Abstract
δ-Catenin is upregulated in human carcinomas. However, little is known about the potential transcriptional factors that regulate δ-catenin expression in cancer. Using a human δ-catenin reporter system, we have screened several nuclear signaling modulators to test whether they can affect δ-catenin transcription. Among β-catenin/LEF-1, Notch1, and E2F1, E2F1 dramatically increased δ-catenin-luciferase activities while β-catenin/LEF-1 induced only a marginal increase. Rb suppressed the upregulation of δ-catenin-luciferase activities induced by E2F1 but did not interact with δ-catenin. RT-PCR and Western blot analyses in 4 different prostate cancer cell lines revealed that regulation of δ-catenin expression is controlled mainly at the transcriptional level. Interestingly, the effects of E2F1 on δ-catenin expression were observed only in human cancer cells expressing abundant endogenous δ-catenin. These studies identify E2F1 as a positive transcriptional regulator for δ-catenin, but further suggest the presence of strong negative regulator(s) for δ-catenin in prostate cancer cells with minimal endogenous δ-catenin expression.
Keywords: δ-Catenin/NPRAP, E2F1, Wnt, Notch, Prostate, catenin, LEF-1, cancer, transcription
Introduction
δ-Catenin was first identified as an interacting molecule with presenilin-1 and is primarily expressed in brain [1; 2]. Overexpressed δ-catenin promotes cellular processes with branching [3], and deletion of δ-catenin leads to severe cognitive and synaptic dysfunction [4]. In addition to presenilin-1, δ-catenin is known to associate with adhesive junction proteins such as E-cadherin [5]. The association of δ-catenin with E-cadherin raises the possibility that δ-catenin may play an important role in epithelial cell function. Recent reports showed that the expression of δ-catenin is upregulated in several human carcinomas including prostate, esophagus, and breast carcinomas, all originating from epithelial cells [6]. Therefore, it is of great importance to identify proteins regulating the expression of δ-catenin in tumors as well as in brain.
Recently, Duparc et al. showed that Pax6, a homeobox transcription factor, is required for δ-catenin expression during retinal, cerebella and cortical development in mice [7]. However, there are no reports for transcription factors affecting δ-catenin expression in cancer. In an effort to identify the potential transcriptional factors for upregulating δ-catenin expression, we analyzed the putative promoter sequences of human δ-catenin using Genomatix program (http://www.genomatix.de/). Many proteins including Pax6 were listed as potential binding factors to the putative promoter region of human δ-catenin. Among these, LEF-1, RBP-Jkappa, and E2F drew our attention for several reasons. LEF-1, a high-mobility-group (HMG) transcription factor, is a well-known transcription factor interacting with β-catenin, a δ-catenin related armadillo protein and a Wnt signaling effector [8]. Wnt signaling is known to be activated in tumors including breast and colon carcinomas, nuclear localization of β-catenin is observed in many different types of human cancers [9; 10]. RBP-Jkappa associates with NICD (Notch Intracellular Domain) in the nucleus, and the activation of Notch signaling pathway also plays an important role in tumorigenesis [11; 12]. E2F, involved in cell cycle regulation, interacts with Rb and p107 protein [13; 14; 15]. The release of E2F from Rb triggers the shift from G1 to S phase in cell cycle, and persistent activation of E2F-driven transcription occurs as results from Rb mutation in many tumors including prostatic carcinoma, retinoblastoma, and bladder cancer [16; 17; 18].
In this report, we tested the hypothesis that β-catenin/LEF-1, Notch1, and E2F1 can regulate the transcription of δ-catenin in human cancer cells. Among these, we demonstrated that E2F1 is a potent positive regulator of δ-catenin transcription. Analyses of 4 human prostate carcinoma cell lines revealed that the regulation of δ-catenin expression in these prostate carcinomas is mainly at the level of transcription.
Experimental procedures
Plasmids and antibodies
The construction of full-length δ-catenin in pEGFP-C1 has been previously described [3]. The antibodies used were as follows: mouse monoclonal anti-δ-catenin (BD Bioscience), rabbit polyclonal anti-δ-catenin (Upstate biotechnology, Millipore), mouse monoclonal anti-actin (Calbiochem); mouse monoclonal anti-retinoblastoma (Rb) (BD Bioscience), and mouse monoclonal anti-HA (12CA5)(Roche Applied Science). Mouse monoclonal anti-δ-catenin/NPRAP/Neurojungin (J19) was a gift from Dr. Werner Franke.
