Abstract
The inner walls of gold nanotubules, prepared by template synthesis in the nanopores of polycarbonate track etch membranes, have been chemically modified with peptide nucleic acid (PNA) and used for label-free quantification of complementary DNA sequences. Selective binding of DNA to the PNA modified nanotubules are shown to decrease the flux of optically detected anionic markers through the nanotubules in a concentration-dependent manner. The strong dependence of the biorecognition-modulated ion transport through the nanopores on the ionic strength suggests a dominantly electrostatic exclusion mechanism of the ion flux decrease as a result of DNA binding to the PNA-modified nanopores.
The extremely low detection limits required for bioanalysis are usually achieved with some chemical or physical amplification mechanism.1,2 In ion channel-mimetic sensors, the target species functions as a stimulus, actuating (opening or closing) ion channels and, thus, modulating the flux of the marker ions to be detected.3 Amplification is achieved because the concentration of markers exceeds the concentration of the target species by many orders of magnitude. In one approach, electrodes are modified with artificial receptors having intramolecular channels that can be blocked by the formation of inclusion complexes with the analyte and, thus, control the heterogeneous redox reactions of electroactive species.4 Another method in these lines makes use of genetically and/or chemically engineered receptors built into cell membranes.5-7 In a third approach based on gold nanopores, pioneered by the group of Martin,8-10 limitations due to the fragility of lipid bilayer membranes, in which biological nanopores are suspended, are eliminated. Modulations of the ionic permeability due to polarity changes (hydrophobic to hydrophilic)11 or steric effects (high molecular weight analytes binding to small molecular weight receptors in the nanopores) have already been explored.12,13 In a previous study, we have demonstrated the feasibility of the ion channel-mimetic biosensing with gold nanoporous membranes modified by biotin. The selective recognition of avidin modulates the Ca2+ ion transport, which is detected with ion-selective potentiometry.13 Later, the protein detection system has been extended by using single conically shaped gold nanopores.12
In this paper, for the first time, gold nanopores are functionalized, with a peptide nucleic acid (PNA)14 in view of a label-free detection of complementary DNA strands. The PNAs are shown to bind their complementary nucleic acid sequence15 with higher affinity and specificity compared with DNA probes.16 The nanopores were prepared by electroless Au plating of track etch polycarbonate membranes.10 In order to obtain regular pore diameters and better control of the plating process, the original recipe was slightly modified in that the membranes were dried in N2 atmosphere before placing them in the Au plating solution of pH 10 kept at 1 °C. As shown by field emission scanning electron micrographs of the resulting membranes, Au deposition times of 200 min gave in regular nanopores with close to uniform thickness of the Au layer (Figure 1B). From the weight of the deposited Au and the surface area of the membrane (calculated using the nominal membrane parameters provided by the manufacturer: 6 × 108 pores/cm2; pore diameter, 50 nm; membrane thickness, 6 μm), the Au pore diameters obtained after a plating time of 200 min were determined to be ∼14 nm (Figure 1B). The pore diameters were much larger (∼37 nm) after a plating time of 100 min (Figure 1A), and, due to the increasing size of the Au grain, too small and irregular at longer plating times (not shown). Based on this, plating times of 200 min were used throughout.
Figure 1.
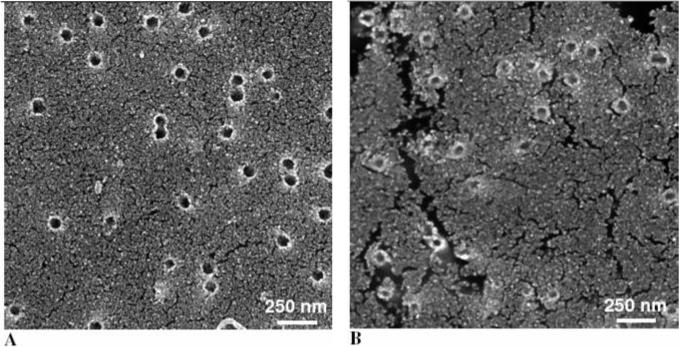
Scanning electron micrographs of Au nanotubule membranes. Polycarbonate track etch membranes 50-nm pore diameter were subjected to electroless Au plating for 100 min (A) or 200 min (B).
