isoform 1 (1,135 amino acids) and S-FILIP1 (965 amino acids) lacks the 247 amino-terminal residues of L-FILIP1 (1,213 amino acids). Furthermore, FILIP1L is shown to be expressed in vessels and smooth muscle cells in human colon tissue (Fig. 1B), and FILIP1 is shown to be expressed in cardiac, skeletal, and smooth muscle, as well as in the nervous system (29). In contrast, the FILIP1L gene is located on chromosome 3q12.1, whereas the FILIP1 gene is located on chromosome 6q14.1. To date, the human FILIP1L gene is known to have 12 orthologues only in mammals, in contrast to the human FILIP1 gene which has 30 orthologues throughout many species.
FILIP1 binds to filamin A and induces degradation of filamentous actin associated filamin A, thereby suppressing cell motility and formation of lamellipodia (29, 30). In this study, we have shown that FILIP1L inhibits cell migration (Fig. 5A and D). In addition, immunofluorescence staining of both FILIP1L (Fig. 2B) and FILIP1(29) reveals a similar punctate distribution in the cytoplasm. Interestingly, LL5β, a PIP3 and γ-filamin–binding protein, shows a punctate distribution in the cytoplasm with colocalization of γ-filamin (31). A novel α-helical coiled-coil domain-containing protein FIP, which is shown to bind Dictyostelium filamin also shows a punctate staining pattern in the cytosol (32). Therefore, we suspect that FILIP1L might be one of FILIP1 family proteins that have a functional relationship to filamin A. Filamin A is an actin-binding protein which stabilizes three-dimensional actin filaments and links them to cellular membranes (reviewed in ref. 33). It is essential for cell motility and fetal development (34–36). Filamin A has been shown to bind a number of membrane receptors and signal transduction intermediates, including some regulatory cofactors of the Rho GTPases family (37–43). Angiogenesis inhibitors, such as endostatin, fumagillin, and EMAP II, have also been shown to regulate cytoskeletal organization to inhibit endothelial cell function (44–47). Therefore, the fact that FILIP1L expression is modulated by these inhibitors and that overexpression of FILIP1L in endothelial cells mimics the effects of these inhibitors further suggests that FILIP1L might exert its functions via the cytoskeleton. For instance, loss of filamin A was shown to result in not only inhibition of cell migration but also induction of apoptosis (48–50). Thus, it is possible that FILIP1L inhibits the function of filamin A after angiogenesis inhibitor treatment, thereby inhibiting endothelial cell function by the induction of apoptosis. Future studies to identify the physiologic binding partner(s) of FILIP1L and examine the potential relationship between FILIP1L and filamin A will be of interest.
In summary, we have characterized FILIP1L as a mediator of apoptosis and an inhibitor of cell migration and proliferation. Targeted delivery of the most potent FILIP1L truncation mutant to tumor vasculature results in tumor growth inhibition in a human xenograft mouse model. Further characterization of FILIP1L may lead to an improved understanding of the role played by agents that affect the cytoskeleton and may lead to the development of more effective anticancer agents.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Acknowledgments
Grant support: Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research.
We thank Drs. Amin Hajitou, Renata Pasqualini, and Wadih Arap for providing us pfUSE5-MCS phage plasmid and pAAV-GFP plasmid and E. coli strains of MC1061 and K91Kan and Dr. Anita Tandle for helpful discussion.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Wary KK. Molecular targets for anti-angiogenic therapy. Curr Opin Mol Ther. 2004;6:54–70. [PubMed] [Google Scholar]
- 2.Ziche M, Donnini S, Morbidelli L. Development of new drugs in angiogenesis. Curr Drug Targets. 2004;5:485–493. doi: 10.2174/1389450043345371. [DOI] [PubMed] [Google Scholar]
- 3.Digtyar AV, Pozdnyakova NV, Feldman NB, Lutsenko SV, Severin SE. Endostatin: current concepts about its biological role and mechanisms of action. Biochemistry (Mosc) 2007;72:235–246. doi: 10.1134/s0006297907030017. [DOI] [PubMed] [Google Scholar]
- 4.Dixelius J, Cross MJ, Matsumoto T, Claesson-Welsh L. Endostatin action and intracellular signaling: β-catenin as a potential target? Cancer Lett. 2003;196:1–12. doi: 10.1016/s0304-3835(03)00267-2. [DOI] [PubMed] [Google Scholar]
- 5.Ferreras M, Felbor U, Lenhard T, Olsen BR, Delaisse J. Generation and degradation of human endostatin proteins by various proteinases. FEBS Lett. 2000;486:247–251. doi: 10.1016/s0014-5793(00)02249-3. [DOI] [PubMed] [Google Scholar]
