SUMMARY
A crucial step in protein translation is the translocation of tRNAs through the ribosome. In the transition from one canonical site to the other, the tRNAs acquire intermediate configurations, so-called hybrid states. At this stage, the small subunits is rotated with respect to the large subunit, and the anticodon stem loops reside in the A and P sites of the small subunit, while the acceptor ends interact with the P and E sites of the large subunit. In this work, by means of cryo-EM and particle classification procedures, we visualize for the first time the hybrid state of both A/P and P/E tRNAs in an authentic factor-free ribosome complex during translocation. In addition, we show how the repositioning of the tRNAs goes hand in hand with the change in the interplay between S13, L1 stalk, L5, H68, H69 and H38 that is caused by the ratcheting of the small subunit.
INTRODUCTION
The protein elongation cycle performed by the ribosome is a multistep sequential process encompassing aa-tRNA selection, peptide bond formation, and translocation. The aminoacyl-tRNA (aa-tRNA) selection by the ribosome is based on the correct Watson-Crick base complementary between the codon of the messenger RNA (mRNA) in the A site of the 30S subunit of the ribosome and the anticodon of the incoming tRNA. As the aa-tRNA enters the ribosome, in complex with EF-Tu and GTP, it assumes a deformed, high-energy conformation (A/T state of tRNA binding (Valle et al., 2003a)). During the decoding process, a signal is transmitted from the codon-anticodon recognition site and received at the GTP-binding site on elongation factor Tu (EF-Tu) (Ogle et al., 2002; Valle et al., 2003a; Cochella and Green, 2005). Following the conformational change of EF-Tu induced by the hydrolysis of GTP (Abel et al., 1996; Polekhina et al., 1996), EF-Tu dissociates from the aminoacyl end of the aa-tRNA and the ribosome. As the tRNA relaxes its distorted conformation, its 3’ end (acceptor arm) is accommodated into the A site of the ribosome, leading to a rapid peptide bond transfer in the peptidyl-transferase center of the 50S subunit. At this stage, the pre-translocational ribosome contains a deacylated tRNA in the P site and a peptidyl-tRNA in the A site. In the last step of the protein elongation cycle, GTPase elongation factor G (EF-G) catalyzes the translocation of the mRNA-tRNA complex through the subunit interface from the A and P sites to the P and E sites, respectively, a process coupled to the ratchet-like rotation of the 30S subunit relative to the 50S subunit (Frank and Agrawal, 2000; Frank et al., 2007). This ratcheting movement was shown to be essential for translocation (Horan and Noller, 2007).
Chemical modification experiments provided evidence that tRNAs can sample a hybrid state of binding from a post-peptidyl transfer, pre-translocation state (Moazed and Noller, 1989). These data provided support for earlier models for a hybrid state of tRNA binding proposed by Bretscher (Bretscher, 1968). Subsequent single molecule FRET studies have detected fluctuations in the positions of tRNAs (Blanchard et al., 2004), leading to the conclusion that the classical (A/A-P/P) and hybrid states (A/P-P/E) of the tRNAs are actually in dynamic equilibrium. Kinetic analysis has also supported the relevance of such intermediates, showing that the translocation driven by EF-G is kinetically more efficient when the tRNAs are bound in the hybrid state (Dorner et al., 2006). However, the exact interaction pattern of the hybrid A-site peptidyl tRNA and Psite deacylated tRNA has remained structurally uncharacterized since the hybrid state of the pre-translocational ribosome has not been observed in previous cryoEM or X-ray studies.
It is well known that the ribosome is very sensitive to buffer conditions, especially to Mg2+ concentration (Agrawal et al., 1999; Muth et al., 2001), as its high negative charge tends to make its conformation especially dependent on ionic conditions (Draper et al., 2005). Along these lines, a recent FRET analysis of the effect of Mg2+ on tRNA dynamics has shown that the Mg2+ concentration dramatically alters the dynamic equilibrium between the classical and hybrid states after peptidyl-transfer in pre-translocational complexes (Kim et al., 2007). In the light of this evidence, we revisit the problem analyzing the structure and dynamics of a pre-translocational ribosome in the presence of low Mg2+ concentration (3.5mM) and thus more physiological-like conditions (Cromie et al., 2007). As the sample corresponds to a homogeneous ribosome preparation showing conformational variability (i.e., equilibrium distribution of conformational states), we have applied supervised classification methodology to separate the initial group of ribosomes into two different subsets. This approach has allowed us to capture both classic and hybrid state of the tRNAs, and furthermore, it has enabled us to correlate these two states with the conformational changes underlying the ratchet-like rotation of the 30S subunit. In particular, we show how the reconfiguration of the tRNAs is based on the concerted action of S13, L1, L5, H68, H69 and H38, structural elements that change their relative position during the movement of the subunit interface. The structural characterization of the classical and hybrid tRNA present in our current work provides a molecular explanation for several previous findings made by other studies using biochemistry, kinetics, structural and single-molecule techniques, and also sheds light on the specific recognition of the pre-translocational ribosomal complex by EF-G. Finally, the results suggest a mechanistic explanation for the sensitivity of observed ribosome conformations to the Mg2+ buffer concentration, as an indirect effect of E-site tRNA trapped on the ribosome.
