Abstract
Lyme disease pathogenesis results from a complex interaction between Borrelia burgdorferi and the host immune system. The intensity and nature of the inflammatory response of host immune cells to B. burgdorferi may be a determining factor in disease progression. Gene array analysis was used to examine the expression of genes encoding cytokines, chemokines, and related factors in the joint tissue of infected C3H/HeJ mice and in a murine macrophage-like cell line in response to a disseminating or attenuated clinical isolate of B. burgdorferi. Both isolates elicited a robust pro-inflammatory response in RAW264.7 cells characterized by an increase in transcript levels of genes encoding CC and CXC chemokines, pro-inflammatory cytokines, and TNF superfamily members. Transcription of genes encoding IL-1β, IL-6, MCP-1, MIP-1α, CXCR4 and TLR2 induced in RAW264.7 cells by either live or heat-killed spirochetes did not differ significantly at any time point over a 24-hour period, nor was there a difference in the protein levels of IL-10, TNF-α, IL-6 and IL-12p70 in culture supernatants. Thus, induction of host macrophage expression of pro-inflammatory mediators by host macrophages does not contribute to the differential pathogenicity of different B. burgdorferi strains.
Keywords: Monocytes/Macrophages, Inflammation, Bacterial
INTRODUCTION
Lyme disease, the most prevalent arthropod-borne infection in the United States (1), is a multisystemic disorder caused by infection with the tick-transmitted spirochete Borrelia burgdorferi (2). Approximately 70–80% of patients develop a characteristic rash, erythema migrans (EM), at the site of inoculation (3). The EM lesion is characterized by an influx of immune cells, predominantly T lymphocytes, macrophages/monocytes and dendritic cells (4), which in most cases eradicate the infection (5). In patients who develop more serious long-term sequelae, the spirochete migrates from the skin via hematogenous dissemination to a variety of secondary sites, most commonly the skin, heart, joints and nervous system (6). Pathological manifestations are of an inflammatory nature and include carditis, arthritis and neurological disorders (6).
Elucidation of spirochetal virulence factors may be facilitated by identification of B. burgdorferi isolates with different pathogenic properties. Clinical isolates of B. burgdorferi sensu stricto can be classified by a variety of typing procedures, including restriction fragment-length polymorphism (RFLP) analysis of the 16S–23S ribosomal DNA spacer (7,8). RST1 isolates are associated with a significantly higher percentage of positive blood cultures as well as an increased incidence of multiple EM in patients, suggesting a greater capacity for hematogenous dissemination relative to RST3 isolates (9). These clinical observations correlate with experimental findings obtained using a murine model of Lyme borreliosis, where infection of C3H/HeJ mice with RST1 strains resulted in significantly higher spirochete loads in tissue, as well as more severe arthritis and aortitis, than did infection with RST3 isolates (10,11).
The host factors contributing to disease pathogenesis have been studied using strains of mice with differing degrees of susceptibility to B. burgdorferi infection (12). Genetically susceptible C3H/HeJ mice develop severe arthritis when infected with as few as 200 spirochetes, whereas C57BL/6N mice develop mild arthritis even at an infectious dose of 2 × 105 spirochetes (13). These distinctly different host responses may depend in part upon the qualitative and quantitative differences in cytokine expression elicited by the spirochete.
A dual role for pro- and anti-inflammatory (Th1/Th2) cytokines in host defense and disease pathogenesis has been established for Lyme disease (14). However, the ratio of these groups of cytokines at different stages of infection appears to be a determining factor in disease outcome. Levels of IFN-γ, IL-4 and IL12p70 produced by B. burgdorferi-stimulated whole blood and PBMCs, or in EM blister fluids, differ between patients with localized or disseminated B. burgdorferi infection (4, 15, 16, 17). Similar observations connecting the nature of the cytokine response with disease outcome have been made using murine models of Lyme borreliosis (18,19). Together, these studies indicate that a strong pro-inflammatory response early in infection mediates host protection. In contrast, a sustained and dominant Th1 cytokine response in serum or in infected target tissues is associated with severe inflammation-induced pathology both in mouse models (20) and in Lyme disease patients (17, 21, 22).
Macrophages have been implicated in the development and progression of Lyme disease pathogenesis. In susceptible laboratory mouse strains, macrophages are the most abundant cell type in the inflammatory infiltrate of Lyme carditis (23, 24) and secrete increased levels of the pro-inflammatory cytokines IL-1β, TNF-α and IL-12 relative to macrophages taken from a site without active disease (25). In several rodent models, macrophages are present in the inflamed synovium of ankle joints and have been shown to be key mediators of severe arthritis (26,27,28). In addition, highly activated macrophages are a predominant component of the cellular infiltrate of EM lesions in Lyme disease patients (4), thereby placing the macrophage among the first cell types to encounter and respond to the invading pathogen.
The nature and/or intensity of the macrophage response to different B. burgdorferi genotypes, as measured by the production of cytokines, may be a decisive factor in survival at the initial inoculation site and subsequent disease progression. Although a number of studies have focused on the expression of selected cytokines and chemokines in various tissues and cell lines in response to B. burgdorferi (4, 29–32), only one study has characterized the global cytokine expression profile elicited by B. burgdorferi (33). We used cytokine gene arrays to assess the B. burgdorferi-induced global cytokine transcriptional profile in the joints of Lyme disease-susceptible mice and compared these results to the cytokine profiles induced in the RAW264.7 gamma NO(−) mouse macrophage-derived cell line in response to two clinical isolates of B. burgdorferi associated with distinctly different disease outcomes.
MATERIALS AND METHODS
Culture of RAW264.7 gamma NO(−) cells
Murine macrophage-like cell line RAW264.7 gamma NO(−) was obtained from the ATCC (CRL-2278, Manassas, VA). RAW264.7 gamma NO(−) cells do not produce nitric oxide upon treatment with IFN-γ alone, but requires LPS for full activation. This property makes its behavior more like that of normal macrophages from some commonly used murine models of Lyme borreliosis (eg: C3H/HeN). Cells were grown in RPMI 1640 medium supplemented with 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mM HEPES, 1.0 mM sodium pyruvate (ATCC cat. no. 30-2001) and 10% FCS (ATCC, 30-2021). Cells were maintained in T-75 cm2 tissue culture flasks (Corning, NY) at 37° C in a humidified 5% CO2 incubator.
B. burgdorferi isolates
Low-passage (passage 3 to 5) B. burgdorferi clinical isolates BL206, B515 and B479, representing the disseminating RST1 genotype; and B356, B331 and B418, representing the non-disseminating RST3A genotype, were used in these studies (10,11). B. burgdorferi was cultured at 33° C in BSK-H medium (Sigma-Aldrich, St. Louis, MO) supplemented with 6% rabbit serum. Spirochetes were grown to late-logarithmic phase and examined for motility by dark-field microscopy. Organisms were quantitated by fluorescence microscopy after mixing 10 µl aliquots of culture material with 10 µl of an acridine orange solution (100 µg/ml). Bacteria were harvested by centrifugation of the culture at 7,000 × g for 15 min, washed twice with sterile PBS (pH 7.4) and diluted in the specified medium to required concentration. Heat-killed B. burgdorferi cells were prepared as described above except for heating at 56° C for 30 min prior to dilution.
Mouse infection model
All animal experiment protocols were reviewed and approved by the Institutional Animal Care and Use Committee of New York Medical College. Four-week-old specific pathogen-free C3H/HeJ mice of either sex were purchased from Jackson Laboratory (Bar Harbor, ME) and maintained in separate cages in the Department of Comparative Medicine at New York Medical College following the NIH guidelines for care and use of laboratory animals. Two groups of 5 mice were inoculated intradermally on the shaved back with 0.1 ml PBS containing 1 × 104 B. burgdorferi BL206 or with PBS alone. Mice were euthanized by exposure to carbon dioxide on day 14 after inoculation (11), and samples of ear biopsy and joint tissue were collected aseptically for culture and total RNA extraction. Culture of B. burgdorferi from ear biopsy was performed as previously described (10,11).
RNA preparation from mouse joint tissues
Individual hind limb ankle joints from which the skin had been removed were frozen in liquid nitrogen, wrapped in aluminum foil, pulverized with a hammer, then placed immediately into a glass tissue homogenizer containing 0.5–1 ml of lysis buffer from the RNA isolation kit (RNAzol™ B, Tel-Test, Inc., Friendswood, TX). RNA was subsequently prepared as described above for the RAW264.7 cells.
Exposure of RAW264.7 gamma NO(−) cells to B. burgdorferi
RAW264.7 gamma NO(−) cells, cultured as described above, were grown to 80–90% confluence in T-75 flasks or in 24-well plates. Medium was aspirated aseptically and replaced with fresh serum-free RPMI 1640 medium containing B. burgdorferi isolates at a multiplicity of infection (MOI) of 10:1 or 1 µg/ml lipopolysaccharide (LPS) from E. coli 0127:B8 (Sigma-Aldrich, cat. # L 3880). Medium was added to controls. Duplicate or triplicate flasks/wells of RAW264.7 cells were harvested for RNA extraction at 2, 8, 16 and 24 hrs after exposure. Two milliliters of supernatant from each flask were also collected and stored at −80° C for measurement of the cytokine protein level by flow cytometry. The experiment was repeated on a later date using a different lot of RAW264.7 gamma NO(−) cells obtained from ATCC.