- 6.O’Reilly MS, Boehm T, Shing Y, et al. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 7.Ingber D, Fujita T, Kishimoto S, et al. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida T, Kaneko Y, Tsukamoto A, Han K, Ichinose M, Kimura S. Suppression of hepatoma growth and angiogenesis by a fumagillin derivative TNP470: possible involvement of nitric oxide synthase. Cancer Res. 1998;58:3751–3756. [PubMed] [Google Scholar]
- 9.Zhang Y, Griffith EC, Sage J, Jacks T, Liu JO. Cell cycle inhibition by the anti-angiogenic agent TNP-470 is mediated by p53 and p21WAF1/CIP1. Proc Natl Acad Sci U S A. 2000;97:6427–6432. doi: 10.1073/pnas.97.12.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger AC, Tang G, Alexander HR, Libutti SK. Endothelial monocyte-activating polypeptide II, a tumor-derived cytokine that plays an important role in inflammation, apoptosis, and angiogenesis. J Immunother. 2000;23:519–527. doi: 10.1097/00002371-200009000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Kao J, Houck K, Fan Y, et al. Characterization of a novel tumor-derived cytokine. Endothelial-monocyte activating polypeptide II. J Biol Chem. 1994;269:25106–25119. [PubMed] [Google Scholar]
- 12.Kao J, Ryan J, Brett G, et al. Endothelial monocyte-activating polypeptide II. A novel tumor-derived poly-peptide that activates host-response mechanisms. J Biol Chem. 1992;267:20239–20247. [PubMed] [Google Scholar]
- 13.Schwarz MA, Kandel J, Brett J, et al. Endothelial-monocyte activating polypeptide II, a novel antitumor cytokine that suppresses primary and metastatic tumor growth and induces apoptosis in growing endothelial cells. J Exp Med. 1999;190:341–354. doi: 10.1084/jem.190.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 15.Mazzanti CM, Tandle A, Lorang D, et al. Early genetic mechanisms underlying the inhibitory effects of endo-statin and fumagillin on human endothelial cells. Genome Res. 2004;14:1585–1593. doi: 10.1101/gr.2552804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tandle AT, Mazzanti C, Alexander HR, Roberts DD, Libutti SK. Endothelial monocyte activating polypep-tide-II induced gene expression changes in endothelial cells. Cytokine. 2005;30:347–358. doi: 10.1016/j.cyto.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Blansfield JA, Caragacianu D, Alexander HR, III, et al. Combining agents that target the tumor microenvironment improves the efficacy of anticancer therapy. Clin Cancer Res. 2008;14:270–280. doi: 10.1158/1078-0432.CCR-07-1562. [DOI] [PubMed] [Google Scholar]
- 18.Giaever I, Keese CR. A morphological biosensor for mammalian cells. Nature. 1993;366:591–592. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- 19.Keese CR, Wegener J, Walker SR, Giaever I. Electrical wound-healing assay for cells in vitro. Proc Natl Acad Sci U S A. 2004;101:1554–1559. doi: 10.1073/pnas.0307588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajitou A, Rangel R, Trepel M, et al. Design and construction of targeted AAVP vectors for mammalian cell transduction. Nat Protoc. 2007;2:523–531. doi: 10.1038/nprot.2007.51. [DOI] [PubMed] [Google Scholar]
- 21.Hajitou A, Trepel M, Lilley CE, et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 22.Schwarze SR, DePrimo SE, Grabert LM, Fu VX, Brooks JD, Jarrard DF. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J Biol Chem. 2002;277:14877–14883. doi: 10.1074/jbc.M200373200. [DOI] [PubMed] [Google Scholar]
- 23.Schwarze SR, Fu VX, Desotelle JA, Kenowski ML, Jarrard DF. The identification of senescence-specific genes during the induction of senescence in prostate cancer cells. Neoplasia. 2005;7:816–823. doi: 10.1593/neo.05250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soghomonyan S, Hajitou A, Rangel R, et al. Molecular PET imaging of HSV1-tk reporter gene expression using [18F]FEAU. Nat Protocol. 2007;2:416–423. doi: 10.1038/nprot.2007.49. [DOI] [PubMed] [Google Scholar]
- 25.Hajitou A, Pasqualini R, Arap W. Vascular targeting: recent advances and therapeutic perspectives. Trends Cardiovasc Med. 2006;16:80–88. doi: 10.1016/j.tcm.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mok SC, Wong KK, Chan RK, et al. Molecular cloning of differentially expressed genes in human epithelial ovarian cancer. Gynecol Oncol. 1994;52:247–252. doi: 10.1006/gyno.1994.1040. [DOI] [PubMed] [Google Scholar]