RESULTS AND DISCUSSION
Structural characteristics of classic and hybrid-state ribosomes
We collected cryo-EM data for pre-translocational ribosomes, originally with fMet-tRNAfMet in the P site (“post-initiation 70S complexes”), that were incubated with cognate Trp-tRNATrp-EF-Tu-GTP ternary complexes. After Trp-tRNATrp accommodation and subsequent peptidyl transfer, the ribosomes thus obtained bear fMet-Trp-tRNATrp and deacylated tRNAfMet (Materials and Methods). Initially, a cryo-EM map at 8.75 Å resolution was calculated using the whole data set (comprising ~216,000 particles). In this initial reconstruction (Supplementary Figure 1) the densities corresponding to the individual tRNAs were fused and inseparable. In addition, certain regions of the 30S subunit showed an inferior definition. The coexistence of different conformations in the initial pool of ribosomes was thus evident. Therefore, the data set was sorted based on the resemblance (as measured by the cross-correlation coefficient) of the data to two reference maps showing the ribosome in two states related by the ratchet motion (Figure 1). In this classification strategy, two 3D references are used that represent the conformations in two different states. Then, the experimental data are compared with projections from the two different references, and the class assignment is made based on the reference that gives the highest correlation value. In preparing these two reference maps, to avoid bias, the densities corresponding to tRNA and EF-G were computationally removed by the application of a soft mask, so that the references differed only by the ratchet rotation of 30S subunit relative to the 50S subunit (see Gao et al., 2004). This strategy allowed us to obtain two different 3D reconstructions corresponding to the different conformations that coexist in the sample. The first map, at 10.5 Å resolution, is seen to correspond to a less populated (~30%) state showing an unratcheted ribosome with tRNAs in the classic A/A-P/P configuration (Figure 2A), while the second map, visualized at 8.9 Å resolution, represents a more populated conformation (~70%) that shows a ratcheted-like rotation of the 30S subunit and tRNAs in a hybrid A/P-P/E configuration (Figure 2B).
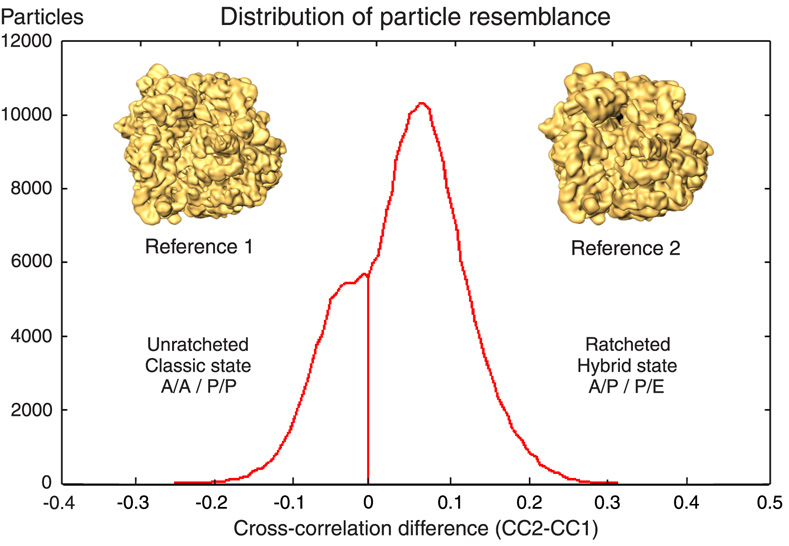
Figure 1. Statistical distribution of particle resemblance.
Histogram of values obtained by forming the difference between correlation coefficients of each experimental projection with respect to two references. A reference line is drawn at the selected cutoff point (ΔCC=0). The data were sorted based on their resemblance to 3D template ribosome maps that differ in the ratcheting rotation of the 30S subunit (insets). Both reference volumes were from a previous study (Valle et al., 2003b). Reference 1 was the cryo-EM density map of a pre-translocational complex (70S-tRNAfMet-fMet-Phe-tRNAPhe), while reference 2 was the cryoEM map of a ribosomal complex bound with EF-G-GDPNP (70S-MFTI-tRNAIle-EF-G-GDPNP + puromycin), which displayed ratchet like rotation. The densities corresponding to tRNAs and EF-G were computationally removed to avoid reference bias. To this end, soft Gaussian masks were used. Using this strategy, a clear bimodal distribution was obtained and two major ribosomal populations were separated. One, composed by 57,603 particles, corresponded to unratcheted ribosomes bearing the classic A/A-P/P tRNA configuration, while the second population, consisting of 158,969 particles, showed the ratched-like rotation of the 30S subunit hand-in-hand with the A/P-P/E hybrid tRNA configuration as shown in Figure 2.