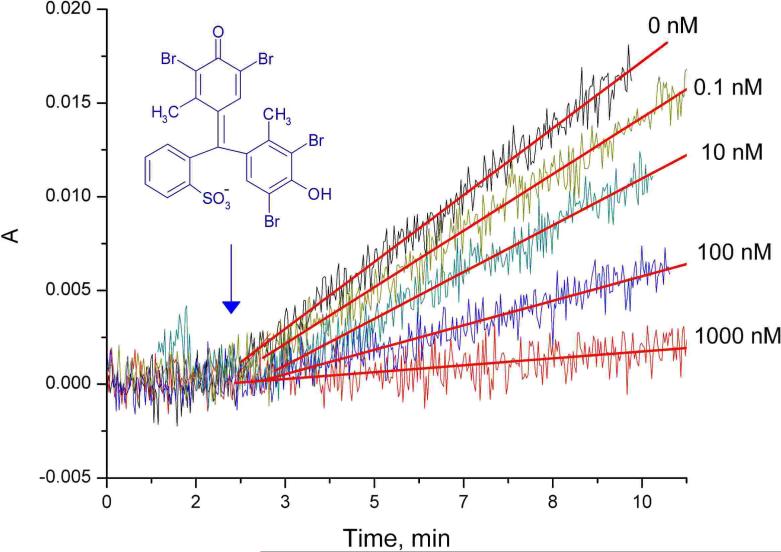
For preliminary experiments, freshly prepared Au nanopores were functionalized with the decamer PNA strand, ACT-CCG-TGA-C-O-Cys (O and Cys denote an ethylene glycol spacer and a cysteine modification at the 3' position), and subsequent blocking with 6-mercapto-1-hexanol (MH). The hybridization was detected by monitoring the flux of a charged dye in a transport cell with two 1-ml compartments separated by a membrane (0.196 cm2) providing 1.17 × 108 nanotubules that connected the two compartments. A concentration gradient of 0.1 or 1.0 mM bromocresol green (BCG) was established between the two stirred compartments containing 0.4 or 4 mM TRIS buffer of pH 8.0. The concentration increase in the acceptor compartment was measured at λmax = 615 nm using a fiber optic transmission probe connected to a spectrophotometer (Avantes BV, Eerbeek, The Netherlands). Typical time responses for the lowest measured BCG fluxes are shown in Figure 2. Ion fluxes were calculated from the corresponding slopes.
Figure 2.
Typical absorbance transients in the acceptor compartment due to the BCG permeation through PNA (ACT-CCG-TGA-C-O-Cys)-functionalized Au nanoporous membranes in contact with different concentrations of complementary ssDNA samples (5’-G-TCA-CGG-AGT-A20-3’). A 0.1 mM BCG concentration difference was established between the two stirred compartments of the transport cell and the absorbance, A, was recorded at λmax = 615 nm.
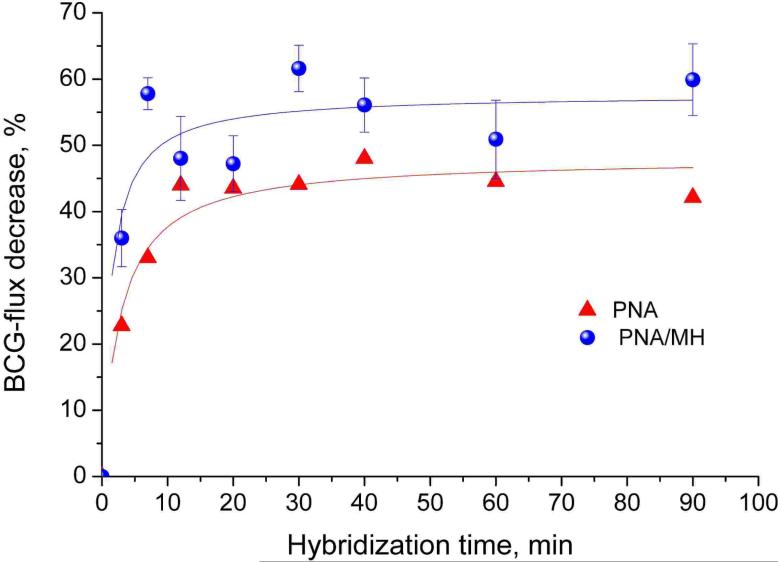
In a first set of experiments, the required hybridization time was determined. As shown in Figure 3, the relative ion fluxes strongly change during the first 10 min and are constant after 15−20 min. According to these results, hybridization times of 15−30 min were used in all subsequent experiments.