Construction of human δ-catenin reporter system
Different lengths of putative human δ-catenin reporter system were generated by five sets of PCR amplification. Primer sequences were as follows: Forward Primers, K1, 5’-GGGGTACCAATGAAATGGGGCACTGAGCAA-3’; K3, 5’-GGGGTACCACTGTGGGGAAGGATCAGGGGA-3’; K4, 5’-GGGGTACCCTAAGGAGAAACCCTCACTACC-3’; K5, 5’-GGGGTACCTGAATTAGGGAAGCGGAGTAAC-3’; and Reverse Primer K7, 5’-CTAGCTAGCCCGGCGGCTTCCTCGCAAACAT-3’. Human genomic DNA isolated from white blood cells were used as DNA template. PCR products were purified and cloned into pCR-XL-TOPO, and positive transformants were selected with KphI and NheI double digestion. Eluted DNAs were subcloned into pGL3-basic reporter vector. After construction, the sequences were confirmed by DNA sequencing.
Luciferase reporter assay
Subconfluent cultures in 24-well plate were transiently transfected by Lipofectamine with expression vector for several genes as indicated in the Result section. Luciferase activity was determined 12 h after transfection using the Luciferase Assay system (Promega). All experiments were performed in triplicate. Luciferase activity of each sample was normalized to β-galactosidase activity, which was used as control for determining transfection efficiency.
RT-PCR
For RT-PCR, up to 1 ug of total RNA was reverse-transcribed using AccuPower RT Premix (Bioneer, Daejeon, Korea) and oligo (dT) primers (Invitrogen) or gene-specific primer sets. A portion of the reverse transcription mixture was subject to PCR (40 cycles of 1 min at 95oC, 1 min at 54oC and 1 min at 72oC) using HotMaster Tag DNA polymerase (Eppendorf, Hamburg, Germany). Primer sequences are as follows: for δ-catenin, forward primer, 5’-ATGTTTGCGAGGAAGCCGC-3’, and reverse primer, 5’GTCTGGTTGCTATGGTAGCTGGC-3’; for E2F1, forward primer, 5’-ACTCCTCGCAGATCGTCATCATCT-3’, and reverse primer, 5’-GGACGTTGGTGATGTCATAGATGCG-3’; and for GAPDH1, forward primer, 5’-GTGGATATTGTTGCCATCAATGACC-3’, and reverse primer, 5’-GCCCCAGCCTTCATGGTGGT-3’. PCR products were analyzed in 0.8% agarose gel. To confirm the even/relative loading, GAPDH1 control primer set was used.
Cell culture, transfection, immunoprecipitation and Western blot
CWR22-Rv1, DU145, PC3, and LNCaP human prostate cancer cell lines, as well as SH-SY5Y human neuroblastoma cells and MDCK cannie kidney epithelial cells, were from ATCC and maintained in DMEM or in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Cells were transfected using Lipofectamine Plus reagent (Invitrogen), according to the manufacturer's instructions, or using the calcium phosphate precipitation method. Transfected cells were harvested with lysis buffer (10% glycerol, 25mM HEPES, pH7.3, 150mM NaCl, 1mM EDTA, 25mM NaF, 1mM Na3VO4, 1% NP40). Equal amounts of protein samples were solubilized and boiled with SDS sample buffer for 2 min and then separated by SDS-PAGE. After proteins were transferred to hydrophobic polyvinylidene difluoride (PVDF) or nitrocellulose membranes (Millipore), Western blots were performed with appropriate antibodies and developed with ECL Western blotting detection reagents (Amersham). Immunoprecipitation was performed as previously described [19].