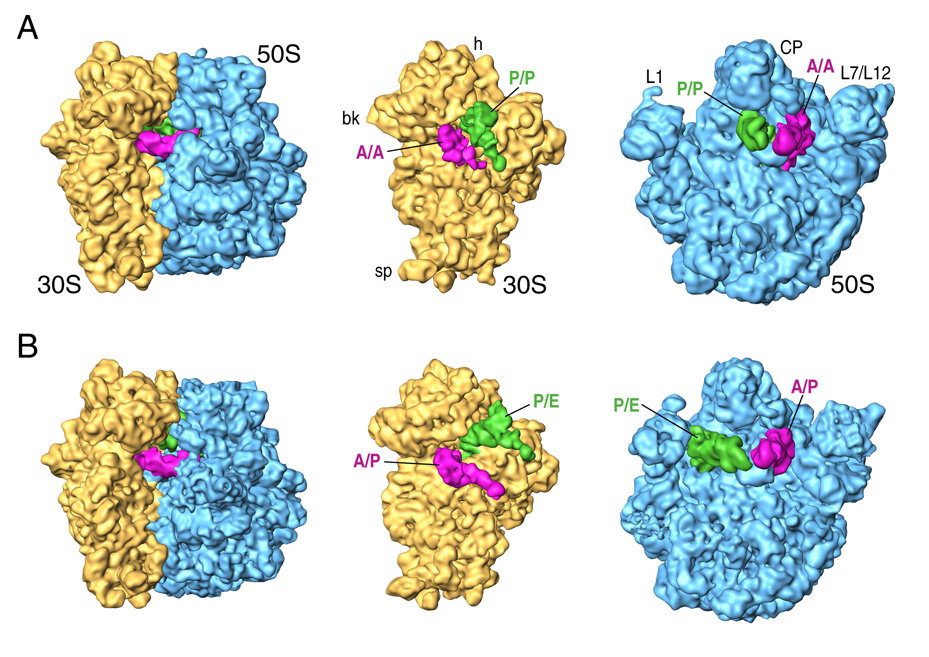
Figure 2. General features of classic and hybrid ribosomes.
Cryo-EM densities are shown for the small (yellow) and large (blue) subunits, and A and P-site tRNAs (magenta and green, respectively). The classic-state ribosome is shown in (A) and the hybrid-state ribosome in (B). Labels and landmarks: sp (spur), bk (beak), h (head), CP (central protuberance), L1 (L1 stalk), L7/L12 (L7/L12 stalk).
It is worth noting that in our reconstruction of the classic state, the A/A tRNA does not show a well-defined acceptor stem end. This is clearly observed when fitting the X-ray structure of the A site tRNA into its cryo-EM density (Supplementary Figure 3). As some degree of flexibility of the 3′ end of the A site tRNA is required for peptide bond formation (the reaction occurs while the tRNAs reside in their canonical A/A-P/P configuration (Schmeing et al., 2002)), the lack of visible density for the CCA-end’s last nucleotides in our reconstruction probably arises from conformational variability due to the mobility of this region.
Besides the different relative configuration of the tRNAs, a set of large-scale rearrangements in the ribosome can be easily noticed when comparing the two maps. A superposition of the large subunits as shown in Figure 3A reveals a clear counter-clockwise rotation of the small subunit when seen from the solvent side of the 30S subunit, as the positions of the spur, beak and head are strongly altered. This rotation causes the displacement of several components placed in the head of the small subunit, and the consequent reconfiguration of several of the original intersubunit bridges (Gavashvili et al., 2000). Bridge B1b, formed by the interaction of L5 and S13, is remodelled, and so is bridge B1a, in which H38’s binding partner S13 is replaced by S19 in the ratcheted state. In this process, H38, as well as the central protuberance region where L5 is located, appear to adapt their new conformations.
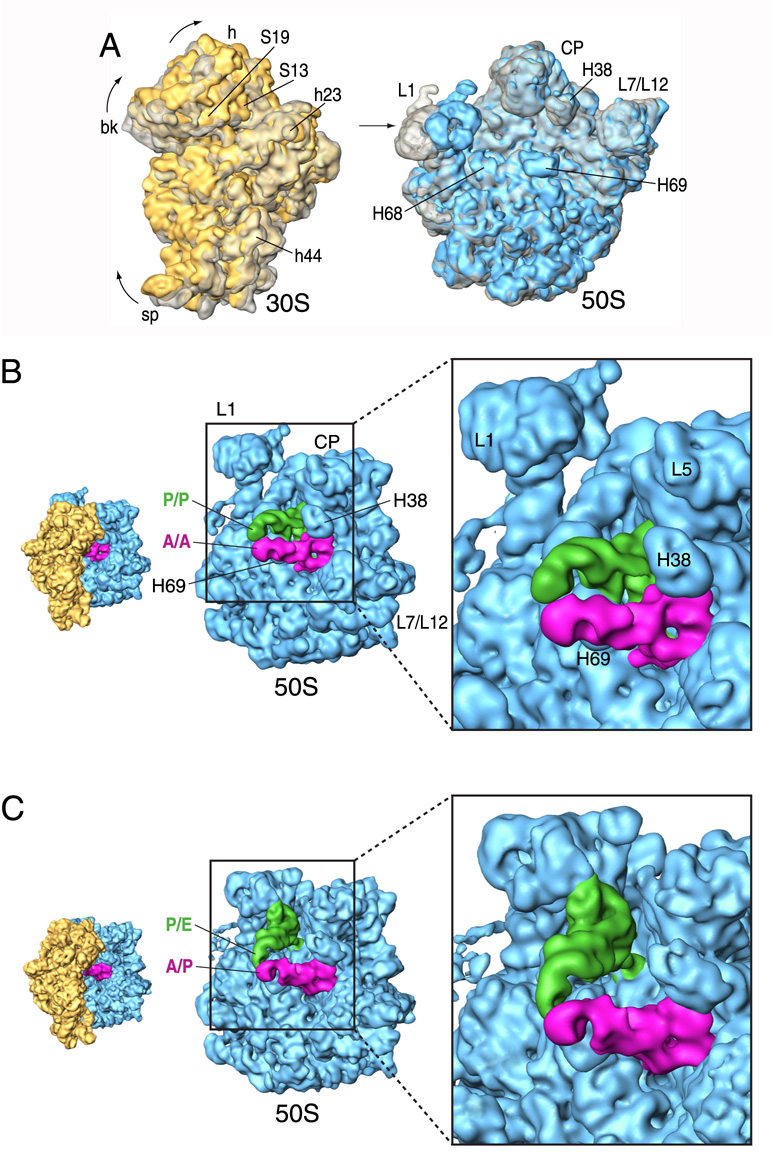
Figure 3. Conformational changes upon ratcheting movement and tRNA reconfiguration.