Figure 3.
Effect of the hybridization time on the BCG flux through PNA functionalized Au nanotubule membranes. PNA- and PNA/MH-modified Au membranes were incubated during different times with 65 μl of 5 μM complementary ssDNA (5’-G-TCA-CGG-AGT-A10-3’) dissolved in hybridization buffer (20 mM phosphate buffer, pH 7.0, 0.1 mM EDTA, 150 mM NaCl).
To exclude the influence of nonspecific absorption, the response of bare Au nanopores and of Au nanopores treated with MH was compared with that of the above described sensing membranes prepared by immobilization of PNA and subsequent blocking with 1 mM MH for 30 min. The nonspecific adsorption of ssDNA on bare Au induces a decrease of 9 % in its flux, as compared with 67% of the PNA-treated nanochannels. On the other hand, no significant flux changes are induced by ssDNA when Au is treated with MH (results not shown).
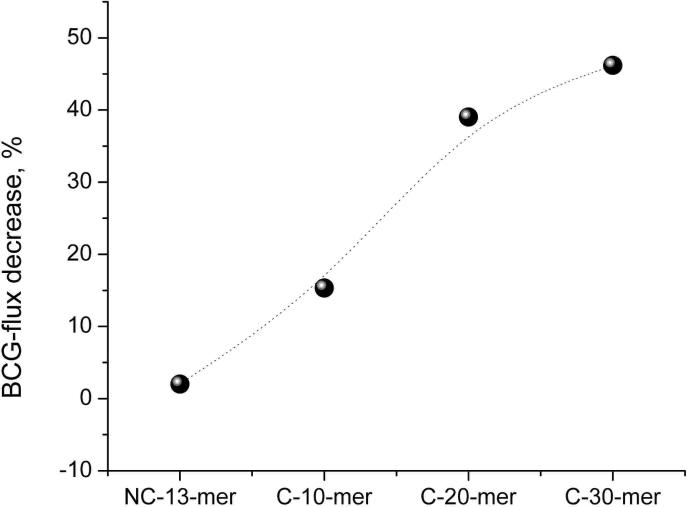
Gold nanotubules functionalized with PNA did not show any response to non-complementary ssDNA (Figure 4) in that the ion fluxes before and after the incubation were not significantly different. On the other hand, the chain length of the complementary ssDNA having 0, 10, or 20 additional adenine-based nucleotides has a significant effect (see Figure 4). The longer the DNA strand, the stronger is the reduction in the ion fluxes. These results indicate that the increased charge density of the nucleotides leads to a stronger reduction in ion fluxes and that the increased DNA strand length does not significantly reduce the efficiency of hybridization.
Figure 4.
Response of PNA-functionalized Au nanotubules to 10 μM complementary ssDNAs of different lengths: C-10-mer (5’-G-TCA-CGG-AGT-3’), C-20-mer (5’-G-TCA-CGG-AGT-A10-3’), C-30-mer (5’-G-TCA-CGG-AGT-A20-3’), and a non-complementary ssDNA, NC-13-mer (5’-GCT-TGT-TGG-TCA-A-3’).