RNA Preparation
Total RNA was prepared from freshly harvested RAW264.7 gamma NO(−) cells using a commercial RNA isolation kit (RNAzol™ B, Tel-Test, Inc., Friendswood, TX). Briefly, cells were lysed by the addition of RNAzol™ B (1 ml/10cm2) to homogenize the cells; subsequently 0.2 ml of chloroform per 2 ml of homogenate was added. After shaking for 15 s and incubation on ice for 5 min, the lysates were centrifuged at 12,000 × g at 4° C for 15 min. The aqueous phase was transferred to a fresh tube and the RNA was precipitated with isopropanol and washed with 70% ethanol. Each dried RNA pellet was resuspended in 20 µl of DEPC-treated, RNase-free water and treated further with DNase (DNase Treatment & Removal Reagents, Ambion, Inc., Austin, TX) to remove any residual DNA. The quality and quantity of RNA samples were determined by gel electrophoresis and spectrophotometry (BioPhotometer, Eppendorf, Westbury, NY). RNA samples were frozen at −80° C until analyzed by gene array or real-time RT-PCR.
Gene Array Hybridization and Data Analysis
The Panorama™ mouse cytokine gene array, consisting of 514 different cytokine-related cDNAs printed as PCR products onto a charged nylon membrane, was purchased from Sigma-Genosys Inc. (The Woodlands, TX). Also included on the array are 8 positive control “housekeeping” genes, mouse genomic DNA and 5 negative controls. 33P-labeled cDNA probes were generated with mouse cytokine cDNA labeling primers (provided by Sigma-Genosys) using 4 µg of total RNA and hybridized with the array membrane overnight at 65° C according to the manufacturer’s protocol. After hybridization, the membrane arrays were washed and exposed to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA) for 16 to 48 hr as described previously (34). Four replicate arrays were prepared for each experimental condition.
The exposed PhosphorImager screen was scanned with a pixel size of 100 µm on a Storm 840 PhosphorImager (Molecular Dynamics). The image files were analyzed with a template containing the spot layout of the array using ArrayVision software (version 8.0; Imaging Research, St. Catharine’s, Ontario, Canada). The raw intensity value, in pixels for each spot (RIVj) on the array, was determined after subtraction of the background intensity and was exported to Microsoft Excel for further analysis. The cytokine microarray data were normalized using a global adjustment of intensity values approach (35). First, the mean intensity value of each array (MIVi) (based on the raw intensity values of 514 cytokine-related genes and control DNAs, each in duplicate) was calculated [MIVi = Σ(RIVij)/the number of spots on the array; i refers to individual array and j refers to individual spot]. Second, a global mean intensity value (GMIV) based on the MIVi of the 12 arrays analyzed was determined (GMIV = Σ(AIVi) /the number of arrays). Third, a normalization factor (NFi) was assigned for each array (NFi = GMIV /MIVi). Finally, the raw intensity value of each spot on an individual array was multiplied with the corresponding NFi to convert it to a normalized intensity value for statistical analysis.
For each gene, normalized intensity values were generated from two duplicate spots on each of four replicate arrays for each experimental condition. A two-tailed, unpaired Student’s t-test was employed to estimate the statistical significance among the experimental groups by comparison of the mean intensity values from all eight replicate spots for each gene. Significance of differential expression was determined at P < 0.05.
Real-time quantitative RT-PCR
The expression of selected cytokine and related genes in mouse tissue and in RAW264.7 cells exposed to either isolate B356 or isolate BL206 was determined by real-time quantitative RT-PCR using SYBR Green technology with the LightCycler (Roche Applied Science, Indianapolis, IN) or the ABI 7900HT SDS (Applied Biosystems, Foster City, CA), as described previously (10,34,36). For each RNA sample, the expression of β-actin was quantified by real-time RT-PCR, and a ΔΔCt method was used to estimate the differential gene expression between samples (37). Oligonucleotide sequences of primers used for RT-PCR using SYBR green technology were as follows: actinb, ACTB-F (5’-TCACCCACACTGTGCCCATCTACGA-3’) and ACTB-R (5’-GGATGCCACAGGATTCCATACCCA-3’); Il1b, IL-betaF (5’-GCCTTGGGCCTCAAAGGAAAGAATC-3’) and IL-betaR (5’-GGAAGACACAGATTCCATGGTGAAG-3’); Il6, IL-6F (5’-TGGAGTCACAGAAGGAGTGGCTAAG-3’) and IL-6R (5’-TCTGACCACAGTGAGGAATGTCCAC-3’); Il10, IL-10F (5’-GTGAAGACTTTCTTTCAAACAAAG-3’) and IL-10R (5’-CTGCTCCACTGCCTTGCTCTTATT-3’); Tnfa, TNFαF (5’-ATAGCTCCCAGAAAAGCAAGC-3’) and TNFαR (5’-CACCCCGAAGTTCAGTAGACA-3’); Mcp1, MCP1F (GGAAAAATGGATCCACACCTTGC-3’) and MCP1R (TCTCTTCCTCCACCACCATGCAG-3’); Mip1a, MIP-1αF (5’-CCCAGCCAGGTGTCATTTTCC-3’) and MIP-αR (5’-GCATTCAGTTCCAGGTCAGTG-3’); Mip2a, MIP-2αF (5’-TCCAGAGCTTGAGTGTGACG-3’) and MIP-2αR (5’-TCAGGTACGATCCAGGCTTC-3’); Tlr2, TLR2F (5’-CTCCTGAAGCTGTTGCGTTAC-3’) and TLR2R (5’-GCTCCCTTACAGGCTGAGTTC-3’); Tlr4, TLR4F (5’-TCGCCTTCTTAGCAGAAACAC-3’) and TLR4R (5’-GCCTTAGCCTCTTCTCCTTC-3’); Cox2, COX-2F (5’-TCTGGAACATTGTGAACAACATC-3’) and COX-2R (5’-AAGCTCCTTATTTCCCTTCACAC-3’); Cxcr4, CXCR-4F (5’-GAAGTGGGGTCTGGAGACTATG-3’) and CXCR-4R (5’-AGGGGAGTGTGATGACAAAGAG-3’); Icam2, ICAM2F (5’-TGCTGTTCTTATTTGTGACATCTG-3’) and ICAM2R (5’-TGTATTGAGGCTAAAAAGGAGAGG-3’); Il2r2, IL-1R2F (5’-TAGTCCCGTGCAAAGTGTTTC-3’) and IL-1R2R (5’-CTGTATGCAGATCCTCCCTTG-3’). The expression levels of selected chemokine, cytokine and tissue remodeling factor genes in RAW264.7 cells exposed to B. burgdorferi BL206, B515, B479, B356, B331 and B418 was determined by real-time PCR using TaqMan gene expression assays (Applied Biosystems). Duplicate or triplicate assays were performed for the following target genes: Il1b (Mm00434228_m1), gdf9 (Mm00433565_m1), Mmp8 (Mm00772335_m1), Bmp-r1b (Mm00432117_m1), Tnfsf6/Fas (Mm00433237_m1), Bmp1 (Mm0080225_m1), Bmp9 (Mm03024080_m1), Tie2 (Mm01256892_m1) and Tgfbr1 (Mm00436971_m1). The β-actin amplicon was used as the endogenous control for normalization of data. All assays were done in 20-µl reaction mixtures containing TaqMan universal PCR master mix (Applied Biosystems), 20X TaqMan gene expression assay mix and cDNA on an Applied Biosystems ABI 7900HT SDS system. The differential gene expression between samples was calculated as described above.
Measurement of cytokine protein levels
The Cytokine Bead Array--Mouse Inflammation kit (BD Biosciences, San Diego, CA) was used for simultaneous measurement of IL-6, TNF-α, IL-10, IL-12, IFN-γ and MCP-1 in the supernatants of RAW264.7 gamma NO(−) cells exposed to live or heat-killed B. burgdorferi isolates, LPS or medium control. 50 µl of each sample were added to an equal volume of the cytokine bead mixture and detection reagent, followed by a 3-hour incubation at room temperature in the dark. Ten additional tubes, each containing equal volumes of beads, detection reagent, and graded amounts of the six cytokines, were prepared in parallel to generate a standard curve for each cytokine. Unstained, FITC-, or phycoerythrin-labeled cytometer setup beads were prepared immediately prior to use. Following incubation, beads were washed with the buffer provided in the kit, centrifuged at 200 × g for 5 min, and the supernatants were carefully aspirated. The pellets were resuspended in 300 µl of the kit wash buffer and assayed immediately on the FACSCalibur (BD Biosciences). Cytokine concentrations were determined using the software provided.