(A) Superimposition of aligned 30S (left) and 50S (right) subunits shown from the intersubunit side. Classic-state subunits are shown in transparent grey, whereas the subunits of the hybrid-state ribosome are shown in solid yellow (30S) and blue (50S). (B) Close-up view of the 50S subunit showing the classic tRNA configuration. (C) Close-up view of the 50S subunit showing the hybrid tRNA configuration. The orientations of the subunits are shown as successive thumbnails on the left. Labels and landmarks: S19 (small subunit/SSU protein S19), S13 (SSU protein S13), h23 (SSU helix 23), h44 (SSU helix 44), H38 (large subunit/LSU helix 38), H68 (LSU helix 68), H69 (LSU helix 69), H76 (LSU helix 76), L5 (LSU protein L5). All other landmarks have been introduced in Figure 1.
Bridge B2a is formed by the interaction of the universally conserved 1915 loop of H69 and the tip of h44, just at the base of the decoding site of the small subunit. Although the bridge remains intact, slight opposite lateral movements of the tip of h44 and H69 are detected after the ratcheting movement (Supplementary Figure 5). H69 is also more inclined toward the small subunit at this stage. H68, which contacts h23 of the 16S rRNA to form bridge B7a, shows a displacement toward the large subunit. In contrast to the 30S subunit, only small-scale, local changes are observed in the 50S subunit with the exception of the L1 stalk (formed by helices 76, 77, 78 and protein L1), which moves toward the intersubunit space by ~20 Å. This rearrangement is facilitated by a large movement of H76 at the base of the stalk.
Similar conformational changes in the ribosome related to the ratcheting movement of the 30S subunit were previously observed in response to the binding of elongation factor G (Valle et al., 2003b), initiation factor IF2 (Allen et al., 2005), class-II release factor RF3 (Klaholz et al., 2004; Gao et al., 2007) and recycling factor RRF (Gao et al., 2005). However, what distinguishes our current observation from the previous ones is that in this case, no factor is present to facilitate ribosomal motions. We will therefore speak of spontaneous ratcheting of the ribosome, in line with data from very recent FRET studies indicating a factor-free rotation of the small subunit (Cornish et al., 2008). Spontaneous ratcheting of the ribosome has previously not been seen in cryo-EM maps of pre-translocational ribosomes, as these complexes invariably showed high occupation of canonical A, P and E sites by tRNAs (e.g., Valle et al., 2003b).
In contrast, our current data differ in two important aspects: first, they show a mixture of two states (“macro-states” – see Frank et al., 2007) in apparent equilibrium, with a preference for the ratcheted state, and second, the reconstruction of the ribosome in the canonical state from the minority population does not show a deacylated tRNA in the E site. Since FRET data have demonstrated that the P-site tRNA adopting the hybrid configuration is the rate-limiting step in the transition between classical and hybrid states (Munro et al., 2007), we argue, much in line with previous suggestions (Zavialov et al., 2005), that the striking difference in the results may result from E-site binding by free deacylated tRNA, which could be in turn controlled by the Mg2+ concentration. The low Mg2+ concentration used in the present experiment could potentially prevent occupation of the E site, and thus favor a more efficient transition of the classic P-site tRNA to the hybrid P/E configuration.
Finally we note that the distribution of particle resemblance as measured by cross-correlation (Figure 1) shows the presence of only two resolved peaks that correspond to two major populations, corresponding to the hybrid/ratcheted and classical/non-ratcheted states of the pre-translocation complex. Along these lines, recent data (Fei et al., 2008; Cornish et al., 2008) support the view that there are no detectable intermediates beyond the two configurations visualized in this work. However, this does not necessarily exclude the possibility of a P/E-A/A intermediate, as independently described by other groups (Munro et al., 2007; Pan et al., 2007), but it does seem that this intermediate, if it exists, it is not very populated and/or stable in wild-type ribosomes.
