Abstract
Loss- and gain-of-function mutations in the broadly expressed gene Lrp5 affect bone formation causing osteoporosis and high bone mass, respectively. Although Lrp5 is viewed as a Wnt coreceptor osteoblast-specific disruption of β-Catenin does not affect bone formation. Instead, we show here that Lrp5 inhibits expression of Tph1, the rate-limiting biosynthetic enzyme for serotonin in enterochromaffin cells of the duodenum. Accordingly, decreasing serotonin blood levels normalizes bone formation and bone mass in Lrp5-deficient mice and gut- but not osteoblast-specific Lrp5 inactivation decreases bone formation in a β-Catenin–independent manner. Moreover, gut-specific activation of Lrp5, or inactivation of Tph1, increases bone mass and prevents ovariectomy-induced bone loss. Serotonin acts on osteoblasts through the Htr1b receptor and CREB to inhibit their proliferation. By identifying duodenum-derived serotonin as a hormone inhibiting bone formation in an Lrp5-dependent manner this study broadens our understanding of bone remodeling and suggests novel therapies to increase bone mass.
Introduction
Bone remodeling, the physiological means whereby vertebrates renew their bones during adulthood, comprises two phases: resorption of preexisting mineralized bone matrix by a specialized cell type, the osteoclast, followed by de novo bone formation by another specialized cell type, the osteoblast. Over the last 15 years, molecular and genetic studies have identified numerous local and systemic regulators of this process and as a result have considerably improved our molecular understanding of both aspects of bone remodeling (Zaidi, 2007).
One of the most intensively studied regulators of bone remodeling is LDL-receptor related protein 5 (LRP5) (Baron and Rawadi, 2007). This interest stems from the fact that LRP5 loss-of-function mutations cause osteoporosis pseudoglioma (OPPG), a rare disease characterized by severe decreased bone formation and persistence of embryonic eye vascularization leading to low bone mass and blindness (Gong et al., 2001). Other, presumably activating, mutations in LRP5 cause the high bone mass syndrome (Boyden et al., 2002). That different mutations in this gene cause two bone diseases of opposite nature underscores the critical importance in the regulation of bone formation of the pathway(s) controlled by Lrp5.
Lrp5 encodes a broadly expressed cell surface molecule sharing sequence homology with Arrow, a coreceptor for the growth factor Wingless in Drosophila (Bhanot et al., 1996). Based on this sequence homology and, among other evidence, cell-based assays in which Lrp5 and Axin interaction is initiated by Wnt proteins it is assumed that Lrp5 is a coreceptor for Wnt proteins, the vertebrate homologs of Wingless (Tamai et al., 2004). As a result OPPG and the high bone mass syndrome are viewed as Wnt-related diseases (Krishnan et al., 2006). Three observations, however, challenge this view. First, contrasting with the developmental function of most Wnt proteins, there is no overt skeletal defect in Lrp5−/− embryos. Second, gain-of-function mutations in Lrp5 do not cause bone tumors as activation of Wnt signaling does in other organs (Moon et al., 2004). Third and more importantly, osteoblast-specific loss- and gain-of-function mutations in β-Catenin (β-Cat), the molecular node of canonical Wnt signaling, do not affect either bone formation or expression of genes dysregulated upon Lrp5 inactivation (Glass et al., 2005). Taken individually none of these observations rules out that Lrp5 functions as a Wnt coreceptor to regulate bone formation. However, when considered together we viewed them as an incentive to search for additional/other mechanisms of action of this gene in osteoblasts.
Serotonin is a bioamine generated in brainstem neurons and enterochromaffin cells of the duodenum, that does not cross the blood-brain barrier (Mann et al., 1992). Thus, it is de facto a molecule with two functional identities depending on its site of synthesis. While brain-derived serotonin is implicated in cognitive functions (Heath and Hen, 1995) the function(s) of gut-derived serotonin (GDS), which accounts for 95% of total serotonin, are still a matter of debate (Gershon and Tack, 2007). GDS biosynthetic pathway involves the rate-limiting enzyme tryptophan hydroxylase 1 (Tph1). GDS is released in the general circulation where most of it is taken up by platelets through a specific transporter (Gershon and Tack, 2007). A small fraction of it, however, remains free in the serum and may conceivably act as a hormone following its binding to serotonin receptors present on target cells (Rand and Reid, 1951). Remarkably, patients taking synthetic serotonin reuptake inhibitors (SSRIs) chronically, a class of drugs increasing extracellular serotonin concentration throughout the body, can have reduced bone mass (Richards et al., 2007).
While searching for molecular mechanisms explaining Lrp5 regulation of bone formation we identified Tph1 as the most highly over-expressed gene in Lrp5−/− bones. Tph1 was also increased in Lrp5−/− duodenal cells, its primary site of expression. Inhibiting serotonin synthesis in Lrp5−/− mice corrects their bone phenotype, and gut- but not osteoblast-specific deletion of Lrp5 recapitulates the bone phenotype of Lrp5−/− mice. The same pathway is affected, in an opposite manner, in the case of Lrp5 gain-of-function mutation thus providing a simple molecular basis for the two diseases. Serotonin, after binding to the Htr1b receptor, determines the extent of bone formation by controlling osteoblast proliferation through the regulation of CyclinD1 expression by CREB. By revealing that Lrp5 regulates bone mass by inhibiting duodenal synthesis of serotonin, a hormone decreasing bone formation, this study points towards adapted therapies for diseases characterized by an impairment of bone formation.
Results
Molecular signature of Lrp5 loss-of-function mutations
The different nature of the cellular events leading to a low bone mass phenotype in Lrp5−/− (decrease in bone formation) and β-Cat(ex3)osb−/− (decrease in bone resorption) mice suggested that distinct molecular mechanisms were at work in these two models. Thus, in an effort to elucidate how Lrp5 regulates bone formation we looked for a specific molecular signature of Lrp5 loss-of-function mutation in bone in vivo. For that purpose we analyzed the expression of genes affecting either cell proliferation (Cyclins), osteoblast differentiation (Runx2, Osx, Atf4), bone matrix deposition (Type I Collagen) or osteoclast differentiation (Osteoprotegerin, RankL) in WT, Lrp5−/− and β-Cat(ex3)osb−/− bones. The only genes whose expression was decreased in Lrp5−/− bones were the regulators of cell proliferation CycD1, D2 and E1 (Figure 1A). Consistent with the fact that osteoblast number is normal in β-Cat(ex3)osb−/− mice, expression of the Cyclin genes was not affected in β-Cat(ex3)osb−/− bones; conversely genes whose expression was affected in β-Cat(ex3)osb−/− bones were normally expressed in Lrp5−/− bones (Figure 1A).
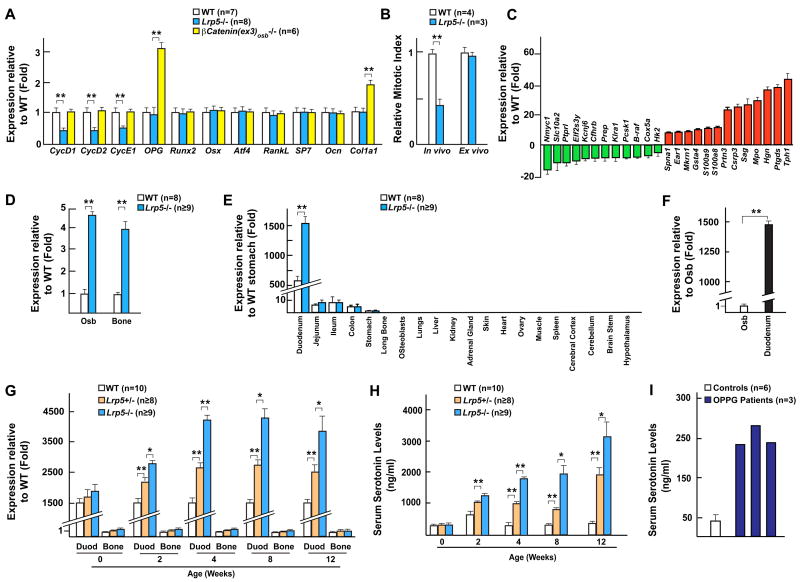
Figure 1. Increased Tph1 expression and serum serotonin levels in Lrp5−/− mice.