RESULTS
B. burgdorferi induces pro-inflammatory cytokine and chemokine gene expression in joints of BL206-infected mice
Although the pathological manifestations of Lyme disease are characterized by inflammation, little is known about the nature of the global cytokine response during early disseminated infection. Therefore, a murine model of Lyme borreliosis was employed to assess the expression of cytokine and related genes in the acutely arthritic joint tissue of disease-susceptible C3H/HeJ mice. This mouse strain develops clinically apparent arthritis approximately 12 days after infection (11). All five mice inoculated intradermally with a dose of 1 × 104 B. burgdorferi BL206 were infected, as confirmed by culture of ear biopsies collected at day 14. RNA was extracted from hind ankle joints on day 14 and analyzed using the Panorama™ mouse cytokine gene array, which consists of 514 different cDNA transcripts representing cytokines, chemokines and other immunomodulatory factors and their receptors. Transcript abundance levels of 46 genes were found to be significantly induced (≥ 2 fold, P < 0.05) in the joints of BL206-infected mice relative to the PBS-inoculated controls (Table 1). The most dramatic induction was observed for CXCL13, CCR9 and IL-1β. Transcription levels of a number of pro-inflammatory mediators were upregulated in the BL206-infected mouse joint tissue, including several chemokines (MCP-1, MCP-3, CCL6, MIP-1γ, MIP-3β) and receptors (CCR2 and CCR9), and IL-1β and its receptor, IL-1R.
Table I.
Genes transcriptionally induced in joint tissue of BL206-infected C3H/HeJ mice two weeks post-infection.
| Function | Gene Name | FC* | P value |
|---|---|---|---|
| Cytokines/Chemokines | CXCL13 | 108.1 | < 0.0001 |
| IL-1β | 41.8 | < 0.0001 | |
| CCL2/MCP-1 | 14.9 | < 0.0001 | |
| CXCL6 | 7.4 | < 0.0001 | |
| CXCL9 | 6.8 | < 0.0001 | |
| CCL7/MCP-3 | 2.8 | < 0.0001 | |
| CCL6 | 2.6 | 0.0005 | |
| CCL9/MIP-1γ | 2.5 | < 0.0001 | |
| CCL19/MIP-3β | 2.4 | 0.0017 | |
| CXCL14/BRAK | 2.3 | < 0.0001 | |
| CXCL15/Lungkine | 2.0 | 0.0002 | |
| CCL3/MIP-1α | 2.0 | 0.0058 | |
| Receptors/Antagonists | CCR-9 | 158.1 | 0.0020 |
| IL-1R antagonist | 26.1 | < 0.0001 | |
| CCR-5 | 17.8 | < 0.0001 | |
| IL-12Rβ1 | 10.2 | 0.0101 | |
| IL-18R | 4.5 | 0.0083 | |
| IL-10Rα | 3.7 | < 0.0001 | |
| IL-2Rγ | 2.9 | 0.0001 | |
| CCR-2 | 2.9 | < 0.0001 | |
| IL-13Rα1 | 2.5 | < 0.0001 | |
| IL-8R | 2.1 | 0.0054 | |
| IL-18R AcP | 2.1 | 0.0026 | |
| IL-1 RI | 2.0 | 0.0025 | |
| Apoptosis | SARP-1 | 2.4 | 0.0002 |
| Cell Adhesion | Integrin-β2 | 4.2 | < 0.0001 |
| Integrin-α4 | 3.7 | 0.0093 | |
| Integrin-αL | 3.4 | < 0.0001 | |
| ICAM-1 | 2.0 | 0.0011 | |
| Cell Proliferation | TP1 | 2.6 | < 0.0001 |
| Cytokine Suppression | CIS3/SOCS3 | 2.1 | 0.0033 |
| Growth/Development | TNFRSF1B | 5.8 | < 0.0001 |
| Cerberus | 3.0 | 0.0010 | |
| c-fos | 2.2 | < 0.0001 | |
| LTBP-2 | 2.1 | 0.0093 | |
| Invasion/Migration | MMP-3 | 8.8 | < 0.0001 |
| TIMP-1 | 3.4 | < 0.0001 | |
| Lymphocyte Activation | CD45 | 2.9 | 0.0002 |
| MD-1 | 2.7 | < 0.0001 | |
| Pro-Inflammatory | Cox-2 | 7.2 | < 0.0001 |
| Stat 1 | 6.5 | < 0.0001 | |
| Caspase-1 | 2.7 | 0.0001 | |
| Stat 4 | 2.1 | 0.0015 | |
| TNF Superfamily | TNFSF6/FasL | 2.7 | < 0.0001 |
| TNFRSF10B | 2.2 | < 0.0001 |
FC = fold change relative to uninfected control.
B. burgdorferi induces pro-inflammatory cytokine and chemokine gene expression in RAW264.7 gamma NO(−) cells
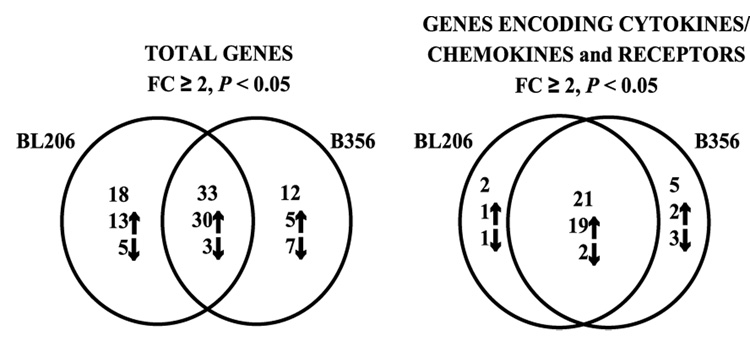
As shown above, isolate BL206 can disseminate to the joints in C3H/HeJ mice. Previous studies have shown that isolate B356 cannot disseminate to the joints in the same mouse strain (10, 11). It was, therefore, of interest to determine whether these two B. burgdorferi isolates elicit different responses in an infected mouse. As isolate B356 is not detected in joint tissue, the murine macrophage-derived cell line RAW264.7 gamma NO(−) was used instead to compare the expression of cytokines and related genes induced by these two B. burgdorferi clinical isolates of differing genotype and pathogenic potential. A combined total of 63 (12.3 %) genes were differentially transcribed (≥ 2-fold change, P < 0.05) upon exposure to the two B. burgdorferi isolates (Fig. 1). Of these, 33 genes (6.4 %; 30 induced and 3 repressed) were differentially transcribed upon exposure to either BL206 or B356 (Table II). The commonly induced genes included CC and CXC chemokines and the pro-inflammatory cytokines IL-1α, IL-1β and IL-6. The three commonly repressed genes included one chemokine, CCL21, a chemokine receptor, CXCR4, and CD28. Although the extent of induction for a number of these genes suggested a trend towards a stronger pro-inflammatory response to isolate BL206 than to B356, these differences did not reach statistical significance.
Figure 1. Differential expression of genes encoding cytokines and receptors is elicited by B. burgdorferi isolates BL206 and B356.
RNA from RAW264.7 gamma NO(−) cells which had been co-cultured for 16 hours with B. burgdorferi isolates BL206 or B356 (MOI = 10:1; spirochetes:RAW264.7 cells) was analyzed by gene array. Differentially expressed transcripts had changes ≥ 2-fold and P < 0.05. Arrows indicate induction or repression relative to the time-matched control.
Table II.
Cytokine and related genes significantly induced or repressed in RAW264.7 gamma NO(−) cells exposed to either B. burgdorferi isolates BL206 or B356 for 16 hours.