Dynamic structural pattern of interactions between the tRNAs and the ribosome during the ratcheting movement, and partial translocation
In the transition from the classic to the hybrid state, the inferred trajectories of the tRNAs are quite different. While the elbow (D and T loops) of the P site tRNA moves by ~40 Å in the P/P to P/E transition, the elbow of the A-site tRNA moves barely. Thus, with the exception of the CCA-end, the interaction of the tRNA with the ribosome remains basically unaltered between classic A/A and hybrid A/P tRNA configurations. On the 50S subunit side, H69 (1915 loop) maintains its contact with the D-stem. The elbow of the tRNA also interacts with H38 in both classic and hybrid states (Figures 3B/3C). Nevertheless, the lateral displacement of H69, in conjunction with the conformational change of H38 accompanying the ratcheting of the small subunit, causes the pivoting of the A site tRNA around its anticodon and the reorientation of the elbow region, allowing the CCA-end to reach the 50S subunit’s active site in the A/P hybrid configuration. Since it is known that the peptidyl tRNA in the A/P configuration is quite unreactive as a peptydil donor in contrast to its high reactivity in the P/P site (Semenkov et al., 1992; Sharma et al., 2004), we can only speculate about the lower reactivity of the hybrid configuration being related to its dynamic and transient nature. On the other hand, in this A/P configuration, the tRNA is brought close to H93, where residue A2602 is situated (Supplementary Figure 4). It has been shown that sparsomycin-dependent rearrangement of this residue decreases the activation energy for EF-G independent translocation (Fredrick and Noller, 2004). These data can be rationalized if the interaction of A2602 with the CCA-end stabilizes the observed A/P hybrid configuration of the tRNA that eventually leads to the repositioning of the peptidyl-tRNA from the A/P to the P/P state. Indeed the original study by Moazed and Noller noted specific changes in the modification pattern for A2602 in the pre- and post-translocation states, likely reporting on conformational rearrangements in the peptidyl transfer center (Moazed and Noller, 1989).
On the other hand, the extensive contacts of the P-site tRNA are drastically remodelled after the P/P to P/E site transition. A remarkable exception is the contact with protein S13. This protein, located in the head region of 30S undergoes a large displacement coupled to the rotation of the small subunit (Valle et al., 2003b; Frank et al., 2007). Despite the size of this movement, the C-terminus of S13 does not lose its original contact with the P site tRNA after ribosomal ratcheting. Along these lines, protein S13 has been shown to be a control element in the EF-G-independent movement, as its omission promotes complete tRNA translocation (Cukras et al., 2003).
On the 50S subunit side, there is an extensive connection of the D stem of the classic-state P/P tRNA with H69, as well as between the T loop and protein L5. However, the map corresponding to the hybrid P/E tRNA indicates that it is no longer bound to H69 (see Li and Frank, 2007). The CCA stem of the P/E tRNA contacts H68 in its new configuration, and the T loop is severed from its original interaction with L5, having established a new contact with H78 situated in the L1 stalk (Figures 3B/3C). This new interaction is only formed after the large displacement of the L1 stalk coupled to the ratcheting of the small subunit. In this hybrid P/E configuration, several residues located in H68, H74, and H11 that are known to be essential for P-site tRNA translocation (Feinberg and Joseph, 2001) are placed in the vicinity of the 3’ acceptor end (Supplementary Figure 4).
In summary, these observations show how the tRNA reconfiguration is promoted by the changes the ratcheting of the small subunit generates in several components involved, not only in tRNA binding interactions, but also in subunit association and reconfiguration of inter-subunit bridges. Some of them, namely S13, L5 and H38, which form the bridges that connect the head of the 30S subunit with the central protuberance of the large subunit, may be instrumental for the control of the rotation as they are located at the periphery where the movement is largest (Frank et al., 2007). The fact that these components are highly dynamic and change their relative position during the movement of the subunit interface reflects the direct relation between the ratcheting movement of the small subunit and the reconfiguration of the tRNAs. Thus, our data suggest that the unlocking of the ribosome (understood as the removal of the physical connection between P-site tRNA and the polypeptide chain by the deacylation of the P site-tRNA (Zavialov and Ehrenberg, 2003; Valle et al., 2003b)) is what releases the inherent ratchet-related reorganization capability of the ribosome which permits the different tRNA environments to be established.
Implications for EF-G based translocation
Kinetic and thermodynamic studies have demonstrated that the hybrid state of tRNA binding makes important energetic contributions to EF-G-dependent translocation (Semenkov et al., 2000; Dorner et al., 2005). These results invite a comparison with results from previous cryo-EM reconstructions of EF-G·GDPNP-bound ribosomes (Agrawal et al., 1998; Valle et al., 2003b).
When superimposing the structures of the 30S subunit, we see that the movement of the small subunit from the hybrid ribosome goes in the same direction as the one observed in the presence of EF-G, although it is smaller in magnitude (Figure 4A, left panel). This fitting result indicates that the inherent ratcheting of the ribosome is less extensive than the one finally stabilized by the action of EF-G, but that it is nevertheless sufficient to position the tRNAs in the hybrid configuration. In Figure 4A (right panel), we show EF-G and its relative position with respect to the 30S subunit in the three complexes. From this juxtaposition, it is seen that the tip of domain IV of EFG (the contact point of the factor with h44) overlaps with h34 in the classic 30S subunit configuration (shown in transparent grey). Therefore, the kinetic efficiency attributed to the hybrid-state tRNA configuration can be explained in structural terms by the fact that the small subunit is already partially ratcheted in the hybrid-state ribosome, thus no longer presenting steric constraints in the vicinity of the 30S head, which would slow down the positioning of EF-G’s domain IV in the 30S subunit’s A-site region.