(A) Real-time PCR analysis of gene expression in WT, Lrp5−/− and β-Cat(ex3)osb−/− bones.
(B) Proliferation analysis (BrdU labeling) in vivo and ex vivo of WT and Lrp5−/− osteoblasts.
(C) Microarray analysis of WT and Lrp5−/− bones.
(D) Increased Tph1 expression in osteoblasts (Osb) and bones of Lrp5−/− mice by real-time PCR analysis.
(E–F) Real-time PCR analysis of Tph1 expression in tissues of WT and Lrp5−/− mice (E), and in primary osteoblasts (Osb) and gut of WT mice (F).
(G–H) Real-time PCR analysis of Tph1 expression in gut versus bone (G), and serum serotonin levels (H) in WT, Lrp5+/− and Lrp5−/− mice at indicated ages.
(I) Serum serotonin levels in two OPPG patients and age matched controls (n=6).
In striking contrast to the paucity of osteoblasts present in Lrp5−/− bones (Kato et al., 2002), Lrp5−/− osteoblasts proliferated as well as WT cells ex vivo (Figure 1B). We interpreted the discrepancy between the in vivo and ex vivo proliferation abilities of the Lrp5−/− osteoblasts as indicating that Lrp5 loss-of-function mutations affect osteoblast proliferation through extracellular signals that may not originate from osteoblasts, in other words that Lrp5-related bone diseases may not originate from bones.
As a first attempt to test the aforementioned hypothesis we reanalyzed results of a microarray experiment performed earlier (Glass et al., 2005). The gene most highly expressed in Lrp5−/− compared to WT bones was Tph1, which encodes an enzyme initiating synthesis, outside the brain of serotonin, a secreted molecule that may affect bone mass (Warden et al., 2005) (Figure 1C). Real-time PCR analysis verified that Tph1 was over-expressed in Lrp5−/− osteoblasts and bones (Figure 1D). Tph1 is predominantly expressed in duodenum in WT mice and it was also vastly over-expressed in this organ in Lrp5−/− mice where its expression was 1500 fold higher than in osteoblasts and 150 fold higher than in any other part of the gastro-intestinal tract (Figures 1E and 1F). Tph1 expression was not affected in β-Cat(ex3)osb−/− bones further differentiating Lrp5 and canonical Wnt signaling regulations of bone mass (Figure S1A). In contrast, neither expression of Tph2, the enzyme initiating serotonin synthesis in the brain, nor brain serotonin content was affected in Lrp5−/− mice (Figures S1B and S1C). This latter observation is consistent with the notion that serotonin does not cross the blood brain barrier (Mann et al., 1992).
Multiple correlative evidence suggested that this increase in Tph1 expression is implicated in the development of the Lrp5−/− mice bone phenotype. First, Tph1 expression was normal in newborn Lrp5−/− mice that do not display overt bone abnormalities but increased steadily thereafter (Figures 1G, S1D and S1E). A significant increase in Tph1 expression was first detected at 2 weeks of age i.e. 2 weeks before the low bone mass phenotype becomes apparent in Lrp5−/− mice. Consistent with this increase in Tph1 expression, blood serotonin levels increased as a function of time in Lrp5−/− mice (Figure 1H). This large increase in blood serotonin levels was not due to more platelets in Lrp5−/− mice (Figure S1F). Second, Tph1 expression and blood serotonin levels were increased in Lrp5+/− mice that also have a low bone mass (Figures 1G and 1H). Third, there was in three OPPG patients tested (Gong et al., 2001; Toomes et al., 2004) a 4 to 5 fold increase in circulating serotonin levels when compared to age matched controls; moreover, the heterozygous parent of one of these patients also had higher circulating serotonin levels than control individuals (Figures 1I and S1G). Thus, Lrp5 regulates blood serotonin level in both mice and humans.
Increased circulating serotonin level as a determinant of the Lrp5 −/− mice bone phenotype
To determine whether the increase in circulating serotonin level induced by the increase in Tph1 expression is a cause of the bone phenotype of the Lrp5−/− mice we relied on cell-based and in vivo experiments.
To elucidate the discrepancy between the paucity of osteoblasts in Lrp5−/− bones but their normal proliferation ability ex vivo we tested whether serotonin is an inhibitor of osteoblast proliferation. When treated with serotonin for 24h proliferation of WT or Lrp5−/− osteoblasts was decreased to the same extent (Figure 2A), in contrast Wnt3A conditioned media did not affect osteoblast proliferation (Figure S2F). Serotonin decreased expression of CycD1, D2 and E1 without affecting expression of Type I Collagen or of other genes characteristic of the osteoblast phenotype (Figures 2B and S2A), a gene expression profile remarkably similar to the one seen in the Lrp5−/− bones (Figure 1A). To further determine whether serotonin inhibits osteoblast proliferation in a Wnt-dependent or -independent manner we treated ROS17 osteoblastic cells transfected with the TOPFLASH reporter construct with increasing amount of serotonin or, as a positive control, with lithium chloride (LiCl) (Clement-Lacroix et al., 2005). While LiCl increased the activity of this reporter vector in all experiments, serotonin always failed to do so indicating that the serotonin and canonical Wnt signaling pathways are distinct in osteoblasts (Figure 2C).
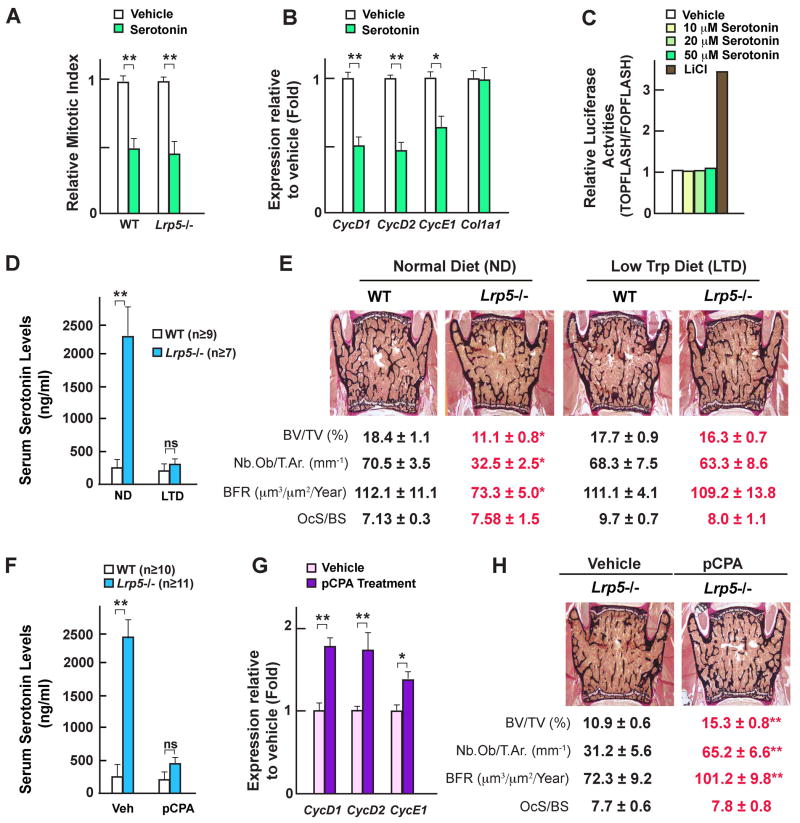
Figure 2. Serotonin inhibits osteoblast proliferation and decreasing serum serotonin levels corrects Lrp5 −/− mice bone phenotype.
(A) Ex vivo proliferation rates of WT and Lrp5−/− osteoblasts treated with serotonin (50μM) for 24 hours.
(B) Real-time PCR analysis of Cyclins and Col1a1 expression in WT osteoblasts treated with serotonin (50μM) for 24 hours.