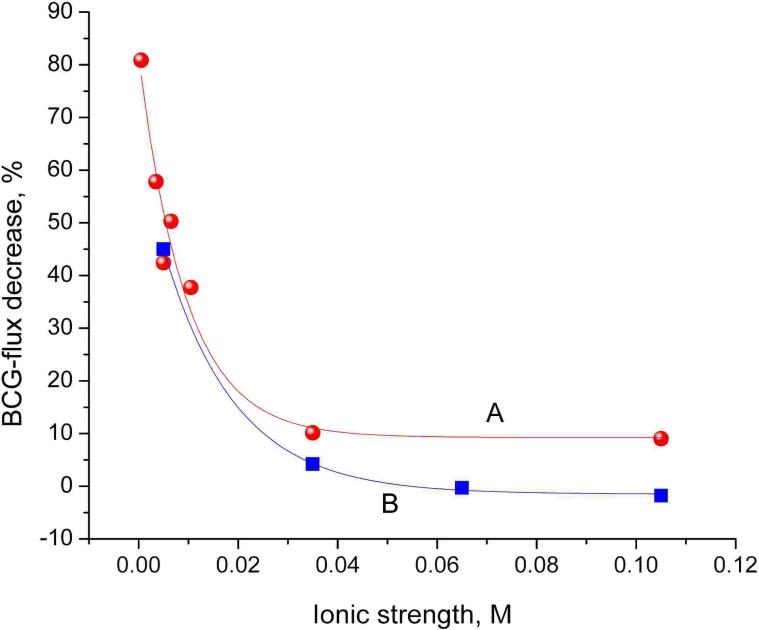
Peptide nucleic acids anchored to the outer surfaces of the membrane would also react with DNA but would not contribute to signal changes. For subsequent measurements with lower concentrations of ssDNA, before modification with PNA, the outer surface of the membranes plated with Au was, therefore, blocked with (1-mercaptoundec-11-yl)hexa(ethylene glycol) (HS-HEG) by microcontact printing using poly(dimethylsiloxane) stamps.13 The influence of the ionic strength on the observed flux changes is shown in Figure 5. First, the flux of BCG ions through PNA-functionalized Au nanotubules was determined at different ionic strengths by varying the concentration of the background electrolyte. Then, the PNA-functionalized membranes were incubated in the solution of the complementary ssDNA and the same experiments were repeated. As shown in Figure 5, at high ionic strengths, the incubation of ssDNA has practically no effect on the ion flux. Thus, the experiment demonstrates that the ion flux decrease is due to the charge repulsion effect. High sensitivity is observed at very low ionic strength. Such experiments are only feasible with PNA as probe since, in contrast to DNA-DNA, the binding strength of PNA-DNA complexes is practically unaffected by the ionic strength of the background.17
Figure 5.
Hybridization-related BCG flux decreases through PNA/MH-functionalized Au nanotubules determined at backgrounds of different ionic strength while establishing a BCG concentration difference of 0.1 mM (A) or 1.0 mM (B) across the membrane. A background of 0.4 mM or 4.0 mM TRIS buffer, with varying concentrations of NaCl, was used to adjust the ionic strength for experiments A and B, respectively. The flux decreases were calculated by using the flux values determined before and after incubating the membranes for 30 min in 1 μM complementary ssDNA (5’-G-TCA-CGG-AGT-A20-3’).
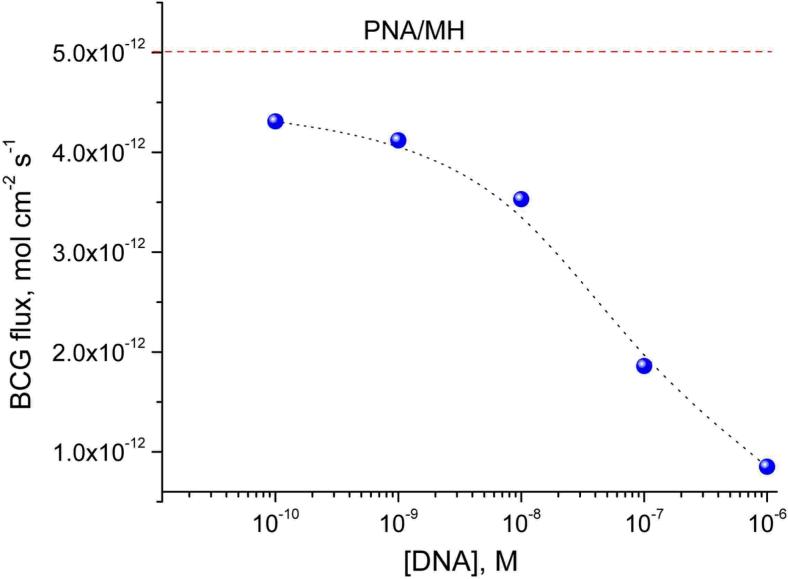
Based on these results, the influence of ssDNA concentration was investigated at a low ionic strength of 0.49 mM. As shown by Figure 6, reliable changes in the complementary ssDNS having 20 additional adenine-based nucleotides are observed down to 10−10 M. Although the flux change at this concentration is significantly higher than without incubation, the reproducibility of the measurements at lower concentrations was insufficient. The lowest amount of DNA that could be reliably determined using the experimental conditions presented before was 6 × 10−15 moles, which corresponds to an average to ∼30 DNA strands/pore. This suggests, on the one hand, that the DNA binding occurs at the nanopore opening hindering the entrance of further DNA strands into the lumen and not along the whole length of the nanotubule, and even so it can efficiently modulate the ion flux. On the other hand, it explains the uncertainties in detecting DNA concentrations below 10−10 M, which can be most likely attributed to small variations in the efficiency of the external membrane surface blocking. It should be noted that for DNA concentrations below 10−6 M, the PNA functionalized nanotubules could be effectively regenerated (within 5%) with 10 mM HCl solution at 70 °C and subsequently used, without apparent loss of activity, for DNA analysis.