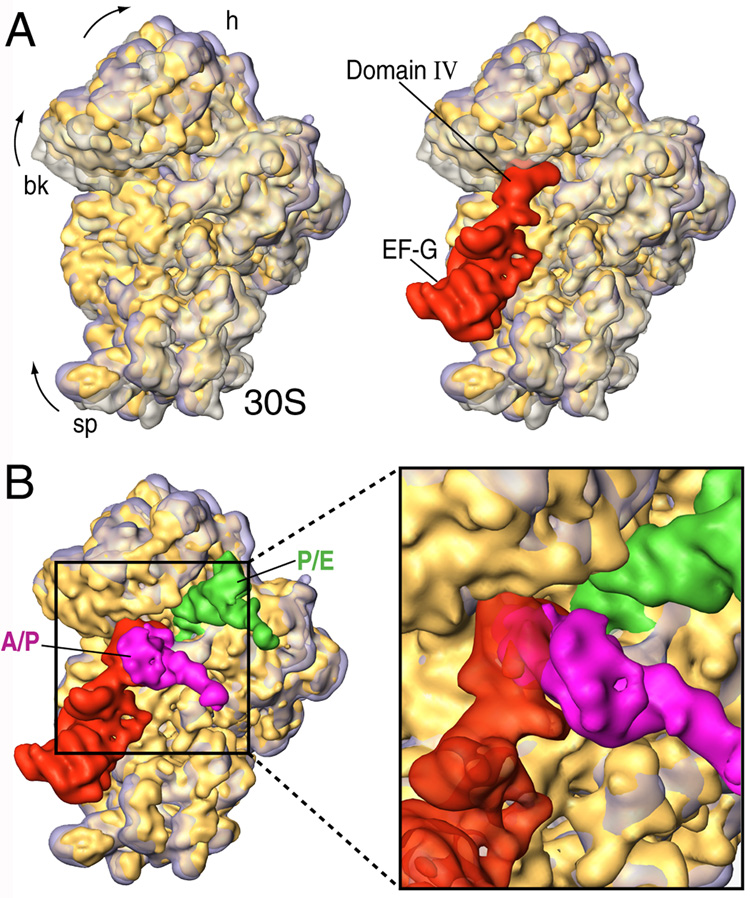
Figure 4. Hybrid configuration of the ribosome and implications for EF-G based translocation.
(A) Superimposition of aligned 30S subunits from hybrid (solid yellow), classic (transparent grey) and EF-G bound (transparent purple) forms (from Valle et al., 2003b). In the right panel, the binding position of EF-G (red) is shown. (B) Close-up view of the superimposition of hybrid (solid yellow) and EF-G bound (transparent purple) forms in the presence of EF-G and hybrid tRNAs. The subunits are shown from the intersubunit side.
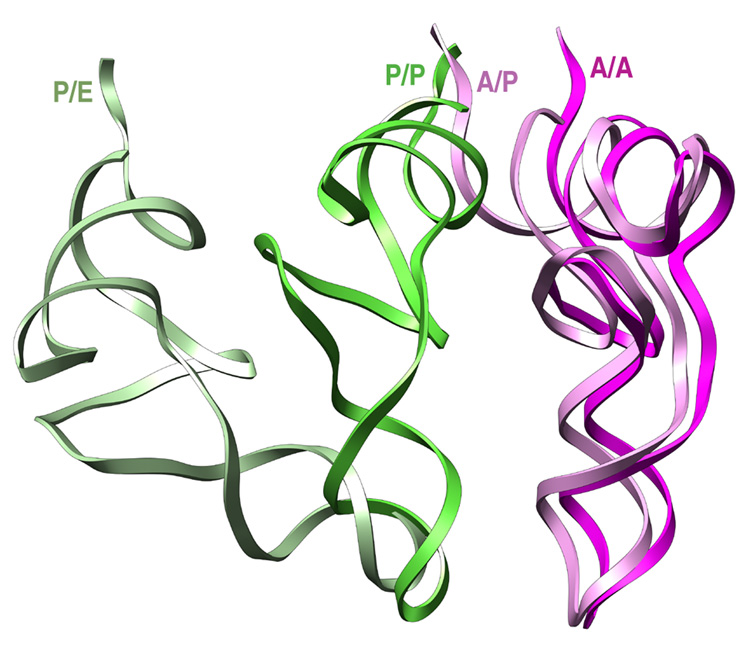
After the initial binding to the ribosome, domain IV of EF-G contacts h44 and the head of the small subunit in the vicinity of the decoding center. It is known that in the post-translocational ribosome, the tip of h44 is shifted toward the P site by ~8 Å (VanLoock et al., 2000). When we compare the position of the tip of h44 from classic, hybrid, and EF-G bound ribosomes (Supplementary Figure 5), we see that this shift is indeed mainly caused by the direct action of EF-G. However, a displacement in h44 (~2 Å) caused by the dynamic behaviour of the ribosome is also noted in the case of the hybrid-state ribosome. In Figure 5 we show the relative position of classic and hybrid-state tRNAs. From this fitting, it is clear that the anticodon stem loop of both classic and hybrid-state tRNAs do not superpose perfectly. The shift observed in the A/A to A/P tRNA transition would be based on this lateral displacement of the decoding center in h44, whereas the shift observed in the P/P to P/E reconfiguration (~4–5 Å), may be generated by the temporary destabilization of the codon anticodon helix shown to occur during the movement of the tRNA into the P/E hybrid state (McGarry et al., 2005).