(C) TOPFLASH reporter activities in osteoblasts in response to serotonin (10–50 μM) or lithium chloride (LiCl, 10mM).
(D–E) Serum serotonin levels (D) and histological analysis of vertebrae (E) of WT and Lrp5−/− mice fed a normal diet or a 75% tryptophan-less diet. Mineralized bone matrix is stained in black by von Kossa reagent. Histomorphometric parameters. BV/TV%, bone volume over trabecular volume; Nb.Ob/T.Ar., number of osteoblasts per trabecular area; BFR, bone formation rate; OcS/BS, osteoclast surface per bone surface.
(F–H) Serum serotonin levels (F), real-time PCR analysis of Cyclin expression in bone (G) and histomorphometric analyses (H) in WT and Lrp5−/− mice treated with the serotonin synthesis inhibitor pCPA (100 mg/kg).
If the increase in blood serotonin level is a cause of the Lrp5−/− mice bone phenotype then normalizing it should also normalize bone mass in these mice. To test this hypothesis we performed two in vivo experiments. Since serotonin is a tryptophan derivative, we first fed WT and Lrp5−/− mice from 3 to 12 weeks of age with a diet containing 75% less tryptophan than a normal diet. This reduction in tryptophan intake decreased circulating serotonin levels 8 to 10-fold without affecting serotonin brain content (Figures 2D and S2B) and normalized bone formation parameters and bone mass in Lrp5−/− mice (Figure 2E). Second, 4 week-old WT and Lrp5−/− mice were administered Para-chlorophenylalanine (pCPA), a drug inhibiting serotonin synthesis, for 8 weeks (Kubera et al., 2000). PCPA decreased circulating serotonin levels 5 to 7-fold in Lrp5−/− mice without affecting their serotonin brain content; it also normalized in Lrp5−/− mice bone mass, bone formation parameters, and Cyclin expression in bones (Figures 2F–H and S2C). That pCPA did not induce regression of the hyaloid vessels in the eyes suggests that the eye phenotype of the Lrp5−/− mice is not caused by this increase in circulating serotonin levels (Figures S2D and S2E).
Taken together results of the cell biology experiments, of the low tryptophan diet and the pCPA-induced serotonin depletion in WT and Lrp5−/− mice indicate that higher blood serotonin levels contribute to the decrease in bone formation and bone mass in Lrp5−/− mice.
Lrp5 regulates serotonin synthesis and bone formation through its duodenal expression
In light of these results we next asked whether Lrp5 regulates Tph1 expression, serotonin synthesis and bone formation through its duodenal or its osteoblasts expression. To that end we generated mice harboring a floxed loss-of-function allele of Lrp5 (Figures S3A and S3B) and crossed them with either Villin-Cre transgenic mice to delete Lrp5 from gut cells (Lrp5gut−/−) or with α1(I)Col-Cre transgenic mice to delete it from osteoblasts only (Lrp5osb−/−) (Dacquin et al., 2002; el Marjou et al., 2004). Real-time PCR analysis demonstrated that efficient recombination occurred in duodenal cells but not in osteoblasts in the Lrp5gut−/− mice while the opposite was true in the Lrp5osb−/− mice (Figures 3A and S3C). Tph1 expression was high in the gut and normal in osteoblasts in Lrp5gut−/− mice, the opposite was true in the Lrp5osb−/− mice (Figure 3B). Circulating serotonin levels were 5–8 fold higher in Lrp5gut−/− than in WT mice but remained normal in Lrp5osb−/− mice (Figure 3C). In both mutant mice, brain serotonin content was normal (data not shown).
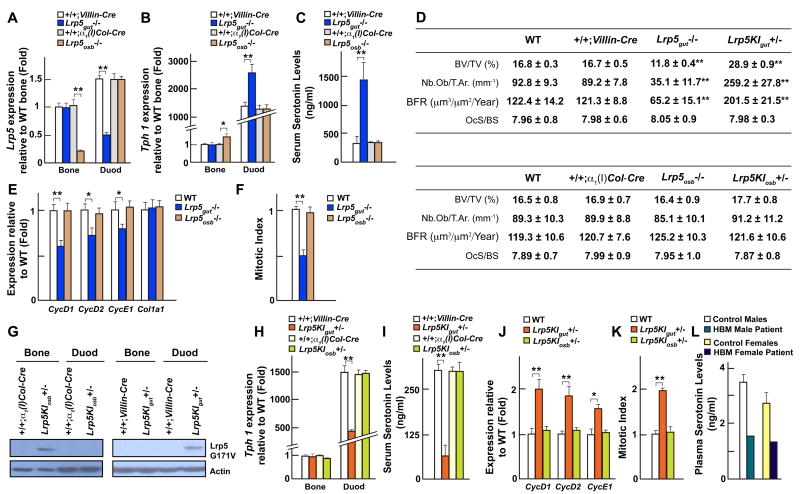
Figure 3. Lrp5 regulates serotonin synthesis through its expression in gut.
(A–B) Real-time PCR analysis of Lrp5 (A) and Tph1 (B) expression in the gut and long bones (Bone) of Lrp5gut−/− and Lrp5osb−/− compared to +/+;Villin-Cre and +/+;α1(I)Col-Cre mice, respectively.
(C) Serum serotonin levels in +/+;Villin-Cre, Lrp5gut−/−, +/+;α1(I)Col-Cre and Lrp5osb−/− mice.
(D) Histomorphometric analysis of vertebrae of WT, +/+;Villin-Cre, Lrp5gut−/−, Lrp5KIgut+/−, +/+;α1(I)Col-Cre, Lrp5osb−/− and Lrp5KIosb+/− mice.
(E) Real-time PCR analysis of Cyclins and Col1a1 expression in long bones of WT, Lrp5gut−/− and Lrp5osb−/− mice.
(F) In vivo osteoblast proliferation in WT, Lrp5gut−/− and Lrp5osb−/− mice.
(G) Western blot analysis of Lrp5 high bone mass (G171V) cDNA expression (Flag) in bone and gut of Lrp5KIosb+/− and Lrp5KIgut+/− compared to +/+;α1(I)Col-Cre and +/+;Villin-Cre mice respectively.
(H–I) Real-time PCR analysis of Tph1 expression (H) in the gut and long bones (Bone) and serum serotonin levels (I) in Lrp5KIosb+/− and Lrp5KIgut+/− compared to +/+;Villin-Cre and +/+;α1(I)Col-Cre mice, respectively.
(J–K) Real-time PCR analysis of Cyclins and Col1a1 expression (J) and in vivo osteoblast proliferation (K) in long bones of WT, Lrp5KIgut+/− and Lrp5KIosb+/− mice.
(L) Plasma serotonin levels in two high bone mass (HBM) patients and age matched controls (n=3).
Histomorphometric analyses showed a decrease in bone mass in Lrp5gut−/− but not in Lrp5osb−/− mice (Figure 3D). This low bone mass was secondary to a decrease in osteoblast numbers and bone formation while osteoclast numbers were not affected (Figure 3D). Expression of CycD1, D2 and E1 was decreased in the bones of Lrp5gut−/− mice but was unaffected in Lrp5osb−/− bones and BrdU incorporation assays showed that osteoblast proliferation was decreased in Lrp5gut−/− but not in Lrp5osb−/− mice (Figures 3E and 3F). Thus, gut- but not osteoblast-specific deletion of Lrp5 recapitulates the low bone mass and the molecular abnormalities observed in the Lrp5−/− mice. As it is the case for Lrp5+/− mice, Lrp5gut+/− mice displayed a low bone formation/low bone mass phenotype (Figure S3D). This data suggests that Lrp5 regulates bone formation through its duodenal expression. In contrast, histological examination failed to detect any persistence of hyaloid vessels of the eyes in either Lrp5gut−/− or Lrp5osb−/− mice (Figure S3E). This latter result, along with the inability of pCPA to prevent appearance of the eye phenotype in the Lrp5−/− mice, demonstrates that Lrp5 uses different mechanisms to regulate bone formation and eye vascularization.