| Function | Gene Name | GenBank Accession Number | BL206 | B356 | ||
|---|---|---|---|---|---|---|
| FC* | P value | FC* | P value | |||
| Cell Cycle Regulation | c-myc | NM_010849 | 3.7 | 0.0118 | 3.3 | 0.0025 |
| Cytokines/Chemokines | CCL20/MIP-3α | NM_016960 | 115.1 | 0.0277 | 78.4 | 0.0444 |
| IL-1β | NM_008361 | 34.0 | 0.0004 | 20.8 | 8.6E-06 | |
| IL-6 | X54542 | 27.8 | 0.0001 | 22.7 | 2.96E-06 | |
| CXCL1/GROα | NM_008176 | 17.1 | 1.07E-05 | 11.6 | 0.0022 | |
| CCL7/MARC/MCP-3 | Z12297 | 16.4 | 0.0006 | 14.4 | 0.0004 | |
| CCL2/JE/MCP-1 | NM_011333 | 13.0 | 0.0002 | 10.5 | 0.0002 | |
| CXCL2/MIP-2α/GROβ | NM_009140 | 8.0 | 0.0009 | 6.0 | 0.0030 | |
| CCL3/MIP-1α | NM_011337 | 5.6 | 0.0032 | 4.7 | 0.0019 | |
| IL-1α | NM_010554 | 4.5 | 0.0040 | 3.3 | 0.0131 | |
| CCL5/RANTES | M77747 | 4.2 | 0.0065 | 3.3 | 0.0026 | |
| CCL6/C10 | NM_009139 | 3.9 | 0.0009 | 3.0 | 0.0001 | |
| CCL22/MDC | NM_009137 | 3.4 | 0.0402 | 2.3 | 0.0072 | |
| CCL9/MIP-1γ | NM_011338 | 2.5 | 0.0023 | 2.4 | 0.0030 | |
| CCL19/MIP-3β | NM_011888 | 2.0 | 0.0034 | 2.4 | 0.0318 | |
| CCL21/C6kine | AF006637 | −26.9 | 2.04E-06 | −10.8 | 7.13E-06 | |
| Receptors and Antagonists | IL-1Ra | M57525 | 7.6 | 0.0001 | 7.5 | 0.0010 |
| CXCR-4 | NM_009911 | −7.3 | 0.0003 | −5.8 | 0.0006 | |
| Cytokine Suppression | CIS3/SOCS3 | NM_007707 | 3.0 | 0.0394 | 3.4 | 0.0095 |
| Growth and Development | Notch-1 | NM_008714 | 5.1 | 0.0087 | 5.1 | 0.0032 |
| BDNF | NM_007540 | 17.7 | 0.0110 | 16.4 | 0.0295 | |
| β-NGF | M35075 | 2.7 | 0.0410 | 2.0 | 0.0244 | |
| Angiopoietin-3/4 | NM_009641 | 2.0 | 0.0396 | 2.3 | 0.0113 | |
| Immune Cell Activation | CD28 | NM_007642 | −2.2 | 0.0360 | −2.2 | 0.0382 |
| Inflammation | Cox-2 | NM_011198 | 9.1 | 0.0002 | 9.8 | 0.0002 |
| iNOS | NM_010927 | 4.9 | 0.0291 | 3.7 | 0.0420 | |
| Caspase-11 | NM_007609 | 3.8 | 0.0106 | 3.2 | 0.0216 | |
| Stat 1 | NM_009283 | 3.6 | 0.0173 | 3.1 | 0.0078 | |
| TNF Superfamily | TNFRSF6/Fas | NM_007987 | 39.3 | 0.0006 | 20.9 | 0.0245 |
| TNF-α | M13049 | 6.8 | 0.0029 | 5.2 | 0.0042 | |
| TNF RII/TNFRSF1B | NM_011610 | 3.8 | 0.0010 | 3.5 | 0.0041 | |
| TNFRSF5/ CD40 | NM_011611 | 3.3 | 0.0007 | 2.5 | 0.0257 | |
| Weight Regulation | MCH | BB175332 | 3.3 | 0.0153 | 2.6 | 0.0489 |
FC = fold change relative to control.
Genes uniquely regulated in RAW264.7 cells exposed to B. burgdorferi isolates BL206 and B356
A surprisingly small number of genes showed patterns of differential transcription unique to either of the two isolates. Eighteen genes were uniquely regulated in RAW264.7 cells exposed to isolate BL206 (Fig. 1 and Table IIIA). These included induction of a number of genes involved in tissue remodeling processes, including growth differentiation factor 9 (GDF-9, 51.2 fold), bone morphogenetic protein 1 (BMP-1, 15.6 fold), bone morphogenetic protein 9 (BMP-9, 4.2 fold), matrix metalloproteinase 8 (MMP-8, 2.2 fold), and transforming growth factor beta receptor 1 (TGF-β R1, 5.4 fold). Transcript abundance of the gene encoding melanocortin receptor subtype 2 (MC2R, 18.5 fold) was also markedly increased. Transcription of five genes were uniquely suppressed by BL206--those encoding IL-18 binding protein (−7.4 fold), E-cadherin (−2.2 fold), a developmental factor (WNT-13, −2.0), an angiogenic factor (TIE-2, −4.8 fold), and a cytokine receptor (IL-6 Rα, −2.1 fold).
Table III.
B. burgdorferi isolate-specific regulation of cytokine and related genes in the murine macrophage-derived cell line RAW264.7 gamma NO(−) after 16 hours co-culture.
| A) Genes Uniquely Regulated by BL206 | ||||
|---|---|---|---|---|
| Gene Function | Gene Name | Genbank Accession Number | FC* | P value |
| Apoptosis | A1 | L16462 | 2.5 | 0.0088 |
| Cell Adhesion | Hip | AF116865 | 3.1 | 0.0474 |
| Integrin-β6 | X69902 | 2.1 | 0.0402 | |
| E-Cadherin | NM_009864 | −2.2 | 0.0029 | |
| Cytokine Receptors | CCR-5 | NM_009917 | 3.3 | 0.0466 |
| IL-6 Rα | X51975 | −2.1 | 0.0237 | |
| IL-18BP | NM_010531 | −7.4 | 0.0250 | |
| Endocrine Stress Response | MC2R | NM_008560 | 18.5 | 0.0074 |
| Growth and Development | GDF-9 | NM_008110 | 51.2 | 0.0332 |
| BMP-1 | L35281 | 15.6 | 0.0320 | |
| TGF-β RI | NM_007394 | 5.4 | 0.0238 | |
| BMP-9 | AF188286 | 4.2 | 0.0359 | |
| Chordin | NM_009893 | 3.6 | 0.0415 | |
| NRG-3 | NM_008734 | 2.1 | 0.0369 | |
| WNT-13 | NM_009520 | −2.0 | 0.0002 | |
| TIE-2 | X71426 | −4.8 | 0.0302 | |
| Invasion/Migration | MMP-8 | NM_008611 | 2.2 | 0.0372 |
| T-cell Regulation | CTLA-4 | NM_009843 | 2.2 | 0.0074 |
| B) Genes Uniquely Regulated by B356 | ||||
|---|---|---|---|---|
| Function | Gene Name | Genbank Accession Number | FC* | P value |
| Apoptosis | Bad | NM_007522 | −2.7 | 0.0295 |
| B-cell Development | EBF | NM_007897 | 4.2 | 0.0211 |
| Chemokines/Cytokines | CCL12/MCP-5 | NM_011331 | 5.6 | 0.0120 |
| CX3CL1 | NM_009142 | −3.1 | 0.0008 | |
| IL-16 | NM_010551 | −3.6 | 0.0047 | |
| CCL28 | NM_020279 | −4.5 | 0.0476 | |
| Cytokine Receptor | IL-8R | NM_009909 | 9.3 | 0.0310 |
| Endocrine Stress Response | CRFR1 | NM_007762 | 2.5 | 0.0446 |
| Growth and Development | TGF-β3 | NM_009368 | 3.8 | 0.0132 |
| FGF-1 | NM_010197 | −3.1 | 0.0373 | |
| BMPRIB | NM_007560 | −10.2 | 0.0087 | |
| Iron Uptake | TransferrinR | X57349 | −2.0 | 0.0248 |
FC = fold change relative to control.
Twelve genes were differentially regulated only by B356 (Fig. 1 and Table IIIB). These included three genes encoding chemokines (MCP-5/CCL12, 5.6 fold; neurotactin/CX3CL1, −3.1 fold; and CCL28, −4.5 fold, IL-16, −3.6 fold), a chemokine receptor (IL-8R, 9.3 fold) and two members of the TGF-β superfamily (TGF-β3, 3.8 fold; BMP-RIB, −10.2 fold).
RT-PCR validation of microarray data
In order to validate the gene array results, transcriptional activity of thirteen genes was analyzed by real-time quantitative RT-PCR. The selected genes included those found to be induced, repressed or unchanged relative to the control by gene array analysis (Table IV). A strong correlation (R2 = 0.71) was found between the fold change values obtained by gene array analysis and real-time RT-PCR.
Table IV.
A comparison of differential expression values of selected genes as determined by microarray and quantitative RT-PCR.