Results
E2F1 is a potent, positive of δ-catenin transcription
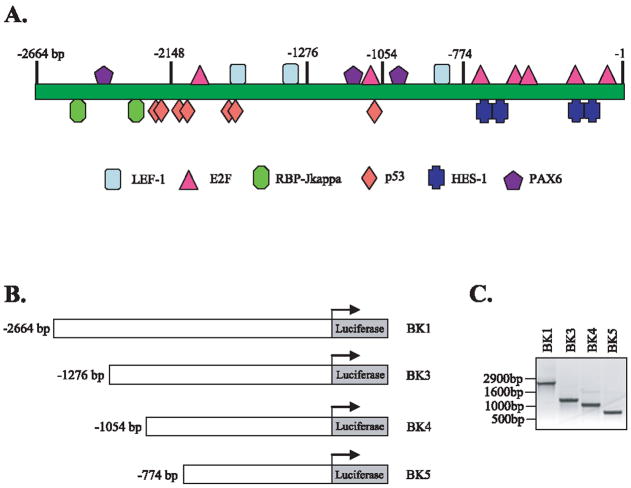
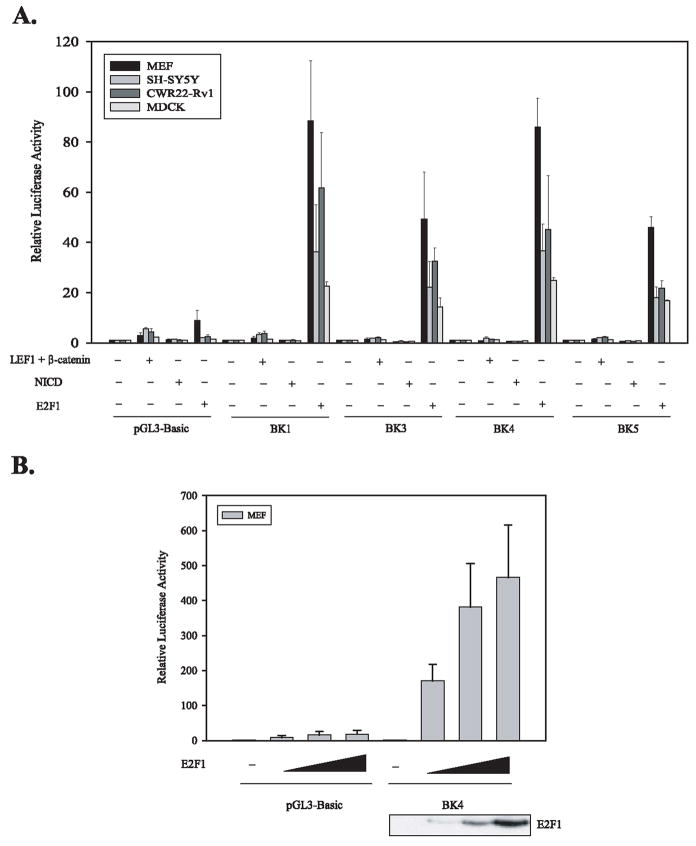
We analyzed 3145 bp DNA sequences upstream of human δ-catenin transcription start site using Genomatix program. We have identified that this human δ-catenin promoter region contains multiple potential binding sites for LEF-1, RBP-Jkappa, and E2F as shown in Figure 1A. Analyses of this human δ-catenin promoter region also revealed the presence of binding sequences for known regulator, Pax6, for δ-catenin expression in addition to many other potential regulators. In order to test the possibility that LEF-1, RBP-Jkappa, and E2F affects the transcription of δ-catenin, we generated 4 different human δ-catenin promoter-reporter systems as illustrated Figure 1B and C. We co-transfected β-catenin/LEF-1, NICD (a binding protein of RBP-Jkappa), and E2F1 together with a human δ-catenin-luciferase vector into 4 different cell lines- mouse embryonic fibroblasts (MEFs), SH-SY5Y human neuroblastoma cells, CWR22-Rv1 human prostate carcinoma cells, and MDCK canine kidney epithelial cells. As shown in Figure 2A, E2F1 induced dramatic increases in δ-catenin-luciferase activities, ranges from 15-to 85-folds over control vector, depending on cell types and reporter vectors used. BK1 (−2664 bp promoter) and BK4 (−1054 bp promoter) δ-catenin-luciferases tended to show higher fold induction than BK3 (−1276 bp promoter) and BK5 (−774 bp promoter) reporters in most cell lines. In contrast to E2F1, NICD did not induce any noticeable increases in δ-catenin-luciferase activities, and β-catenin/LEF-1 induced only 2–3 fold increases. E2F1 increased δ-catenin-luciferase activities in a concentration-dependent manner as shown in Figure 2B. The expression of β-catenin, LEF-1, NICD, and E2F1 was confirmed using Western blot analyses (E2F1- bottom panel, Fig. 2B; β-catenin, LEF-1, and NICD-data not shown).