Figure 6.
Calibration curve of complementary ssDNA (5’-G-TCA-CGG-AGT-A20-3’) using PNA/MH-functionalized nanotubules with the external membrane surface blocked with HS-HEG by microcontact printing. The BCG fluxes were measured at a concentration difference of 0.1 mM BCG across the membrane using 0.4 mM TRIS buffer as background. The membranes were incubated for 20 min in the complementary ssDNA followed by thorough washing in hybridization buffer before transport measurements.
The detection limit compares favorably with other label-free DNA detection technologies using PNA having a similar number of bases as capture probes, such as impedance spectroscopy (1 nM),18 quartz crystal microbalance (ca. 0.2 μM),16 and ion channel-mimetic sensors based on permeability changes of mixed PNA monolayers (∼1 μM19, 5 × 10−10 M).20 In the case of electrochemical ion channel-mimetic sensors, the self-assembled monolayer serves also as electric insulation (with the ionic transport perpendicular to the electrode surface), whereas in the case of synthetic nanopores, the chemical functionalization has merely the role to induce selectivity by attaching specific receptors (the ionic flux is parallel to the nanopore surface). Therefore, the chemically modified synthetic nanopores are less affected by contingent deterioration of the self-assembled monolayer, while for electrochemical ion channel-mimetic sensors, any defect in the self-assembled monolayer immediately increases the background signal.
It has been shown recently that, due to the increased stability of the corresponding DNA/PNA hybridization complexes, the use of longer PNA probes improves the detection limit of ion channel-mimetic sensors that are based on permeability changes in PNA monolayers, down to the femtomolar range.21 The same performance has been achieved also by miniaturized nanoFETs employing boron-doped single Si nanowires and decamer PNA probes.22 We expect that the miniaturization of the present setup, i.e. a lower number of pores integrated in the microfluidic system as well as longer PNA probes, would further enhance the sensitivity of the method.
Acknowledgement
This work has been supported by the Hungarian Scientific Fund (OTKA). REGy and GyJ gratefully acknowledge the Bolyai János and Sanofi Aventis research fellowships, respectively. The authors thank the National Institutes of Health (grants EB002189 and GM07178), the National Science Foundation, and the Swiss National Foundation for financial support of their electrochemical sensor research and Dr. D. Wegmann for careful reading of the manuscript.