| Gene | BL206 | B356 | ||
|---|---|---|---|---|
| Microarray | qRT-PCR | Microarray | qRT-PCR | |
| IL-1β | 34.0 | 24.4 | 20.8 | 21.6 |
| Il-6 | 27.8 | 12.5 | 22.7 | 10.5 |
| IL-10 | 1.1 | 4 | 0.8 | 1.9 |
| IL-1RII | 1.1 | 1.4 | −1.7 | −5 |
| TNFα | 6.8 | 2.7 | 5.2 | 3 |
| CCL3/MIP-1α | 5.6 | 5.5 | 4.7 | 5.6 |
| CXCL2/MIP-2α | 8.0 | 12.6 | 6.0 | 13.7 |
| CCL2/MCP-1 | 13.0 | 2.3 | 10.5 | 2.2 |
| CXCR-4 | −7.3 | −7.7 | −5.8 | −7.3 |
| Cox-2 | 9.1 | 11.7 | 9.8 | 13.3 |
| ICAM2 | 1.8 | −2.5 | −1.7 | −2.5 |
| TLR2 | 1.3 | 1.3 | 1.1 | 1.3 |
| TLR4 | −1 | −0.5 | −1.7 | −0.7 |
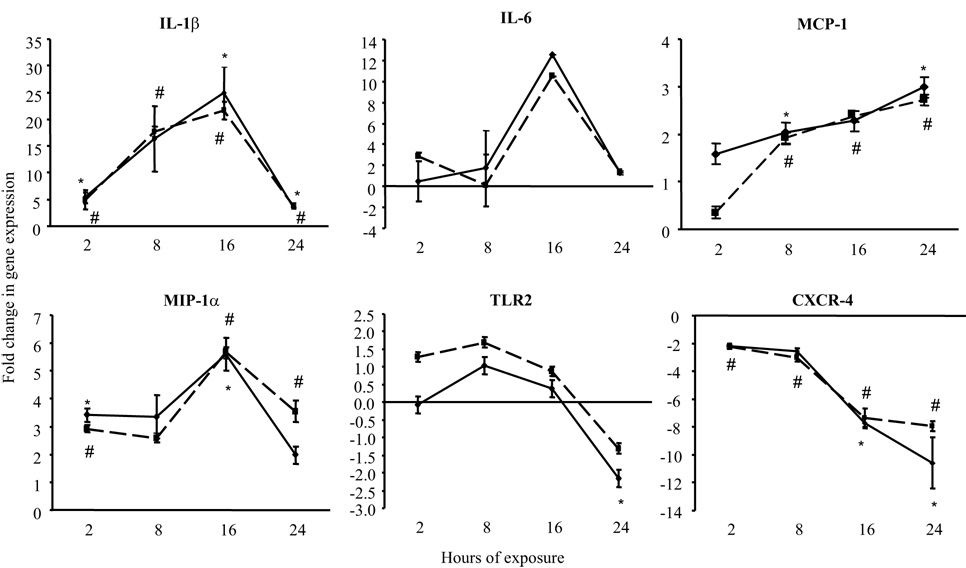
B. burgdorferi isolates induce identical transcription kinetics for selected cytokines and chemokines
Real-time RT-PCR was employed to analyze the transcriptional expression kinetics of selected genes in response to B. burgdorferi isolates B356 and BL206 over a 24-hour time period. Nearly identical patterns of transcription for genes encoding IL-1β, IL-6, MCP-1, MIP-1α, CXCR-4 and TLR2 were elicited by both isolates over this time period (Fig. 2). Relative transcript abundance of IL-1β, IL-6, and MIP-1α increased over time, peaking at 16 hours, and then falling to 2-hour levels by 24 hours. This latter decrease may be attributable to an increased death rate of the RAW264.7 gamma NO(−) cells at 24 hours of exposure, although this parameter was not quantitatively assessed. Transcript levels of MCP-1 increased steadily over 24 hours to maximum increases of 3.0-fold (BL206) and 2.7-fold (B356). In contrast, transcription of CXCR-4 was repressed 2-fold by both isolates relative to the control by 2 hours, and continued to decrease over time, reaching a maximal decline of −10.6 (BL206) and −7.9 (B356) at 24 hours. For comparison, the transcriptional profiles of these genes in response to E. coli LPS, a potent stimulator of cytokine expression produced by gram-negative bacteria (38), were also examined. At 16 or 24 hours, transcription levels of CXCR-4, IL-6 and IL-1β were 6-, 7- and 46-fold higher, respectively, in RAW264.7 cells exposed to 1µg/ml LPS (not shown). There was no significant difference at any time point in the transcript abundance of these cytokines in RAW264.7 cells exposed to either live or heat-killed BL206 (data not shown).
Figure 2. Gene expression profiles of RAW264.7 gamma NO(−) cells after exposure to B. burgdorferi clinical isolates BL206 and B356 as determined by real-time RT-PCR.
RNA was isolated from the murine macrophage-like cell line after incubation for 2, 8, 16 or 24 hours with either BL206 or B356 clinical isolate of B. burgdorferi (MOI = 10:1, spirochetes:RAW264.7 cells). Samples were assayed in duplicate, with the exception of 16- and 24-hr time points for IL-6, for which a single value was available. * P < 0.05, BL206-exposed RAW264.7 cells versus medium control (unpaired Student’s t-test). # P < 0.05, B356-exposed RAW264.7 cells versus medium control.
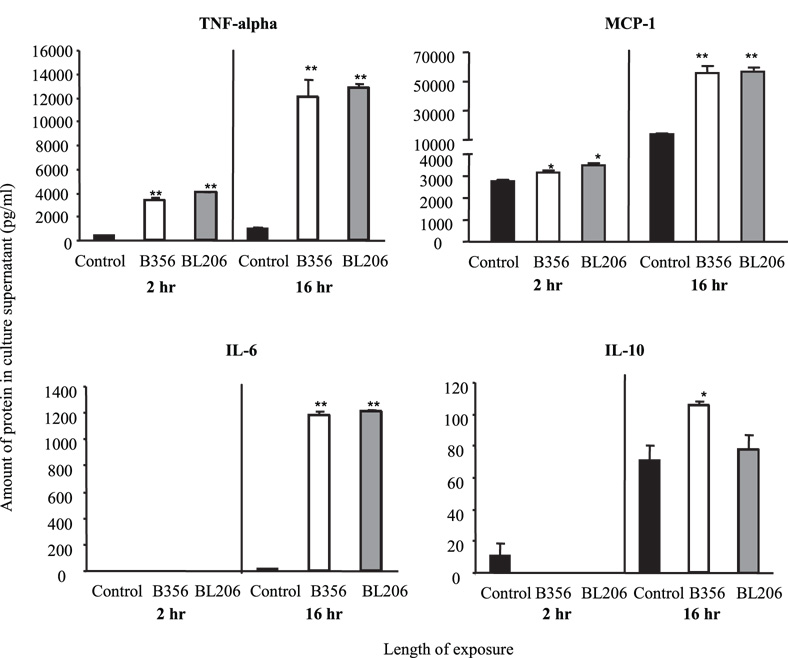
B. burgdorferi strains induce similar cytokine protein expression profiles
The protein levels for selected cytokines (TNF-α, MCP-1, IFN-γ, IL-6, IL-10 and IL-12p70) in the culture supernatants of RAW264.7 cells were measured after co-culture with B. burgdorferi strains BL206 or B356 for 2 or 16 hrs (Fig. 3). Strikingly, no statistically significant differences were detected in protein levels induced by B356 or BL206 for any parameter tested. Both isolates rapidly induced a pro-inflammatory cytokine response by 2 hrs, characterized by significant production of TNF-α and MCP-1 relative to the medium controls (P < 0.01 and P < 0.05, respectively). Levels of these cytokines increased substantially by 16 hrs. Production of the pro-inflammatory cytokine IL-6 was delayed relative to that of TNF-α and MCP-1; protein was not detected at 2 hrs but was measured at 1100 pg/ml by 16 hours (P < 0.01 vs control). The anti-inflammatory cytokine, IL-10, was not detected at 2 hrs, and by 16 hrs was produced at levels which differed significantly from the control only in RAW264.7 cells which had been co-cultured with B356. However, there was no significant difference in IL-10 levels induced by either isolate B356 or BL206. Neither of the B. burgdorferi isolates induced protein expression of IL-12p70 at levels which differed significantly from the medium control at either of the two time points tested (data not shown). IFN-γ was included as a negative control, as this cytokine is not expressed by macrophages under most conditions, and no expression of IFN-γ was observed, as expected (data not shown). As a positive control, RAW264.7 cells were incubated with 1 µg/ml E. coli LPS. With the exceptions of IL-12p70 and IFN-γ, LPS induced production of all cytokines at levels significantly higher than the medium controls (not shown).
Figure 3. Cytokine protein levels in supernatants of RAW264.7 gamma NO(−) cells exposed to B. burgdorferi isolates BL206 or B356 for 2 hr or 16 hr.
RAW264.7 cells grown to 80–90% confluence were incubated with live spirochetes (MOI = 10:1, spirochetes:RAW264.7 cells). Cytokine proteins were measured by flow cytometry using the Cytokine Bead Array, as described in Materials and Methods. Values represent the average of 2 samples ± SD. * P < 0.05 relative to control (unpaired Student’s t-test). ** P < 0.01 relative to control.