Fig. 1.
Construction of human δ-catenin reporter systems. (A) A schematic diagram showing putative transcription factor binding regions on δ-catenin promoter. Numbers on top indicates the distance in base pairs from the transcription start site. (B) Generation of 4 δ-catenin-Luciferase constructs containing human δ-catenin promoter in different length in pGL3-basic reporter vector. (C) PCR products of human δ-catenin DNA, used for reporter construction, confirm different δ-catenin promoters at the predicted sizes.
Fig. 2.
E2F1 is a potent positive regulator of δ-catenin transcription. (A) β-catenin/LEF-1, NICD (a binding protein of RBP-Jkappa), and E2F1, together with a different human δ-catenin-lucferase vector (BK1, 3, 4, 5), were co-transfected into 4 different cell lines (MEFs, SH-SY5Y, CWR22-Rv1, and MDCK cells) as indicated. E2F1 induced dramatic increases in δ-catenin-luciferase activity regardless of cell types and reporter vectors used. (B) E2F1 increased δ-catenin-luciferase activities in a concentration-dependent manner. The expression of E2F1 was confirmed using Western blot analysis (bottom panel).
Rb suppressed the upregulation of δ-catenin-luciferase activities induced by E2F1 but did not interact with δ-catenin
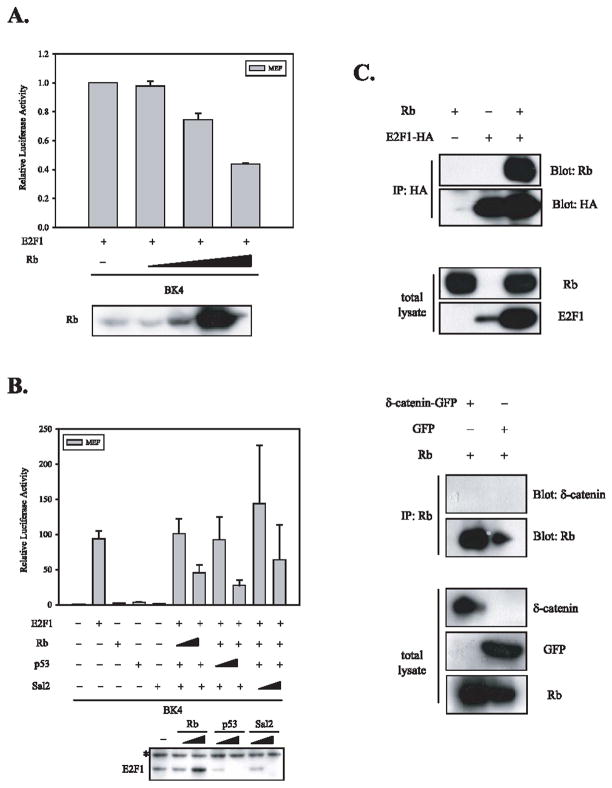
As Rb interacts with E2F1 and suppresses E2F1-driven transcription, we tested the possibility that Rb suppresses the upregulation of δ-catenin reporter activity induced by E2F1. As shown in Figure 3, the increased doses of Rb (Fig 3A, bottom panel) suppressed the upregulation of δ-catenin-luciferase activities induced by E2F1 (Fig 3A), while it stabilized the level of E2F1 (Fig 3B, bottom panel). Interestingly, two other tumor suppressors-p53 and Sal2 [20], implicated in the regulation of E2F, also suppressed the upregulation of δ-catenin reporter activity induced by E2F1 (Fig. 3B). However, unlike Rb, the levels of E2F1 were dramatically reduced by increased doses of p53 and Sal2 (Fig 3B, bottom panel), suggesting that the regulation mechanism of δ-catenin expression by p53 and Sal2 is different from that of Rb.
Fig. 3.