- 27.Poole LJ, Yu Y, Kim PS, Zheng QZ, Pevsner J, Hayward GS. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi’s sarcoma-associated herpesvirus. J Virol. 2002;76:3395–3420. doi: 10.1128/JVI.76.7.3395-3420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klener P, Szynal M, Cleuter Y, et al. Insights into gene expression changes impacting B-cell transformation: cross-species microarray analysis of bovine leukemia virus tax-responsive genes in ovine B cells. J Virol. 2006;80:1922–1938. doi: 10.1128/JVI.80.4.1922-1938.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagano T, Yoneda T, Hatanaka Y, Kubota C, Murakami F, Sato M. Filamin A-interacting protein (FILIP) regulates cortical cell migration out of the ventricular zone. Nat Cell Biol. 2002;4:495–501. doi: 10.1038/ncb808. [DOI] [PubMed] [Google Scholar]
- 30.Nagano T, Morikubo S, Sato M. Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J Neurosci. 2004;24:9648–9657. doi: 10.1523/JNEUROSCI.2363-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paranavitane V, Coadwell WJ, Eguinoa A, Hawkins PT, Stephens L. LL5β is a phosphatidylinositol (3,4,5)-trisphosphate sensor that can bind the cytoskeletal adaptor, γ-filamin. J Biol Chem. 2003;278:1328–1335. doi: 10.1074/jbc.M208352200. [DOI] [PubMed] [Google Scholar]
- 32.Knuth M, Khaire N, Kuspa A, Lu SJ, Schleicher M, Noegel AA. A novel partner for Dictyostelium filamin is an α-helical developmentally regulated protein. J Cell Sci. 2004;117:5013–5022. doi: 10.1242/jcs.01366. [DOI] [PubMed] [Google Scholar]
- 33.Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham CC. Actin structural proteins in cell motility. Cancer Metastasis Rev. 1992;11:69–77. doi: 10.1007/BF00047604. [DOI] [PubMed] [Google Scholar]
- 35.Fox JW, Lamperti ED, Eksioglu YZ, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 36.Robertson SP, Twigg SR, Sutherland-Smith AJ, et al. Localized mutations in the gene encoding the cytoskel-etal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 37.Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2:888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- 38.Calderwood DA, Huttenlocher A, Kiosses WB, et al. Increased filamin binding to β-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 39.Jimenez-Baranda S, Gomez-Mouton C, Rojas A, et al. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat Cell Biol. 2007;9:838–846. doi: 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- 40.Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U. Physical and functional interact-tion of filamin (actin-binding protein-280) and tumor necrosis factor receptor-associated factor 2. J Biol Chem. 2000;275:271–278. doi: 10.1074/jbc.275.1.271. [DOI] [PubMed] [Google Scholar]
- 41.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8:803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 42.Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates trans-forming growth factor-β signaling. J Biol Chem. 2001;276:17871–17877. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- 43.Vadlamudi RK, Li F, Adam L, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 44.Dixelius J, Cross M, Matsumoto T, Sasaki T, Timpl R, Claesson-Welsh L. Endostatin regulates endothelial cell adhesion and cytoskeletal organization. Cancer Res. 2002;62:1944–1947. [PubMed] [Google Scholar]
- 45.Keezer SM, Ivie SE, Krutzsch HC, Tandle A, Libutti SK, Roberts DD. Angiogenesis inhibitors target the endothelial cell cytoskeleton through altered regulation of heat shock protein 27 and cofilin. Cancer Res. 2003;63:6405–6412. [PubMed] [Google Scholar]
- 46.Wickstrom SA, Alitalo K, Keski-Oja J. Endostatin associates with lipid rafts and induces reorganization of the actin cytoskeleton via down-regulation of RhoA activity. J Biol Chem. 2003;278:37895–37901. doi: 10.1074/jbc.M303569200. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz MA, Zheng H, Liu J, Corbett S, Schwarz RE. Endothelial-monocyte activating polypeptide II alters fibronectin based endothelial cell adhesion and matrix assembly via α5 β1 integrin. Exp Cell Res. 2005;311:229–239. doi: 10.1016/j.yexcr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Bachmann AS, Howard JP, Vogel CW. Actin-binding protein filamin A is displayed on the surface of human neuroblastoma cells. Cancer Sci. 2006;97:1359–1365. doi: 10.1111/j.1349-7006.2006.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kainulainen T, Pender A, D’Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. J Biol Chem. 2002;277:21998–22009. doi: 10.1074/jbc.M200715200. [DOI] [PubMed] [Google Scholar]
- 50.Kim H, Sengupta A, Glogauer M, McCulloch CA. Filamin A regulates cell spreading and survival via β1 integrins. Exp Cell Res. 2008;314:834–846. doi: 10.1016/j.yexcr.2007.11.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).