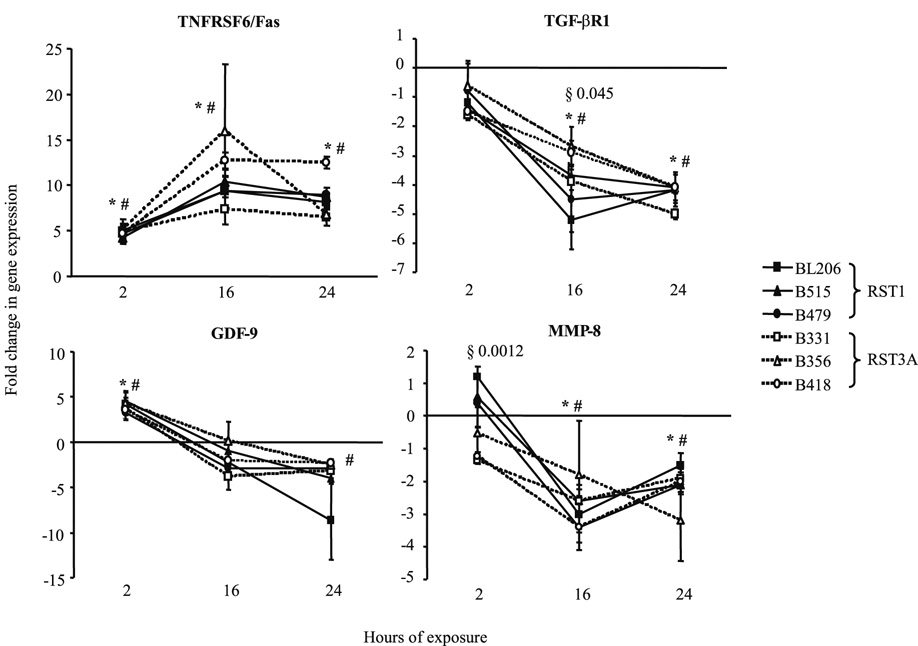
Virulent and attenuated isolates of B. burgdorferi induce similar transcription kinetics for selected tissue remodeling factors
Microarray analysis identified eighteen genes uniquely regulated in RAW264.7 cells by isolate BL206 (Table IIIA). Notably, many of these genes have annotated functions in growth and development or invasion/migration. In order to determine whether the ability to effect changes in the extracellular matrix is associated with invasive potential, RAW264.7 cells were co-cultured with B. burgdorferi clinical isolates BL206, B515 or B479, which have been shown to disseminate in C3H/HeJ mice (11), or isolates B356, B331 or B418, which do not disseminate. Total RNA collected after 2, 16 and 24 hrs was analyzed by real-time RT-PCR for the expression genes encoding TGF-βR1, MMP-8 and TNFSF6/Fas. Surprisingly, transcriptional expression profiles were strikingly similar for all isolates (Fig. 4). Transcript levels of TGF-βR1 and MMP-8 were significantly repressed at 16 and 24 hrs upon exposure to either disseminating or attenuated isolates. The difference between the two groups of isolates was statistically significant only for TGF-βR1 at 16 hrs (P = 0.045). Transcript levels of GDF-9 were significantly induced by 2 hrs, then declined. Transcript levels of TNFRSF6/Fas were significantly induced by either disseminating and attenuated isolates by 2 hours and reached maximum increase at 16 hours with mean fold changes of 9.7 and 12.0, respectively. As a control, transcription of IL-1β was also measured and was found to be significantly induced at all time points by all six isolates (data not shown). Transcription of BMP-1, BMP-9, BMP-RIβ and TIE-2 was also explored but could not be quantitated, as the expression levels of these genes were below the detection threshold even after 40 amplification cycles.
Figure 4. Transcriptional expression profiles of tissue remodeling genes in RAW264.7 gamma NO(−) cells exposed to disseminating (RST1) or non-disseminating (RST3A) B. burgdorferi isolates.
RNA was isolated from the murine macrophage-like cell line after incubation for 2, 16 or 24 hours with RST1 (BL206, B515, B479) or RST3A (B356, B331, B418) clinical isolates of B. burgdorferi (MOI = 10:1, spirochetes:RAW264.7 cells). Samples were assayed in triplicate. Mean fold-change values between groups were compared. * P < 0.05, RST1-exposed RAW264.7 cells versus medium control. # P < 0.05, RST3A-exposed RAW264.7 cells versus medium control. § P < 0.05, RST1-exposed RAW264.7 cells vs RST3A-exposed RAW264.7 cells.
DISCUSSION
B. burgdorferi clinical isolates BL206 and B356 represent genotypes which are associated with distinctly different disease profiles in both humans and mice; BL206 infection results in severe, disseminated disease in C3H/HeJ mice, whereas B356 does not disseminate from the inoculation site (10,11). Gene expression profiling of the acutely arthritic joint tissue of BL206-infected C3H/HeJ mice during the early disseminated stage (14 days post-infection) revealed strong transcriptional induction of genes encoding pro-inflammatory cytokines, CC and CXC chemokines and receptors, inflammatory mediators such as COX-2 and tissue remodeling factors MMP-3 and TIMP-1. Transcription of genes encoding both IL-1β and its receptor antagonist, IL-1ra, was induced by 41.8- and 26.1-fold, respectively (Table I), which is consistent with previously reported findings connecting a higher IL-1β/IL-1ra ratio with Lyme arthritis (39,40).
While these experiments were in progress, a study was published which used array analysis to profile the global gene expression in the B. burgdorferi-infected joints of arthritis-susceptible C3H/HeNCr mice and arthritis-resistant C57BL/6 mice (33). Despite the differences in the mouse strains (C3H/HeJ vs C3H/HeNCr), the array methodologies (cytokine-specific membrane array vs Affymetrix whole gene array) and the B. burgdorferi strains (BL206 vs N40) employed, the induction of a similar subset of cytokines and chemokines was observed in the present study. By 2 weeks post-infection, a number of transcripts encoding factors involved in host defense and inflammation were induced in the joint tissue of both mouse strains. These included chemokines and cytokines (CXCL13, CCL9, CXCL14, CCL2 and IL-1β) and factors involved in invasion and migration (MMP3, TIMP1). The gene encoding CXCL13, a proposed diagnostic marker for Lyme neuroborreliosis (41) which was induced 108-fold in joint tissue by isolate BL206 in the present study, was found by Crandall et al. to be associated with greater numbers of spirochetes in the joints. One notable difference between the studies was the complete absence of differentially regulated chemokine and cytokine receptor genes observed in the joints of N40-infected C3H/HeNCr mice, whereas ten of these genes were induced in the joints of C3H/HeJ mice by isolate BL206 in the present study. Crandall et al. also observed induction of a number of Type I and/or Type II interferon-responsive genes and concurrent repression of genes involved in epidermal differentiation in the joints of N40-infected C3H/HeNCr mice at 2 weeks post-infection. Transcriptional activity of these interferon-responsive and epidermal differentiation genes could not be assessed in the current study because the Panorama™ mouse cytokine gene array does not contain transcripts for these genes.
We hypothesized that the differences in pathogenic potential of disseminating and attenuated isolates may be attributable, at least in part, to differences in the nature of the inflammatory response induced in host cells. However, as strain B356 and other RST3A isolates do not disseminate to joint tissue, an in vitro model was used to test this hypothesis. Host macrophages are prolific producers of cytokines and chemokines and are among the first cell types to encounter the spirochete at the site of inoculation (4). The expression of 514 cytokine and related genes by the murine macrophage RAW264.7 cell line was analyzed by gene array after 16 hours of co-culture with the B. burgdorferi isolates B356 and BL206. The results yielded several interesting observations. Remarkably, over half (33/63) of the total number of genes differentially regulated transcriptionally in response to B. burgdorferi were commonly regulated by both genotypes. This group consisted primarily of genes encoding potent mediators of inflammation, including CC and CXC chemokines and their receptors, and the pro-inflammatory cytokines IL-1α and IL-1β. Transcription of three genes was commonly repressed in RAW264.7 cells by both B. burgdorferi isolates. The most striking repression was observed for the gene encoding CCL21. CCL21 is reportedly upregulated in different skin inflammatory conditions and is proposed to function in the recruitment of dendritic cells and T-lymphocytes to inflammatory foci (42,43). Down-regulation of CCL21 expression by B. burgdorferi may therefore be a means of suppressing its clearance by skin dendritic cells.
Real-time quantitative RT-PCR analysis of selected genes over a 24-hour time course of co-culture with isolates B356 and BL206 revealed that levels of transcriptional expression of IL-1β, IL-6, MCP-1, MIP-1α and CXCR-4 did not significantly differ between the two strains. These data support the initial observation of similar cytokine induction by both genotypes at 16 hours and expand it to include indistinguishable expression kinetics of key inflammatory mediators from as early as 2 hours after pathogen contact. No difference in transcription of these cytokines upon exposure to live or heat-killed BL206 was observed, which is consistent with other studies that found no difference in the production of selected cytokines elicited by live spirochetes or by heat-killed or sonicated B. burgdorferi extracts in human (44) and canine cells (45). This suggests that the elicited host cytokine responses are most likely induced by cell membrane components rather than by secreted or cytosolic factors. While it is possible that incubation in serum-free medium may have affected the expression of spirochetal virulence factors which distinguish the two isolates, we believe that this is unlikely as B. burgdorferi cultures were grown in BSK medium and only exposed to serum-free RPMI for a maximum time of 24 hours. Even after only 2 hours of incubation in the serum-free medium, there were no differences in cytokine mRNA or protein levels elicited by the two isolates, suggesting that the similar cytokine responses of RAW264.7 cells to these isolates are not due to the effects of serum starvation on the expression of B. burgdorferi virulence factors.
Measurement of the protein levels of selected cytokines by RAW264.7 cells yielded results which were consistent with the transcriptional data. Both B. burgdorferi isolates stimulated rapid and robust production of TNF-α, MCP-1 and IL-6. There was no significant difference in protein production between isolates BL206 and B356 at any time point. The barely significant induction of IL-10 by B356 which was observed is not entirely consistent with published data which report B. burgdorferi-stimulated production of IL-10 by human PBMCs, a human monocytic cell line and murine macrophages (20,46,47). However, while the protein concentration of induced IL-10 in several of these studies was comparable to that observed here, the background production of IL-10 was very low. In the current study, high background production of both IL-10 (71 pg/ml) and MCP-1 (14,101 pg/ml) was observed at 16 hours for RAW264.7 cells exposed to medium alone. This has not been reported with other cell types and may be a characteristic of RAW264.7 gamma (NO)- cells. The ability of RAW264.7 gamma (NO)- cells to secrete IL-10 in response to a stimulus was established by co-culture with E. coli LPS which resulted in IL-10 production at levels significantly higher than the control (164.9 ± 18.5 pg/ml; P < 0.05; data not shown).