References
- 1.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 2.Chang MMC, Cuda G, Bunimovich YL, Gaspari M, Heath JR, Hill HD, Mirkin CA, Nijdam AJ, Terracciano R, Thundat T, Ferrari M. Nanotechnologies for biomolecular detection and medical diagnostics. Curr. Opin. Chem. Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Buhlmann P, Aoki H, Xiao KP, Amemiya S, Tohda K, Umezawa Y. Chemical sensing with chemically modified electrodes that mimic gating at biomembranes incorporating ion-channel receptors. Electroanalysis. 1998;10:1149–1158. [Google Scholar]
- 4.Sugawara M, Kojima K, Sazawa H, Umezawa Y. Ion-Channel Sensors. Anal. Chem. 1987;59:2842–2846. doi: 10.1021/ac00151a004. [DOI] [PubMed] [Google Scholar]
- 5.Movileanu L, Howorka S, Braha O, Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 2000;18:1091–1095. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Quesada J, Ghadiri MR, Bayley H, Braha O. Cyclic peptides as molecular adapters for a pore-forming protein. J. Am. Chem. Soc. 2000;122:11757–11766. [Google Scholar]
- 7.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat. Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 8.Nishizawa M, Menon VP, Martin CR. Metal Nanotubule Membranes With Electrochemically Switchable Ion-Transport Selectivity. Science. 1995;268:700–702. doi: 10.1126/science.268.5211.700. [DOI] [PubMed] [Google Scholar]
- 9.Hulteen JC, Jirage KB, Martin CR. Introducing chemical transport selectivity into gold nanotubule membranes. J. Am. Chem. Soc. 1998;120:6603–6604. [Google Scholar]
- 10.Jirage KB, Hulteen JC, Martin CR. Effect of thiol chemisorption on the transport properties of gold nanotubule membranes. Anal. Chem. 1999;71:4913–4918. doi: 10.1021/ac990615i. [DOI] [PubMed] [Google Scholar]
- 11.Steinle ED, Mitchell DT, Wirtz M, Lee SB, Young VY, Martin CR. Ion channel mimetic micropore and nanotube membrane sensors. Anal. Chem. 2002;74:2416–2422. doi: 10.1021/ac020024j. [DOI] [PubMed] [Google Scholar]
- 12.Siwy Z, Trofin L, Kohli P, Baker LA, Trautmann C, Martin CR. Protein biosensors based on biofunctionalized conical gold nanotubes. J. Am. Chem. Soc. 2005;127:5000–5001. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- 13.Gyurcsányi RE, Vigassy T, Pretsch E. Biorecognition-modulated ion fluxes through functionalized gold nanotubules as a novel label-free biosensing approach. Chem. Commun. 2003:2560–2561. doi: 10.1039/b307393a. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-Selective Recognition Of Dna By Strand Displacement With A Thymine-Substituted Polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 15.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Nielsen PE, Jiang M, Cai XH, Fernandes JR, Grant DH, Ozsoz M, Beglieter A, Mowat M. Mismatch sensitive hybridization detection by peptide nucleic acids immobilized on a quartz crystal microbalance. Anal. Chem. 1997;69:5200–5202. doi: 10.1021/ac9706077. [DOI] [PubMed] [Google Scholar]
- 17.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen PE, Norden B, Graslund A. Ionic effects on the stability and conformation of peptide nucleic acid complexes. J. Am. Chem. Soc. 1996;118:5544–5552. [Google Scholar]
- 18.Liu JY, Tian SJ, Nielsen PE, Knoll W. In situ hybridization of PNA/DNA studied label-free by electrochemical impedance spectroscopy. Chem. Commun. 2005:2969–2971. doi: 10.1039/b419425j. [DOI] [PubMed] [Google Scholar]
- 19.Aoki H, Buhlmann P, Umezawa Y. Electrochemical detection of a one-base mismatch in an oligonucleotide using ion-channel sensors with self-assembled PNA monolayers. Electroanalysis. 2000;12:1272–1276. [Google Scholar]
- 20.Aoki H, Umezawa Y. High sensitive ion-channel sensors for detection of oligonucleotides using PNA modified gold electrodes. Electroanalysis. 2002;14:1405–1410. [Google Scholar]
- 21.Aoki H, Umezawa Y. Trace analysis of an oligonucleotide with a specific sequence using PNA-based ion-channel sensors. Analyst. 2003;128:681–685. doi: 10.1039/b300465a. [DOI] [PubMed] [Google Scholar]
- 22.Hahm J, Lieber CM. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 2004;4:51–54. [Google Scholar]