Duodenal-specific Lrp5 activating mutation causes high bone mass
The demonstration that Lrp5 loss-of-function affects bone mass by enhancing serotonin synthesis in the enterochromaffin cells raised the prospect that, conversely, Lrp5 gain-of-function mutations might affect bone mass by decreasing serotonin synthesis in these cells. To test this hypothesis we generated mice harboring a floxed allele of Lrp5 that included the mutation causing high bone mass in humans (Boyden et al., 2002) (Figures S3F and S3G). These mice were crossed with either Villin-Cre or α1(I)Col-Cre transgenic mice to generate mice expressing this Lrp5 gain-of-function mutation in gut cells (Lrp5KIgut+/−) or in osteoblasts (Lrp5KIosb+/−). We studied heterozygous mutant mice because the high bone mass syndrome is observed in patients heterozygous for the mutation (Boyden et al., 2002).
Lrp5KIgut+/− mice expressed the mutated protein in gut but not in osteoblasts, the opposite was true for Lrp5KIosb+/− mice (Figures 3G, S3H and S3I). Tph1 expression in gut and circulating serotonin levels were low in Lrp5KIgut+/− mice (Figures 3H and 3I) while serotonin content in the brain was not affected (data not shown). Histomorphometric analyses showed that bone formation parameters and bone mass were high in Lrp5KIgut+/− but normal in Lrp5KIosb+/− mice (Figure 3D). Expression of Cyclins and in vivo osteoblast proliferation were increased in Lrp5KIgut+/− bones; none of these parameters were modified in Lrp5KIosb+/− mice (Figures 3J, 3K and S3J). To validate our findings obtained in mice we studied two patients with high bone mass (HBM) caused by the same activating mutation (G171V) in LRP5 (Boyden et al., 2002) and observed a 50% decrease in circulating serotonin levels (Figure 3L).
In summary, based on the analyses of cell-specific loss- and gain-of-function mutations of Lrp5 models it appears that this gene regulates bone formation through its duodenal but not its bone expression.
Gut-derived serotonin regulates bone formation
To further demonstrate that it is duodenal-derived serotonin that regulates bone formation we generated mice harboring a floxed loss-of-function allele of Tph1 and crossed them with Villin-Cre or α1(I)Col-Cre mice to generate Tph1gut−/− or Tph1osb−/− mice (Figures S4A–C). Since Tph1 is over-expressed in Lrp5−/− mice its deletion in the appropriate cell type should result in a bone phenotype opposite to the one of the Lrp5−/− mice (i.e. high bone mass). Tph1gut−/− mice had a 10-fold reduction in Tph1 expression in gut while Tph1 expression in bone and osteoblasts was not affected; conversely Tph1osb−/− mice had a 10-fold reduction in Tph1 expression in bones and osteoblasts but a normal Tph1 expression in gut (Figures 4A and 4B). Circulating serotonin levels were low in Tph1gut−/− but normal in Tph1osb−/− mice (Figure 4C).
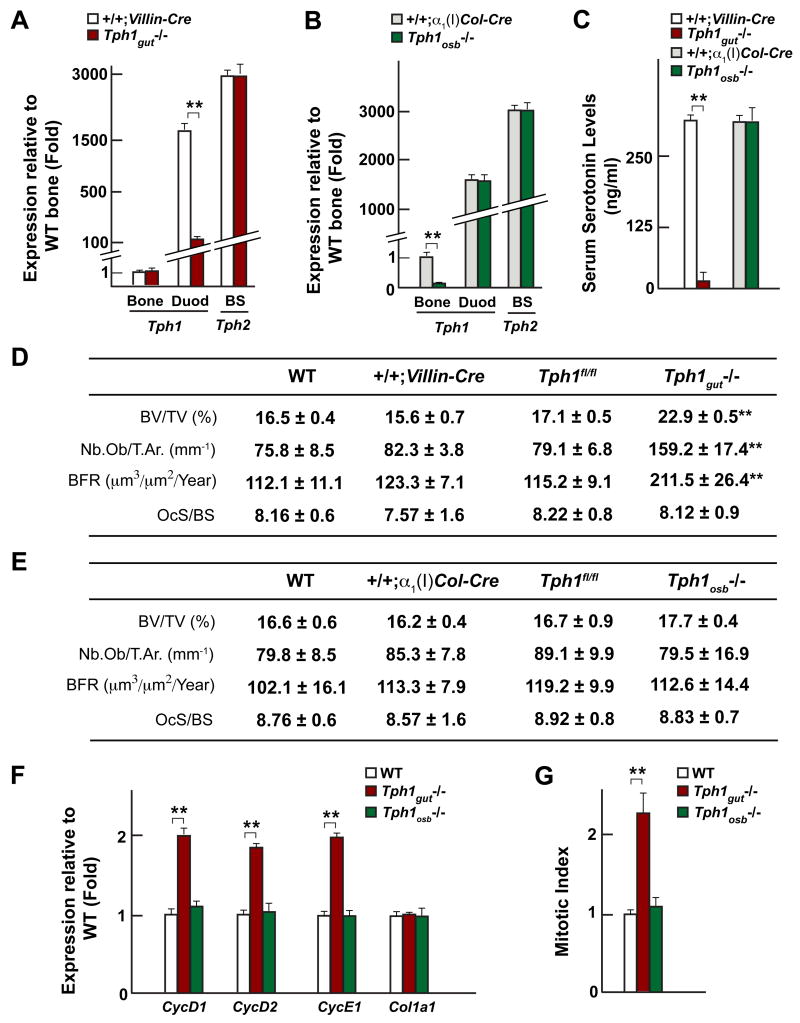
Figure 4. Duodenal-derived serotonin regulates bone formation.
(A–B) Real-time PCR analysis of Tph1 expression in gut and long bones (Bone) and Tph2 expression in the brainstem (BS) of Tph1gut−/− and Tph1osb−/−compared to +/+;Villin-Cre and +/+;α1(I)Col-Cre mice respectively.
(C–E) Serum serotonin levels (C) and bone histomorphometric analysis (vertebrae) (D–E) in WT, +/+;Villin-Cre, Tph1gut−/−, +/+;α1(I)Col-Cre and Tph1osb−/− mice.
(F) Real-time PCR analysis of Cyclins and Col1a1 expression in long bones of WT, Tph1gut−/− and Tph1osb−/− mice.
(G) In vivo osteoblast proliferation in WT, Tph1gut−/− and Tph1osb−/− mice.
As anticipated, mice lacking Tph1 in the gut developed a severe high bone mass phenotype secondary to an increase in osteoblast number and bone formation rate, bone resorption parameters were not affected (Figure 4D). This phenotype, that is the mirror image of the one seen in the Lrp5−/− mice, was present in 4, 6 and 12 week-old Tph1gut−/− mice (Figures 4D and S4D). In contrast, mice lacking Tph1 in osteoblasts had only a marginal, non-significant, increase in bone mass (Figure 4E). Expression of CycD1, D2 and E1, that was decreased in the absence of Lrp5, was increased in Tph1gut−/− but normal in Tph1osb−/− bones and BrdU incorporation assays showed that osteoblast proliferation was increased in Tph1gut−/− but not in Tph1osb−/− mice (Figures 4F and 4G).
The Htr1b receptor mediates serotonin regulation of osteoblast proliferation
If circulating serotonin is an hormone inhibiting bone formation it must act through a specific receptor(s) present on osteoblasts whose inactivation should lead to an increase in bone formation and high bone mass.