Rb suppresses the upregulation of δ-catenin-luciferase activities induced by E2F1 but does not interact with δ-catenin. (A) The increased doses of Rb suppressed the upregulation of δ-catenin-luciferase activities induced by E2F1 in a concentration dependent manner. The expression of Rb was confirmed using Western blot analysis (bottom panel). (B) Two other tumor suppressors-p53 and Sal2, implicated in the regulation of E2F, also suppress the upregulation of δ-catenin reporter activity induced by E2F1. Unlike Rb, the levels of E2F1 were dramatically reduced by increased doses of p53 and Sal2 (bottom panel). The asterisk indicates a non-specific band detected by anti-HA antibody. (C) E2F1, a well-known binding partner of Rb, was co-immunoprecipitated with Rb (top 2 panels). However, δ-catenin was not co-immunoprecipitated with Rb (bottom 2 panels). The relative expression of Rb, E2F1, δ-catenin, and GFP in total lysates was confirmed by Western blot analyses using specific antibodies against each protein.
Both mouse and human δ-catenin contain putative Rb binding sites. In human δ-catenin, there were two putative Rb binding sites- 793LxCxE797 and 81ExCxL85 - which are conserved in mouse δ-catenin. Therefore, Rb can not only suppress the transcription of δ-catenin through its association with E2F1 but may also modify δ-catenin expression and function via its direct interaction. In order to investigate whether Rb interacts with δ-catenin, we performed immunoprecipitation assays. As shown in Figure 3C, E2F1, a well-known binding partner of Rb, was co-immunoprecipitated with Rb. However, δ-catenin was not co-immunoprecipitated with Rb, ruling out the possibility that Rb inhibits δ-catenin expression and function via its direct interaction at the protein level.
δ-Catenin expression in human prostate carcinoma cell lines is controlled primarily at the transcriptional level
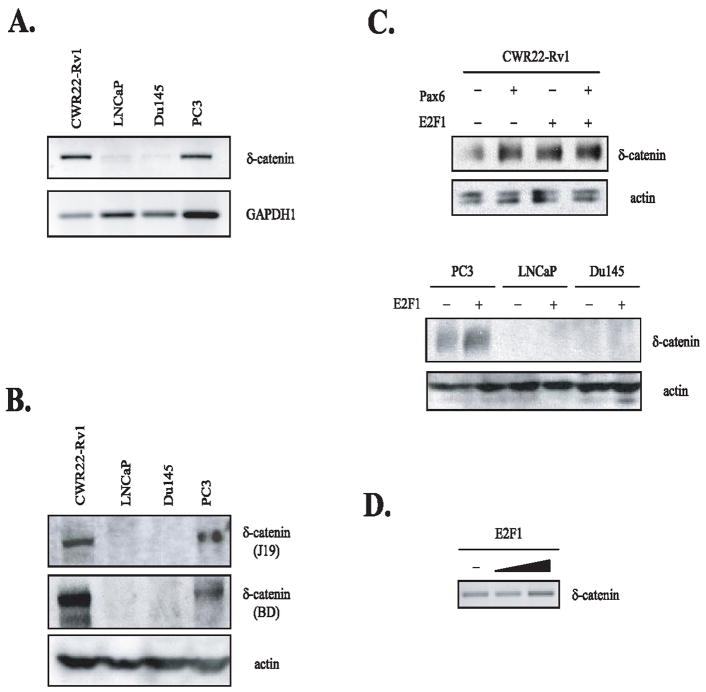
In order to demonstrate that E2F1 can indeed increase the endogenous δ-catenin expression in cells, we performed RT-PCR and Western blot analyses. First, we performed RT-PCR and Western blot analyses in order to detect the basal, endogenous levels of human δ-catenin RNA and protein, respectively. As shown in Figure 4A and B, the expression levels of δ-catenin can be demonstrated in the following order: CWR22-Rv1 > PC3 > DU145 and LNCaP. These results strongly suggest that regulation of δ-catenin expression in these 4 cancer cells is mainly controlled at the level of transcription. DU145 and LNCaP prostate carcinoma cells contained barely detectable levels of δ-catenin RNAs. δ-Catenin protein expression was undetectable despite using 2 different anti-δ-catenin antibodies directed against different binding epitopes. We then transfected E2F1 into these 4 cell lines and analyzed the expression of δ-catenin by Western blot analysis (Fig. 4C). Pax6, a known regulator of δ-catenin expression, was used as a positive control and also for comparison. Interestingly, the effects of E2F1 on δ-catenin expression were observed in human tumor cell lines expressing abundant levels of endogenous δ-catenin, most notably in CWR22-Rv1, but not in DU145 and LNCaP prostate carcinoma cells having undetectable levels of basal, endogenous δ-catenin. The effects of E2F1 on δ-catenin RNA expression in CWR22-Rv1 cells were also confirmed by RT-PCR (Fig. 4D). These data suggest the presence of strong negative regulator(s) for δ-catenin transcription in some cancer cell lines.