Figure 5. Comparison between the positions and orientations of classic and hybrid tRNAs deduced from the cryoEM maps.
The fitting of the experimental densities with the X-ray structures of the tRNAs by real space refinement using rigid body fitting. Although the CCA-end of the classic A/A tRNA was not well-defined in the experimental density map, the complete quasi-atomic structure is shown for clearer visualization and perception of the relative positions of the different configuration tRNAs. The structures are displayed in ribbons.
On the other hand, in Figure 4B we show the superposition of 30S subunits from hybrid and EF-G bound forms in the presence of hybrid A/P and P/E tRNAs. According to this fitting, it is clear that before the complete ratcheting has occurred, and h44 is further shifted by the action of EF-G, the ASL from the A/P tRNA and domain IV of EF-G occupy similar positions. As it is known that EF-G, even in the absence of GTP hydrolysis, accelerates the translocation rate by 1000-fold (Rodnina et al., 1997), the sole binding of EF-G·GTP likely facilitates the complete translocation of the hybrid A/P tRNA to the P site (Pan et al., 2007), displacing both the decoding center (the tip of h44) and the mRNA-tRNA complex toward the P site. Thus, the potential steric clash between the ASL of the A/P tRNA and EF-G explains why all previous cryo-EM reconstructions of ribosomes bearing EF-G failed to show the simultaneous presence of hybrid A/P and P/E tRNAs. Only the hybrid P/E tRNA was visualized in a ribosome bound with EF-G, as it was in the post-termination state lacking A-site bound tRNA (Valle et al., 2003b).
Concluding Remarks
Chemical footprinting experiments followed the movement of the tRNAs through the intersubunit space and showed that after the peptidyl transfer, the tRNAs reside in intermediate hybrid positions (Moazed and Noller, 1989). The proposed driving force for this partial translocation is based on the well established different affinities the CCA-end shows for 50S subunit A, P, and E sites (Lill et al., 1989; Feinberg and Joseph, 2001; Virumae et al., 2002; reviewed in Noller et al., 2002). However, the ratcheting movement of the ribosome has never been integrated in this scheme. Based on our new data, we demonstrate how the repositioning of the tRNAs in the 50S subunit is based not only on the affinity the CCA-ends show for the different sites, but also on the structural reconfiguration of S13, L1 stalk, L5, H68, H69 and H38 that is caused by the rotation of the small subunit and the new environment thus created which favours transitions from A/A-P/P to A/P-P/E.
MATERIALS AND METHODS
Sample preparation
The ribosomal complex was prepared using an in vitro translation system in HiFi buffer (50 mM Tris-HCl pH:7.5, 70mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 0.5 mM spermidine, 8mM putrescine, 2 mM DTT) (Gromadski and Rodnina, 2004). Ribosomes from strain MRE600 were obtained as previously described (Moazed and Noller, 1989). EF-Tu and tRNATrp expression and purification was performed as previously described (Cochella and Green, 2005). A 70S initiation mix (500 µl) containing 1 µM ribosomes, 1.5 µM fMet-tRNAfMet, 1 mM GTP, 1 µM each IF (1, 2 and 3) and 8 µM MWstop-mRNA was incubated at 37 °C for 70 min. After a sucrose cushion in buffer D (with MgCl2 added to 10 mM final concentration), IC-s were pelleted, washed once and resuspended in HiFi buffer (100 µl). 35S-labeled fMet-tRNAfMet containing complexes were made in parallel with the unlabeled complexes to check the concentrations by scintillation counting. The yield of IC formation was 65%, and thus, the final concentration, 3.2 µM. This complex was then diluted with the same buffer to 1 µM concentration. The ternary complex was prepared separately by incubating (in 80 µl) 2 µM Trp-tRNATrp, 40 µM EF-Tu, 1 mM GTP, 67 mM L-Trp, 8 µM TrpRS and energy regeneration system (0.025 mg/ml PK, 1.5 mM ATP, 1.5 mM PEP) at 37 °C for 45 min. Elongation was started by mixing equal volumes (40 µl) of the initiation and ternary complex. After 5 min incubation at 37 °C, the mixtures (80 µl) were frozen in liquid nitrogen immediately. 35S-labeled fMet-tRNAfMet containing complexes were checked for dipetide formation and release activity (A site occupancy). The release activity was determined as the amount of peptide that could be released by RF1 in a stop codon dependent reaction after fMet-Trp-tRNATrp translocation with addition of EF-G. Elongation complexes were incubated with EF-G and RF1 for 1 min at 37 °C and quenched with 10% formic acid. For peptide bond formation measurement, complexes were hydrolyzed with 0.75 N KOH. Products from both assays were separated by TLC electrophoresis in pyridine-acetic acid pH:2.8. The complexes formed 82% fMet-Trp dipeptide and released 65% fMet-Trp upon addition of EF-G and RF1.