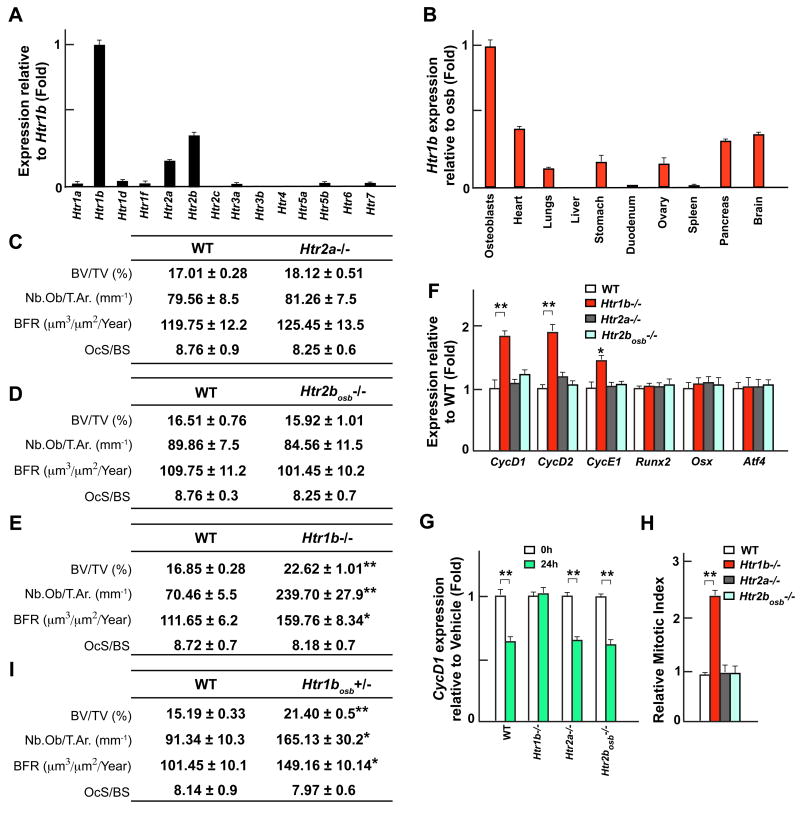
Among the 14 known serotonin receptors only 3 are expressed in osteoblasts, Htr1b, the most highly expressed, Htr2b and Htr2a (Figure 5A). Htr1b is preferentially expressed in bone and osteoblasts outside the brain while Htr2b and Htr2a have a broader pattern of expression (Figures 5B and S5A). Analysis of Htr2a−/− mice failed to detect any abnormalities in osteoblast number, bone formation, bone resorption and bone mass at 1 or 3 month of ages (Figures 5C and S5B), two time points at which Lrp5−/− mice already display a severe bone phenotype. The same was true for, mice lacking in osteoblasts only, Htr2b (Figures 5D and S5B–E). In contrast, mice lacking either one or the two Htr1b alleles displayed a similar increase in osteoblast number, bone formation rate and bone mass at those ages (Figures 5E and S5B). Molecularly, expression of CycD1, D2 and E1, that is downregulated in Lrp5−/− bones, was upregulated in Htr1b−/− but not in Htr2a−/− or Htr2bosb−/− bones and serotonin treatment decreased CycD1 expression in WT, Htr2a−/− and Htr2bosb−/− but not in Htr1b−/− osteoblasts (Figures 5F and 5G). BrdU incorporation assays confirmed that osteoblast proliferation was increased in Htr1b−/− but not in Htr2a−/− and Htr2bosb−/− bones (Figure 5H). Other osteoblast-specific genes such as Osteoprotegerin, a Wnt target gene (Glass et al., 2005), were normally expressed in Htr1b−/− bones further differentiating canonical Wnt and serotonin signaling pathways in osteoblasts (Figure S5F).
Figure 5. Serotonin inhibits osteoblasts proliferation through Htr1b.
(A–B) Real-time PCR analysis of serotonin receptors expression in primary osteoblasts (A) and of Htr1b expression in different tissues of WT mice (B).
(C–E) Histomorphometric analysis of vertebrae of 3 month-old WT, Htr2a−/− (C), Htr2bosb−/− (D) and Htr1b−/− (E) mice.
(F) Real-time PCR analysis of Cyclins, Runx2, Osx, and Atf4 expression in long bones of WT, Htr1b−/−, Htr2a−/− and Htr2bosb−/− mice.
(G) Real-time PCR analysis of CycD1 expression in primary osteoblasts of WT, Htr1b−/−, Htr2a−/− and Htr2bosb−/− mice treated with serotonin (50μM).
(H) In vivo osteoblast proliferation in WT, Htr1b−/−, Htr2a−/− and Htr2bosb−/− mice.
(I) Histomorphometric analysis of vertebrae of 6 week-old WT and Htr1bosb+/− mice.
To ascertain that Htr1b regulates bone formation through its expression in osteoblasts we generated mice lacking Htr1b only in osteoblasts (Htr1bosb-deficient mice; Figure S5G–H). As shown in Figure 5I Htr1bosb+/− mice display the same high bone formation/high bone mass phenotype than the one observed in the Htr1b+/− mice. Taken together these results indicate that serotonin uses one predominant receptor, Htr1b, to affect osteoblast biology.
Serotonin regulates osteoblast proliferation through CREB
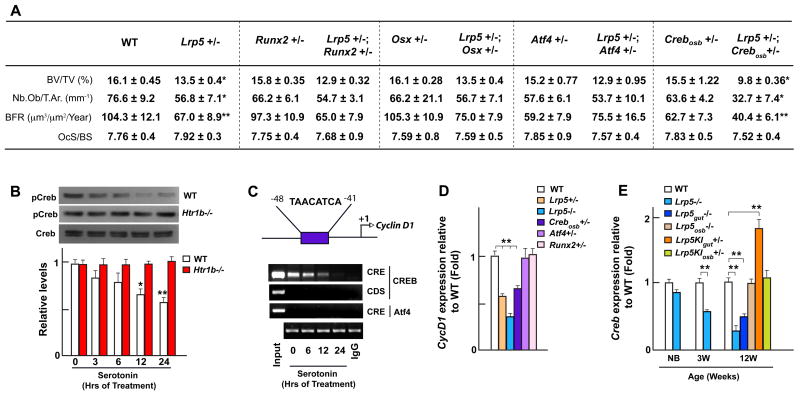
We next searched for the transcriptional mediator(s) of serotonin signaling in osteoblasts. To address this question, since loss-of-function mutations in most of the key transcription factors governing osteoblast differentiation are embryonic lethal, we generated compound mutant mice lacking one allele of Lrp5 and one allele of each transcription factor of interest. We reasoned that if Lrp5 and a particular transcription factor are in the same genetic cascade, compound heterozygous mice may reproduce the bone phenotype of the Lrp5−/− mice. Removing one allele of Runx2 or Osx from Lrp5+/− mice did not worsen the bone phenotype of these latter mutant mice, a result consistent with the normal expression of Runx2 and Osx in Lrp5−/− osteoblasts (Figure 1A). These observations strongly suggest that neither of these two factors act downstream of serotonin (Figure 6A and S6).
Figure 6. Lrp5 mediates its effect through CREB.
(A) Histomorphometric analysis of vertebrae of WT, single (Lrp5+/−, Runx2+/−, Osx+/−, Atf4+/− or Crebosb+/−) and double (Lrp5+/−;Runx2+/−, Lrp5+/−;Osx+/−, Lrp5+/−;Atf4+/− or Lrp5+/−;Crebosb+/−) heterozygous mutant mice.
(B–C) Western blot analysis of CREB phosphorylation (B) and chromatin-immunoprecipitation assay of CREB binding to the consensus cAMP response element (CRE) in CycD1 promoter (C) at 0, 3, 6, 12 and 24 hours upon serotonin (50μM) treatment. Coding sequence (CDS) PCR, IgG pulldown and ATF4 binding to the same site were used as negative controls.
(D) Real-time PCR analysis of CycD1 expression in the long bones of WT, Lrp5+/−, Lrp5−/−, Crebosb+/−, Osx+/− and Runx2+/− mice.
(E) Real-time PCR analysis of Creb expression in the long bones of WT, Lrp5−/−, Lrp5gut−/−, Lrp5osb−/−, Lrp5KIgut+/− and Lrp5KIosb+/− mice.