B. burgdorferi has been shown to induce cytokine production in monocytes/macrophages (48, 46) primarily by signaling through TLR-2 (32, 49). We did not observe any differential expression of the gene encoding TLR-2 over a 24-hr period of exposure to either B. burgdorferi isolate (Fig. 2), although transcript levels of genes encoding pro-inflammatory cytokines and chemokines increased significantly. This contrasts with other reports of increased TLR-2 protein expression in B. burgdorferi-stimulated human monocytes and PBMCs (4, 50). Several factors may account for this apparent disparity. We used a macrophage-derived cell line, while other studies used samples obtained from human subjects and likely contained populations of activated cells. Secondly, Cabral et al. measured surface protein expression of TLR-2 after 48 hrs (50), whereas we did not measure protein levels but quantitated mRNA transcript abundance for up to 24 hrs. However, both Cabral et al. (50) and the present study concur in detecting significantly elevated levels of IL-1β and IL-6.
The strikingly similar chemokine and cytokine gene expression profiles of the B356- and BL206-stimulated RAW264.7 cells suggests that modulation of other host factors by B. burgdorferi contributes to the development and severity of clinical disease. The array results indicated that the disseminating isolate BL206 uniquely induced transcription of a number of genes involved in tissue remodeling processes. These genes consisted of MMP-8 and members of the TGF-β superfamily, including BMP-1, BMP-9, GDF-9 and TGF-β RI. The role of host MMPs in the pathogenesis of Lyme arthritis has been well established (51), and their presence in EM skin lesions has been hypothesized to facilitate spirochete dissemination through the extracellular matrix (52). BMPs cleave collagen and other extracellular matrix components and have been implicated in Salmonella pathogenesis (53). We therefore examined the transcriptional expression of these genes by real-time RT-PCR in RAW264.7 cells co-cultured with additional disseminating and attenuated isolates. In all samples, transcript levels of BMP-1 and BMP-9 were too low to be detected even after 40 PCR cycles. With the exception of one timepoint, there was no significant difference in the mean fold changes in transcript levels of GDF-9, MMP-8 and TGF-β RI elicited by the RST1 or RST3 isolates. The apparent disparity between the array and RT-PCR results may be explained by the degree of variability between the replicate samples of RNA used in array analysis. Our criteria for differential regulation of a gene consisted of both a fold change ≥ 2 and P < 0.05 using a two-tailed, unpaired Student’s t-test. Many of the genes meeting these criteria for one isolate also displayed a fold change of similar magnitude by the other isolate but, due to sample variability, had a P value > 0.05. These genes were therefore considered uniquely induced by either BL206 or B356. In contrast, RNA used for RT-PCR analysis of these genes was obtained from a separate experiment using multiple isolates assayed in triplicate. The larger sample size resulted in greater statistical robustness and, consequently, significant differences in expression relative to the control for both experimental groups.
In summary, transcriptional profiling of a murine macrophage cell line co-cultured with disseminating or non-disseminating clinical isolates of B. burgdorferi revealed no differences in the pro- and anti-inflammatory cytokine and chemokine responses. Further, there was no difference in the expression of selected factors involved in remodeling of the extracellular matrix. We conclude that the differential pathogenicity of disseminating and non-disseminating isolates of B. burgdorferi does not result from differences in the induction or repression of host macrophage-mediated inflammation.
Footnotes
This work was supported by National Institutes of Health grant AI45801 and grant 5UO1CI000160 from the Centers for Disease Control and Prevention.
Disclosures The authors report no financial conflicts of interest.
References
- 1.Orlowski KA, Hayes EB, Campbell GL, Dennis DT. MMWR CDC Surveill. Summ. 2000;49:1–11. [PubMed] [Google Scholar]
- 2.Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 3.Steere AC, Sikand VK. The presenting manifestations of Lyme disease and the outcomes of treatment. N. Engl. J. Med. 2003;348:2472–2474. doi: 10.1056/NEJM200306123482423. [DOI] [PubMed] [Google Scholar]
- 4.Salazar JC, Pope CD, Sellati TJ, Feder HM, Jr, Kiely TG, Dardick KR, Buckman RL, Moore MW, Caimano MJ, Pope JG, Krause PJ, Radolf JD. Coevolution of markers of innate and adaptive immunity in skin and peripheral blood of patients with erythema migrans. J. Immunol. 2003;171:2660–2670. doi: 10.4049/jimmunol.171.5.2660. [DOI] [PubMed] [Google Scholar]
- 5.Georgilis K, Steere AC, Klempner MS. Infectivity of Borrelia burgdorferi correlates with resistance to elimination by phagocytic cells. J. Infect. Dis. 1991;163:150–155. doi: 10.1093/infdis/163.1.150. [DOI] [PubMed] [Google Scholar]
- 6.Steere AC. Lyme disease. N. Engl. J. Med. 2001;345:115–125. doi: 10.1056/NEJM200107123450207. [DOI] [PubMed] [Google Scholar]
- 7.Liveris D, Wormser GP, Nowakowski J, Nadelman RB, Bittker S, Cooper D, Varde S, Moy FH, Forseter G, Pavia CS, Schwartz I. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 1996;34:1306–1309. doi: 10.1128/jcm.34.5.1306-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liveris D, Gazumyan A, Schwartz I. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 1995;33:589–595. doi: 10.1128/jcm.33.3.589-595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wormser GP, Liveris D, Nowakowski J, Nadelman RB, Cavaliere LF, McKenna D, Holmgren D, Schwartz I. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J. Infect. Dis. 1999;180:720–725. doi: 10.1086/314922. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Ojaimi C, Iyer R, Saksenberg V, McClain SA, Wormser GP, Schwartz I. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 2001;69:4303–4312. doi: 10.1128/IAI.69.7.4303-4312.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Ojaimi C, Wu H, Saksenberg V, Iyer R, Liveris D, McClain SA, Wormser GP, Schwartz I. Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J. Infect. Dis. 2002;186:782–791. doi: 10.1086/343043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wooten RM, Weis JJ. Host-pathogen interactions promoting inflammatory Lyme arthritis: use of mouse models for dissection of disease processes. Curr. Opin. Microbiol. 2001;4:274–279. doi: 10.1016/s1369-5274(00)00202-2. [DOI] [PubMed] [Google Scholar]
- 13.Ma Y, Seiler KP, Eichwald EJ, Weis JH, Teuscher C, Weis JJ. Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infect. Immun. 1998;66:161–168. doi: 10.1128/iai.66.1.161-168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weis JJ. Host-pathogen interactions and the pathogenesis of murine Lyme disease. Curr. Opin. Rheumatol. 2002;14:399–403. doi: 10.1097/00002281-200207000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Glickstein L, Moore B, Bledsoe T, Damle N, Sikand V, Steere AC. Inflammatory cytokine production predominates in early Lyme disease in patients with erythema migrans. Infect. Immun. 2003;71:6051–6053. doi: 10.1128/IAI.71.10.6051-6053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksi J, Savolainen J, Pene J, Bousquet J, Laippala P, Viljanen MK. Decreased interleukin-4 and increased gamma interferon production by peripheral blood mononuclear cells of patients with Lyme borreliosis. Infect. Immun. 1996;64:3620–3623. doi: 10.1128/iai.64.9.3620-3623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widhe M, Jarefors S, Ekerfelt C, Vrethem M, Bergstrom S, Forsberg P, Ernerudh J. Borrelia-specific interferon-gamma and interleukin-4 secretion in cerebrospinal fluid and blood during Lyme borreliosis in humans: association with clinical outcome. J. Infect. Dis. 2004;189:1881–1891. doi: 10.1086/382893. [DOI] [PubMed] [Google Scholar]
- 18.Zeidner N, Dreitz M, Belasco D, Fish D. Suppression of acute Ixodes scapularis-induced Borrelia burgdorferi infection using tumor necrosis factor-alpha, interleukin-2, and interferon-gamma. J. Infect. Dis. 1996;173:187–195. doi: 10.1093/infdis/173.1.187. [DOI] [PubMed] [Google Scholar]