Electron microscopy and image processing
To prepare the sample for cryoEM, an aliquot of the ribosomal complex at 1 µM concentration was thawed on ice and diluted with HiFi buffer to a final concentration of ribosome of 30nM. A carbon-coated Quantifoil 2/4 grid (Quantifoil Micro Tools GmbH, Jena, Germany) was prepared following standard cryo procedures (Graccussi et al., 2007). Images were recorded on a single-port 4K × 4K CCD camera (TVIPS TemCam-F415) mounted on an FEI Tecnai Polara operating at 300 kV and a nominal magnification of 59,000× in low dose conditions (~20 e−/Å2) by using the automated data collection system AutoEMation (Lei and Frank, 2005). Because of the post-magnification ratio, the effective magnification is 100,000×, corresponding to a pixel size of 1.5 Å on the object scale. The AutoEMation session lasted 44 hours and was completely left unattended including an overnight period, except for refilling of liquid nitrogen 1–2 times day. 2,073 high-magnification CCD images were acquired from 691 holes, which were automatically selected from 16 grid squares as the system can automatically collect multiple high-magnification low-dose images in each hole. After visual inspection and evaluation of micrographs and their power spectra, 1,996 CCD images were selected for further analysis. Particles were chosen via automated particle picking (Rath and Frank, 2004) followed by visual selection. The total number of particles used was 216,517. The 3D reconstructions then followed the standard SPIDER protocols for reference-based reconstruction (Frank et al., 1996) including a 3D projection alignment procedure with correction of the contrast transfer function (CTF) using defocus groups, varying the defocus of the selected images from 1.2 to 4 µm, followed by multiple rounds of angular refinement with angular increments. The map corresponding to the accommodated state of Phe-tRNAPhe was used as initial 3D reference (EMD-1056). Supervised classification methodology was applied as described in Supplementary Figure 1. The final resolution for the maps was 8.75 Å, 8.9 Å and 10.5 Å, using a cutoff of 0.5 in the Fourier Shell Correlation (Supplementary Figure 2), and the maps were filtered accordingly.
The fitting of the cryo-EM maps with the atomic models was done using the real space refinement program RSREF (Chapman, 1995). The initial atomic model was based on the X-ray structure of an empty E. coli 70S ribosome (PDB codes: 2AVY, 2AW4; Schuwirth et al., 2005). The P-site fMet-tRNA used was the one described in the 70S structure of Thermus Thermophilus (2j00; Selmer et al., 2006), whereas the A-site tRNA used corresponds to Phe-tRNA described in Yusupov et al., 2001 (1GIX). The 7 nt fragment from the tip of h44 (nt 1989–1996) corresponds to the X-ray structure of the 30S subunit in the presence of cognate Phe-tRNA and paramomycin (1lBK; Ogle et al, 2001). The cryo-EM densities and the atomic models were visualized using the programs Amira (http:amira.zib.de) and Chimera (Pettersen et al., 2005), respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Derek Taylor and Wen Li for discussion of the manuscript, Jayati Sengupta and Ruben L. Gonzalez for valuable suggestions, Robert A. Grassucci for assistance with the image collection and Michael Watters for assistance with the preparation of the illustrations. Supported by HHMI and NIH grants R01 GM55440 and R37 GM29169 (to J.F.) and NIH grant R01 GM059425 (to R.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS.
The EM maps have been deposited in the 3D-EM database, EMBL-European Bioinformatics Institute, Cambridge under the corresponding accession numbers: EMD-6309 (classic-70S·fMet-tRNAfMet·Trp-tRNATrp) and EMD-6308 (hybrid-70S·fMet-tRNAfMet·Trp-tRNATrp).
REFERENCES
- Abel KM, Yoder MD, Hilgenfeld R, Jurnak F. An α to β conformational switch in EF-Tu. Structure. 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. Effect of buffer conditions on the position of tRNA on the 70S ribosome as visualized by cryoelectron microscopy. J. Biol. Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- Chapman MS. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron density function. Acta Crystallogr. A. 1995;51:69–80. [Google Scholar]
- Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Feinberg JS, Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc. Natl. Acad. Sci. USA. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc. Natl. Acad. Sci. USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Gao H, Valle M, Ehrenberg M, Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J. Struct. Biol. 2004;147:283–290. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, Sanyal S, Ehrenberg M, Frank J, Song H. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc. Natl. Acad. Sci. USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys. J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- Lei J, Frank J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 2005;150:69–80. doi: 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Li W, Frank J. Transfer RNA in the hybrid P/E state: correlating molecular dynamics simulations with cryo-EM data. Proc. Natl. Acad. Sci. USA. 2007;104:16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Robertson JM, Wintermeyer W. Binding of the 3' terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth GW, Chen L, Kosek AB, Strobel SA. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA. 2001;10:1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Polekhina G, Thirup S, Kjeldgaard M, Nissen P, Lippmann C, Nyborg J. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure. 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]
- Rath BK, Frank J. Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: a case study. J. Struct. Biol. 2004;145:84–90. doi: 10.1016/j.jsb.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Seila AC, Hansen JL, Freeborn B, Soukup JK, Scaringe SA, Strobel SA, Moore PB, Steitz TA. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Shapkina T, Makhno V, Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 1992;296:207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanLoock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC. Movement of the decoding region of the 16S ribosomal RNA accompanies tRNA translocation. J. Mol. Biol. 2000;304:507–515. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003a;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov AV, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Virumae K, Saarma U, Horowitz J, Remme J. Functional importance of the 3'-terminal adenosine of tRNA in ribosomal translation. J.Biol.Chem. 2002;277:24128–24134. doi: 10.1074/jbc.M200393200. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Hauryliuk VV, Ehrenberg M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J. Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.