Since Htr1b is linked to the Gαi protein that inhibits cAMP production and PKA-dependent phosphorylation (Heath and Hen, 1995), we studied transcription factors that, like CREB, are phosphorylated by PKA. Removing one allele of Atf4, a CREB-related osteoblast-enriched transcription factor (Yang et al., 2004), from Lrp5+/− mice did not affect the bone phenotype of these latter mice arguing against a role for ATF4 in mediating serotonin signaling in osteoblasts (Figure 6A and S6). In contrast, compound mutant mice lacking one allele of Lrp5 and, in osteoblast only, one allele of Creb displayed a low bone formation/low bone mass phenotype undistinguishable from the one observed in the Lrp5−/− mice (Figure 6A and S6). This result suggested that CREB is a major transcriptional mediator of serotonin signaling in osteoblasts. In support of this contention, serotonin treatment of primary osteoblasts decreased phosphorylation of CREB on Ser133, the target of PKA, in WT but not in Htr1b−/− cells (Figure 6B) and abolished binding of CREB to the promoter of CycD1, a known target gene of CREB (Figure 6C) (Fu et al., 2005). That CycD1 expression is decreased in Crebosb+/− but not in Atf4+/− mice, used here as a negative control, is in full agreement with the ex vivo assays presented above (Figure 6D). Furthermore, Creb expression was decreased in Lrp5gut−/− long bones, increased in Lrp5KIgut+/− long bones but unaffected in either Lrp5osb−/− or Lrp5KIosb+/− long bones (Figure 6E) a finding that may mean that serotonin regulates Creb expression in vivo.
Genetic validation of the interactions between Lrp5, Tph1, Htr1b and Creb
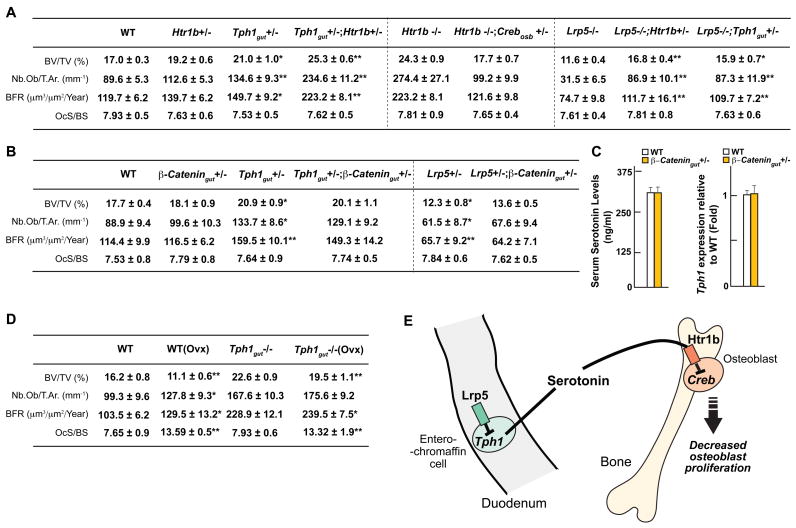
To demonstrate that the gut-derived serotonin/osteoblasts/CREB pathway described here mediates Lrp5-dependent regulation of osteoblast proliferation and bone formation we generated various compound mutant mice. First, we showed that mutant mice lacking one allele of Htr1b and one allele of Tph1 in gut cells developed the same high bone mass phenotype than Htr1b−/− or Tph1gut−/− mice (Figure 7A and S7A). Moreover, removal of one allele of Creb, in an osteoblast-specific manner, from the Htr1b−/− mice rescued the increase in bone formation parameters and the high bone mass of these mutant mice (Figure 7A and S7A). These experiments indicated that Tph1 in gut cells, Htr1b and Creb in osteoblasts are in the same genetic cascade regulating bone formation. Next, to ascertain that Tph1 in the gut and Htr1b mediate Lrp5-dependent regulation of bone mass, we removed from the Lrp5−/− mice one allele of Tph1 in gut cells or one allele of Htr1b (Figure 7A and S7A). Both these manipulations could normalize the bone abnormalities observed in the Lrp5−/− mice indicating that serotonin synthesis in duodenal enterochromaffin cells, and serotonin binding to Htr1b are downstream of Lrp5 (Figure 7A and S7A). Thus the results presented in this study indicate that the main mechanism whereby Lrp5 favors bone formation and bone mass accrual is by inhibiting Tph1 expression in and serotonin synthesis by enterochromaffin cells of the gut. Serotonin in turn prevents bone formation following its binding to Htr1b by inhibiting Creb expression and function, CycD1 expression and osteoblast proliferation.
Figure 7. Genetic interaction between Lrp5 and serotonin.
(A) Bone histomorphometric analysis (vertebrae) of WT, Htr1b+/−, Tph1gut+/−, Htr1b+/−;Tph1gut+/−, Lrp5−/−, Lrp5−/−;Htr1b+/− and Lrp5−/−;Tph1gut+/− mice.
(B) Bone histomorphometric analysis (vertebrae) of WT, β-Catgut+/−, Tph1gut+/−;β-Catgut+/−, Lrp5+/− and Lrp5+/−;β-Catgut+/− mice.
(C) Serum serotonin levels and real-time PCR analysis of Tph1 expression in the gut of WT and β-Catgut+/− mice.
(D) Bone histomorphometric analysis (vertebrae) of WT, WT(Ovx), Tph1gut−/− and Tph1gut−/−(Ovx) mice.
(E) Model of the Lrp5-dependent regulation of bone formation. Lrp5 favors bone formation and bone mass accrual by inhibiting Tph1 expression and serotonin synthesis in enterochromaffin cells. Following its binding to Htr1b serotonin inhibits Creb expression and function, this results in a decrease in CycD1 expression and osteoblast proliferation.
Lrp5, β-Cat expression in gut and bone mass
Since Lrp5 has been proposed to be a Wnt co-receptor we next asked if it was regulating Tph1 expression in a β-Cat-dependent or -independent manner. To that end we generated mice lacking one allele of Lrp5 and one allele of β-Cat in gut cells (Figures S7B and S7C) reasoning that if Lrp5 and β-Cat were in the same genetic pathway then these compound heterozygous mice should have the same low bone formation and low bone mass phenotype than the Lrp5−/− mice. This strategy has been used by us to demonstrate genetic interaction between β-Cat and other genes in osteoblasts (Glass et al., 2005).
As shown in Figure 7B while Lrp5+/− mice had a low bone formation and low bone mass phenotype β-Catgut+/− mice did not; more importantly the compound heterozygous Lrp5+/−;β-Catgut+/− did not have a more severe low bone formation and low bone mass than the Lrp5+/− mice. Molecularly, there was no change in Tph1 expression in the gut and in serum serotonin levels in β-Catgut+/− compared to WT mice (Figure 7C). We also generated compound mutant mice lacking one allele of Tph1 and one allele of β-Cat in gut. Here again we did not see in Tph1gut+/−;β-Catgut+/− any worsening of the bone phenotype observed in the Tph1gut+/− mice (Figures 7B). Because of their negative nature these results have to be interpreted cautiously, yet that β-Cat haploinsufficieny can in other cell types lead to phenotypic abnormalities indicate that Lrp5 regulates Tph1 expression and serotonin synthesis independently of β-Cat expression in gut (Glass et al., 2005).
Decreasing duodenal serotonin synthesis protects mice from ovariectomy-induced bone loss
To assess the biological importance of the serotonin regulation of bone mass we tested whether the increase in bone formation parameters observed in Tph1gut−/− mice was sufficient to protect them from ovariectomy-induced bone loss. WT and Tph1gut−/− were ovariectomized (Ovx) at 6 weeks of age and analyzed at 3 months of age. Histomorphometric analyses showed an increase in bone resorption parameters in both WT and Tph1gut−/− mice following Ovx (Figure 7D). Nevertheless, while Ovx WT mice developed a low bone mass phenotype Ovx Tph1gut−/− mice still harbored a high bone mass phenotype which was due to their increase in bone formation parameters (Figure 7D). Thus decreasing serotonin production in enterochromaffin cells of the gut protects mice from ovariectomy-induced osteoporosis.