- 19.Zeidner N, Mbow ML, Dolan M, Massung R, Baca E, Piesman J. Effects of Ixodes scapularis and Borrelia burgdorferi on modulation of the host immune response: induction of a TH2 cytokine response in Lyme disease-susceptible (C3H/HeJ) mice but not in disease-resistant (BALB/c) mice. Infect. Immun. 1997;65:3100–3106. doi: 10.1128/iai.65.8.3100-3106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown JP, Zachary JF, Teuscher C, Weis JJ, Wooten RM. Dual role of interleukin-10 in murine Lyme disease: regulation of arthritis severity and host defense. Infect. Immun. 1999;67:5142–5150. doi: 10.1128/iai.67.10.5142-5150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross DM, Steere AC, Huber BT. T helper 1 response is dominant and localized to the synovial fluid in patients with Lyme arthritis. J. Immunol. 1998;160:1022–1028. [PubMed] [Google Scholar]
- 22.Harjacek M, az-Cano S, Alman BA, Coburn J, Ruthazer R, Wolfe H, Steere AC. Prominent expression of mRNA for proinflammatory cytokines in synovium in patients with juvenile rheumatoid arthritis or chronic Lyme arthritis. J. Rheumatol. 2000;27:497–503. [PubMed] [Google Scholar]
- 23.Armstrong AL, Barthold SW, Persing DH, Beck DS. Carditis in Lyme disease susceptible and resistant strains of laboratory mice infected with Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 1992;47:249–258. doi: 10.4269/ajtmh.1992.47.249. [DOI] [PubMed] [Google Scholar]
- 24.Ruderman EM, Kerr JS, Telford SR, III, Spielman A, Glimcher LH, Gravallese EM. Early murine Lyme carditis has a macrophage predominance and is independent of major histocompatibility complex class II-CD4+ T cell interactions. J. Infect. Dis. 1995;171:362–370. doi: 10.1093/infdis/171.2.362. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery RR, Wang XM, Malawista SE. Murine Lyme disease: no evidence for active immune down-regulation in resolving or subclinical infection. J. Infect. Dis. 2001;183:1631–1637. doi: 10.1086/320703. [DOI] [PubMed] [Google Scholar]
- 26.Brown CR, Blaho VA, Loiacono CM. Susceptibility to experimental Lyme arthritis correlates with KC and monocyte chemoattractant protein-1 production in joints and requires neutrophil recruitment via CXCR2. J. Immunol. 2003;171:893–901. doi: 10.4049/jimmunol.171.2.893. [DOI] [PubMed] [Google Scholar]
- 27.Du Chateau BK, England DM, Callister SM, Lim LC, Lovrich SD, Schell RF. Macrophages exposed to Borrelia burgdorferi induce Lyme arthritis in hamsters. Infect. Immun. 1996;64:2540–2547. doi: 10.1128/iai.64.7.2540-2547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DuChateau BK, Jensen JR, England DM, Callister SM, Lovrich SD, Schell RF. Macrophages and enriched populations of T lymphocytes interact synergistically for the induction of severe, destructive Lyme arthritis. Infect. Immun. 1997;65:2829–2836. doi: 10.1128/iai.65.7.2829-2836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebnet K, Simon MM, Shaw S. Regulation of chemokine gene expression in human endothelial cells by proinflammatory cytokines and Borrelia burgdorferi. Ann. N. Y. Acad. Sci. 1996;797:107–117. doi: 10.1111/j.1749-6632.1996.tb52953.x. [DOI] [PubMed] [Google Scholar]
- 30.Habicht GS, Beck G, Benach JL, Coleman JL, Leichtling KD. Lyme disease spirochetes induce human and murine interleukin 1 production. J. Immunol. 1985;134:3147–3154. [PubMed] [Google Scholar]
- 31.Sprenger H, Krause A, Kaufmann A, Priem S, Fabian D, Burmester GR, Gemsa D, Rittig MG. Borrelia burgdorferi induces chemokines in human monocytes. Infect. Immun. 1997;65:4384–4388. doi: 10.1128/iai.65.11.4384-4388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wooten RM, Morrison TB, Weis JH, Wright SD, Thieringer R, Weis JJ. The role of CD14 in signaling mediated by outer membrane lipoproteins of Borrelia burgdorferi. J. Immunol. 1998;160:5485–5492. [PubMed] [Google Scholar]
- 33.Crandall H, Dunn DM, Ma Y, Wooten RM, Zachary JF, Weis JH, Weiss RB, Weis JJ. Gene expression profiling reveals unique pathways associated with differential severity of lyme arthritis. J. Immunol. 2006;177:7930–7942. doi: 10.4049/jimmunol.177.11.7930. [DOI] [PubMed] [Google Scholar]
- 34.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YH, Speed T. Normalization. In: Bowtell D, Sambrook J, editors. DNA Microarrays, a Molecular Cloning Manual. CSHL Press; 2003. pp. 536–543. [Google Scholar]
- 36.Wang G, Ma Y, Buyuk A, McClain S, Weis JJ, Schwartz I. Impaired host defense to infection and Toll-like receptor 2-independent killing of Borrelia burgdorferi clinical isolates in TLR2-deficient C3H/HeJ mice. FEMS Microbiol. Lett. 2004;231:219–225. doi: 10.1016/S0378-1097(03)00960-1. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Ulevitch RJ, Tobias PS. Recognition of gram-negative bacteria and endotoxin by the innate immune system. Curr. Opin. Immunol. 1999;11:19–22. doi: 10.1016/s0952-7915(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 39.Miller LC, Isa S, Vannier E, Georgilis K, Steere AC, Dinarello CA. Live Borrelia burgdorferi preferentially activate interleukin-1 beta gene expression and protein synthesis over the interleukin-1 receptor antagonist. J. Clin. Invest. 1992;90:906–912. doi: 10.1172/JCI115966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller LC, Lynch EA, Isa S, Logan JW, Dinarello CA, Steere AC. Balance of synovial fluid IL-1 beta and IL-1 receptor antagonist and recovery from Lyme arthritis. Lancet. 1993;341:146–148. doi: 10.1016/0140-6736(93)90006-3. [DOI] [PubMed] [Google Scholar]
- 41.Rupprecht TA, Pfister HW, Angele B, Kastenbauer S, Wilske B, Koedel U. The chemokine CXCL13 (BLC): a putative diagnostic marker for neuroborreliosis. Neurology. 2005;65:448–450. doi: 10.1212/01.wnl.0000171349.06645.79. [DOI] [PubMed] [Google Scholar]
- 42.Eberhard Y, Ortiz S, Ruiz LA, Kuznitzky R, Serra HM. Up-regulation of the chemokine CCL21 in the skin of subjects exposed to irritants. BMC. Immunol. 2004;5:7–14. doi: 10.1186/1471-2172-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serra HM, Eberhard Y, Martin AP, Gallino N, Gagliardi J, Baena-Cagnani CE, Lascano AR, Ortiz S, Mariani AL, Uguccioni M. Secondary lymphoid tissue chemokine (CCL21) is upregulated in allergic contact dermatitis. Int. Arch. Allergy Immunol. 2004;133:64–71. doi: 10.1159/000076129. [DOI] [PubMed] [Google Scholar]
- 44.Diterich I, Harter L, Hassler D, Wendel A, Hartung T. Modulation of cytokine release in ex vivo-stimulated blood from borreliosis patients. Infect. Immun. 2001;69:687–694. doi: 10.1128/IAI.69.2.687-694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Straubinger RK, Straubinger AF, Summers BA, Erb HN, Harter L, Appel MJ. Borrelia burgdorferi induces the production and release of proinflammatory cytokines in canine synovial explant cultures. Infect. Immun. 1998;66:247–258. doi: 10.1128/iai.66.1.247-258.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giambartolomei GH, Dennis VA, Lasater BL, Philipp MT. Induction of pro- and anti-inflammatory cytokines by Borrelia burgdorferi lipoproteins in monocytes is mediated by CD14. Infect. Immun. 1999;67:140–147. doi: 10.1128/iai.67.1.140-147.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murthy PK, Dennis VA, Lasater BL, Philipp MT. Interleukin-10 modulates proinflammatory cytokines in the human monocytic cell line THP-1 stimulated with Borrelia burgdorferi lipoproteins. Infect. Immun. 2000;68:6663–6669. doi: 10.1128/iai.68.12.6663-6669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radolf JD, Arndt LL, Akins DR, Curetty LL, Levi ME, Shen Y, Davis LS, Norgard MV. Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides activate monocytes/macrophages. J. Immunol. 1995;154:2866–2877. [PubMed] [Google Scholar]
- 49.Hirschfeld M, Kirschning CJ, Schwandner R, Wesche H, Weis JH, Wooten RM, Weis JJ. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J. Immunol. 1999;163:2382–2386. [PubMed] [Google Scholar]
- 50.Cabral ES, Gelderblom H, Hornung RL, Munson PJ, Martin R, Marques AR. Borrelia burgdorferi lipoprotein-mediated TLR2 stimulation causes the down-regulation of TLR5 in human monocytes. J. Infect. Dis. 2006;193:849–859. doi: 10.1086/500467. [DOI] [PubMed] [Google Scholar]
- 51.Hu LT, Eskildsen MA, Masgala C, Steere AC, Arner EC, Pratta MA, Grodzinsky AJ, Loening A, Perides G. Host metalloproteinases in Lyme arthritis. Arthritis Rheum. 2001;44:1401–1410. doi: 10.1002/1529-0131(200106)44:6<1401::AID-ART234>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Z, Chang H, Trevino RP, Whren K, Bhawan J, Klempner MS. Selective up-regulation of matrix metalloproteinase-9 expression in human erythema migrans skin lesions of acute Lyme disease. J. Infect. Dis. 2003;188:1098–1104. doi: 10.1086/379039. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 2000;164:5894–5904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]