Discussion
This study uncovers an unanticipated molecular mechanism accounting for the Lrp5 regulation of bone formation; our findings shift the emphasis from a paracrine to a novel endocrine regulation of bone mass, and from bone cells to enterochromaffin cells of the gut (Figure 7E). Moreover, given the cell-specific expression of Tph1 this study points toward novel therapeutic avenues for diseases characterized by a relative or absolute decrease in bone formation.
Lrp5-dependent serotonin synthesis in the gut and regulation of bone formation
Regardless of the mode of action of Lrp5, a hallmark of the Lrp5−/− bones is a paucity of osteoblasts, a feature contrasting strikingly with the fact that Lrp5−/− osteoblasts proliferate normally ex vivo. One possible explanation of this discrepancy could be that osteoblast proliferation in the Lrp5−/− mice is affected by an extracellular signal that is not originating from osteoblasts themselves.
In full support of the hypothesis that the extracellular signal regulating osteoblasts proliferation in absence of Lrp5 does not emanate from osteoblasts, cell-specific gene inactivation or gain-of-function studies established that it is through its expression in enterochromaffin cells of the gut and not through its expression in osteoblasts that Lrp5 controls Tph1 expression, serotonin synthesis, osteoblast proliferation and bone formation. The differences in the severity of the bone phenotype observed in Tph1gut−/− and Lrp5KIgut+/− mice respectively raises the hypothesis that Lrp5 regulates expression in enterochromaffin cells of other gene/s regulating bone mass. Finally, we cannot rule out that Lrp5 may have an accessory role in osteoblasts. By showing that OPPG and high bone mass syndrome are more gut- than bone-originating diseases these results put a new light on these two conditions. This work does provide an explanation for the osteoporosis often observed in patients with autism that have high blood serotonin levels (Hediger et al., 2007), however, it does not explain why patients taking chronically SSRIs often develop osteoporosis (Richards et al., 2007).
LRPs, bone remodeling, canonical Wnt signaling
Lrp5 is a member of the family of lipoprotein receptor related proteins (Lrps), a small group of single-pass trans-membrane proteins (Tamai et al., 2000). Lrp5 has a significant degree of sequence homology with the drosophila protein arrow, a wingless coreceptor (Pinson et al., 2000; Tamai et al., 2004). Based on this sequence homology and multiple forced expression experiments in cell culture and in vivo it has been assumed that Lrp5 is a coreceptor for Wnt proteins, the mammalian homologues of wingless, and that it is in this capacity that it regulates bone formation (Gong et al., 2001; Kato et al., 2002).
However, at the present time all in vivo evidence available indicate that Lrp5 regulates gut-derived serotonin production and bone formation in a canonical Wnt signaling pathway-independent manner. First, expression of classical Wnt target genes is normal in Lrp5−/− mice gut (Figure S7D); second, serotonin does not affect the activity of a TOPFLASH reporter construct or the expression of Osteoprotegerin in osteoblasts nor does it affect osteoclast differentiation as β-Cat inactivation does (Glass et al., 2005). Third, neither Lrp5+/−;β-Catgut+/− or Tph1gut+/−;β-Catgut+/− mice show the bone phenotype of Lrp5−/− or Tph1gut−/− mice respectively indicating that Lrp5 and Tph1 on one hand and β-Cat on the other hand are not acting in the same pathways. It has been shown before that LiCl could correct the bone phenotype of the Lrp5−/− mice (Clement-Lacroix et al., 2005). This does not necessarily contradict this contention since LiCl does not act exclusively on Wnt signaling and can affect many pathways in cells including serotonin in the brain where its function, if any, in regulating bone mass is for now unknown (Basselin et al., 2005).
The homology of Arrow with Lrp5 extends to other Lrp proteins. Indeed, closest Lrp5 relative, Lrp6, also has a high degree of homology with arrow. This is important since, all genetic evidence obtained before the function of Lrp5 during bone formation was discovered, suggested that Lrp6 was a better candidate than Lrp5 to be a Wnt coreceptor. For instance, Lrp6 but not Lrp5 is required for Wnt signaling in xenopus and in mice (Pinson et al., 2000; Tamai et al., 2004; Wehrli et al., 2000). Likewise, Wnt proteins fused to Frizzled could interact with Lrp6 but not with Lrp5 to activate a Wnt-responsive reporter gene (Holmen et al., 2002), Lrp6 inactivation affects embryonic development while Lrp5 inactivation does not (Pinson et al., 2000), more decisively, Lrp6 inactivation affects bone resorption in a TCF-dependent manner like β-Cat osteoblast-specific inactivation does but does not impair bone formation as Lrp5 loss-of-function mutation does (Glass et al., 2005; Kato et al., 2002; Kubota T., 2008).
Our results do not rule out, however, that Lrp5, in other organs and/or in cell types other than osteoblasts, may transduce a Wnt signal. In particular that the eye phenotype of the Lrp5−/− mice is not observed in the case of a duodenal- and osteoblast-specific deletion of Lrp5 and is not corrected by decreasing extracellular serotonin concentration rules out a serotonin involvement in this abnormality. This observation provides indirect support to a previous work showing an involvement of Lrp5 in Wnt signaling in that case (Lobov et al., 2005).
Serotonin signaling in osteoblasts
Three serotonin receptors are expressed in osteoblasts. Two of them Htr2b and Htr2a, are broadly expressed while Htr1b is, outside the brain, a fairly osteoblast-enriched gene. According to gene deletion experiments, Htr1b is the receptor responsible for the effect of serotonin on osteoblasts, while inactivation of either Htr2b or Htr2a does not affect bone mass. Our results regarding Htr2b are different from those of Collet et al. (Collet et al., 2008). However, in their case the bone phenotype was observed in mice lacking the gene in all cells while in our case Htr2b was deleted only in osteoblasts. Thus our results relate more tightly to the function (or lack of) of this gene in osteoblasts. It is possible that the low bone mass phenotype they noted in the classical Htr2b−/− mice could be secondary to the decrease in heart function observed in these mice (Nebigil et al., 2000).
Until recently the prevailing view of the transcriptional control of osteoblast biology is that it involves one of 3 players that are osteoblast-specific: Runx2, Osx and ATF4. Yet, none of these 3 transcription factors appears implicated in serotonin signaling in osteoblasts. Instead, CREB a factor already known to affect osteoblast proliferation but that is broadly expressed (Fu et al., 2005), is the main transcriptional effector of serotonin signaling in osteoblasts. This observation along with the emerging role in osteoblasts of other transcription factors, such as among others Schnurri-3 or FOXO that are not osteoblast-specific (Jones et al., 2006; Manolagas and Almeida, 2007) suggest that besides cell-specific transcription factors, other ones, more broadly expressed, will influence significantly differentiation and/or functions of the osteoblasts.
Remaining questions
The demonstration that Lrp5 belongs to a novel endocrine axis regulating bone formation illustrates the power of an integrative approach to bone physiology. It also raises novel questions. For instance and from a biological point of view we do not know yet how Lrp5 expression affects Tph1 expression in enterochromaffin cells of the gut. The structure of the Lrp5 protein raises the prospect that it may be a signaling receptor for a yet to be identified serotonin synthesis inhibiting molecule. From a therapeutic point of view the fact that ovariectomized Tph1gut−/− mice do not display bone loss holds great promise given Tph1 cell-specific expression. Further experiments will be needed to determine whether pharmacologically inhibiting Tph1 or use of selective peripheral Htr1b receptor antagonist(s) is a valid approach in the treatment of post-menopausal osteoporosis.
Experimental Procedures
Mice Generation
To generate osteoblast-specific and gut-specific gene-deficient mice for Lrp5, Tph1, Lrp5KI mutation and Htr2b (osb−/−, and gut−/−) targeting vectors harboring LoxP sites as well as a floxed neomycin resistance cassette were electroporated into ES cells (For details see Figures S3–5). ES cells containing the floxed allele (after NeoR removal) were injected in 129Sv/EV blastocysts to generate chimeric mice. flox/+ mice were crossed with α1(I)Col-Cre mice or Villin-Cre (obtained from national cancer institute mouse repository) mice to generate osb+/− or gut+/− mice, and their progeny was intercrossed to obtain osb−/− or gut−/− mice. Generation of Lrp5−/−, β-Catfl/fl, β-Cat(ex3)fl/fl, Htr1b−/−, Htr2a−/−, Crebfl/fl, Atf4−/−, Runx2−/−, Osx−/−, Villin-Cre and α1(I)Col-Cre mice was previously reported (Dacquin et al., 2002; el Marjou et al., 2004; Glass et al., 2005; Heath and Hen, 1995; Kato et al., 2002; Mantamadiotis et al., 2002; Weisstaub et al., 2006; Yang et al., 2004).
Histology, protein expression and proliferation assays
Immunohistochemistry was performed according to standard protocols on specimens embedded in paraffin and sectioned at 6μm (Ducy et al., 2000). In vivo osteoblast proliferation assays (Zymed Laboratories, Inc.) were performed on 4 week-old mice injected with BrdU (0.4 mg) and sacrificed 4 hr later. Static and dynamic histomorphometric analyses were performed on vertebral column specimens collected from 3-month old mice using undecalcified sections according to standard protocols using the Osteomeasure Analysis System (Osteometrics, Atlanta). Six to 12 animals were analyzed for each group.
Cell Cultures and Bioassays
Primary osteoblasts were cultured as previously described (Ducy et al., 2000). Drug treatments were performed in 0.1% FBS. Cell proliferation was quantified by BrdU or MTT assays (Kato et al., 2002). Serum serotonin levels were quantified by ELISA (Serotonin kit, Fitzgerald) while serotonin levels in brain regions were quantified by HPLC as described previously (Mann et al., 1992).
Molecular studies
RNA isolation and Real-time PCR was performed following standard protocols. Genotypes of all the mice were determined by PCR (See Figures S3–5). All primer sequences are available upon request.
Statistical Analysis
Results are given as means ± standard deviations. Statistical Analysis was performed by Student’s t test. All panels in Figures 1–7 and S1–S6 * p < 0.05 and ** p < 0.01 versus WT or control.
Supplementary Material
Acknowledgments
We thank Drs. JP Bilezikian, S. Cremers, DA Mackey, M Zacharin R. Lang, S. Rao, H. Schubert and J. Tosi for patient samples and eye analysis; G. Ren and Yung-yu Huang for superb technical assistance and Dr. M. Gershon for his critical reading of this manuscript. This work was supported by grants from the March of Dimes foundation and the National Institutes of Health (to G.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baron R, Rawadi G. Targeting the Wnt/beta-catenin pathway to regulate bone formation in the adult skeleton. Endocrinology. 2007;148:2635–2643. doi: 10.1210/en.2007-0270. [DOI] [PubMed] [Google Scholar]
- Basselin M, Chang L, Seemann R, Bell JM, Rapoport SI. Chronic lithium administration to rats selectively modifies 5-HT2A/2C receptor-mediated brain signaling via arachidonic acid. Neuropsychopharmacology. 2005;30:461–472. doi: 10.1038/sj.npp.1300611. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Clement-Lacroix P, Ai M, Morvan F, Roman-Roman S, Vayssiere B, Belleville C, Estrera K, Warman ML, Baron R, Rawadi G. Lrp5-independent activation of Wnt signaling by lithium chloride increases bone formation and bone mass in mice. Proc Natl Acad Sci U S A. 2005;102:17406–17411. doi: 10.1073/pnas.0505259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet C, Schiltz C, Geoffroy V, Maroteaux L, Launay JM, de Vernejoul MC. The serotonin 5-HT2B receptor controls bone mass via osteoblast recruitment and proliferation. FASEB J. 2008;22:418–427. doi: 10.1096/fj.07-9209com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, Shen J, Vinson C, Rueger JM, Karsenty G. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell. 2000;100:197–207. doi: 10.1016/s0092-8674(00)81558-5. [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi: 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Glass DA, 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL Receptor-Related Protein 5 (LRP5) Affects Bone Accrual and Eye Development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Heath MJ, Hen R. Serotonin receptors. Genetic insights into serotonin function. Curr Biol. 1995;5:997–999. doi: 10.1016/s0960-9822(95)00199-0. [DOI] [PubMed] [Google Scholar]
- Hediger ML, England LJ, Molloy CA, Yu KF, Manning-Courtney P, Mills JL. Reduced Bone Cortical Thickness in Boys with Autism or Autism Spectrum Disorder. J Autism Dev Disord. 2007 doi: 10.1007/s10803-007-0453-6. [DOI] [PubMed] [Google Scholar]
- Holmen SL, Salic A, Zylstra CR, Kirschner MW, Williams BO. A novel set of Wnt-Frizzled fusion proteins identifies receptor components that activate beta -catenin-dependent signaling. J Biol Chem. 2002;277:34727–34735. doi: 10.1074/jbc.M204989200. [DOI] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubera M, Kenis G, Bosmans E, Scharpe S, Maes M. Effects of serotonin and serotonergic agonists and antagonists on the production of interferon-gamma and interleukin-10. Neuropsychopharmacology. 2000;23:89–98. doi: 10.1016/S0893-133X(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Kubota TMT, Sakaguchi N, Kokubu C, Suzuki A, Namba N, Sakai N, Nakajima S, Imai K, Ozono K. An Lrp6 hypomorphic mutation affects bone mass through bone resorption in mice and impairs interaction with Mesd. Journal of Bone and Mineral Research. 2008 doi: 10.1359/jbmr.080512. [DOI] [PubMed] [Google Scholar]
- Lobov IB, Rao S, Carroll TJ, Vallance JE, Ito M, Ondr JK, Kurup S, Glass DA, Patel MS, Shu W, et al. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 2005;437:417–421. doi: 10.1038/nature03928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, McBride PA, Brown RP, Linnoila M, Leon AC, DeMeo M, Mieczkowski T, Myers JE, Stanley M. Relationship between central and peripheral serotonin indexes in depressed and suicidal psychiatric inpatients. Arch Gen Psychiatry. 1992;49:442–446. doi: 10.1001/archpsyc.1992.01820060022003. [DOI] [PubMed] [Google Scholar]
- Manolagas SC, Almeida M. Gone with the Wnts: beta-catenin, T-cell factor, forkhead box O, and oxidative stress in age-dependent diseases of bone, lipid, and glucose metabolism. Mol Endocrinol. 2007;21:2605–2614. doi: 10.1210/me.2007-0259. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Nebigil CG, Choi DS, Dierich A, Hickel P, Le Meur M, Messaddeq N, Launay JM, Maroteaux L. Serotonin 2B receptor is required for heart development. Proc Natl Acad Sci U S A. 2000;97:9508–9513. doi: 10.1073/pnas.97.17.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535– 538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Rand M, Reid G. Source of ‘serotonin’ in serum. Nature. 1951;168:385. doi: 10.1038/168385b0. [DOI] [PubMed] [Google Scholar]
- Richards JB, Papaioannou A, Adachi JD, Joseph L, Whitson HE, Prior JC, Goltzman D. Effect of selective serotonin reuptake inhibitors on the risk of fracture. Arch Intern Med. 2007;167:188–194. doi: 10.1001/archinte.167.2.188. [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- Toomes C, Bottomley HM, Jackson RM, Towns KV, Scott S, Mackey DA, Craig JE, Jiang L, Yang Z, Trembath R, et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004;74:721–730. doi: 10.1086/383202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden SJ, Robling AG, Sanders MS, Bliziotes MM, Turner CH. Inhibition of the serotonin (5-hydroxytryptamine) transporter reduces bone accrual during growth. Endocrinology. 2005;146:685–693. doi: 10.1210/en.2004-1259. [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D. Arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Weisstaub NV, Zhou M, Lira A, Lambe E, Gonzalez-Maeso J, Hornung JP, Sibille E, Underwood M, Itohara S, Dauer WT, et al. Cortical 5-HT2A receptor signaling modulates anxiety-like behaviors in mice. Science. 2006;313:536–540. doi: 10.1126/science.1123432. [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.