Fig. 4.
Regulation of δ-catenin expression in human prostate carcinoma cell lines is mainly at the transcriptional level. (A) RT-PCR analysis of δ-catenin in 4 different human prostate carcinoma cell lines (CWR22-Rv1, LNCaP, DU145 and PC3). Control primer sets for GAPDH1 were used to show relative loadings in these carcinoma cell lines. (B) Western blot analysis of human δ-catenin in 4 different human prostate carcinoma cell lines. Two different anti-δ-catenin antibodies were used to confirm the relative expression of δ-catenin in these cell lines. Anti-actin antibody was used to show relative loadings in these carcinoma cell lines. (C) E2F1 overexpression induced increased expression of δ-catenin in CWR22-Rv1 to the similar extent as that of Pax6 (top panel). However, the effects of E2F1 on δ-catenin expression were not observed in DU145 and LNCaP prostate carcinoma cells, which showed undetectable levels of endogenous δ-catenin (bottom panel). (D) The effects of E2F1 on δ-catenin RNA expression in CWR22-Rv1 cells were also confirmed by RT-PCR analysis.
Discussion
Aberrant expression of δ-catenin is associated with different pathologic conditions in human. For example, the hemizygous loss of the chromosomal 5p15.2 region, which contains the human δ-catenin gene, results in severe mental retardation associated with Cri-du-Chat syndrome [21]. On the other hand, the overexpression of human δ-catenin has been reported in several human tumors including breast and prostate carcinomas [6]. These studies strongly suggest that the expression of δ-catenin is tightly regulated as shown by various δ-catenin expression levels in different tissues [1]. However, except for Pax6 identified recently as a positive regulator for δ-catenin expression in the central nervous system [7], no reports are available about the potential transcription factor(s) responsible for up-regulating δ-catenin expression in human tumors.
Our promoter analysis using Genomatix program indicated that putative promoter region of human δ-catenin contains binding sites for many transcriptional factors. For control of neuronal expression of δ-catenin, it is of great interest to note that there are Pax6 binding sites as previously reported [7], NGF-induced protein C binding sites, and neural-restrictive-silencer-element binding sites. For tumor expression, we focused on the potential effects of β-catenin/LEF-1, NICD/RBP-Jkappa, and E2F on δ-catenin expression because Wnt, Notch and E2F/Rb signaling pathways play a pivotal role in tumorigenesis.
Due to the presence of multiple putative transcription factor binding sites in the δ-catenin promoter region for these molecules, as illustrated in Figure 1A, it is difficult to pinpoint a critical binding site for each transcription factor. Instead, we generated 4 human-δ-catenin reporter systems containing different lengths of putative δ-catenin promoter (BK1: −2,664 bp; BK3: −1,276 bp; BK4: −1,054 bp; BK5: −774 bp). Even though there were two putative RBP-Jkappa and 4 Hes-1 binding sites in BK1 human δ-catenin-luciferase system, the overexpression of NICD did not induce any noticeable increases in δ-catenin-luciferase activity, ruling out the possibility that Notch signaling regulates the transcription of δ-catenin. In spite of 3 putative LEF-1 binding sites, the overexpression of β-catenin and LEF-1 showed only 2–3 fold increases in δ-catenin-luciferase activity, suggesting that the canonical Wnt signaling may not play a major role in the regulation of δ-catenin transcription. However, we clearly demonstrated that E2F1 is a potent effector upregulating δ-catenin expression. The dose-response of E2F1 and suppression by Rb protein support the possibility that E2F1 indeed upregulates the expression of human δ-catenin at the transcriptional level. Two other tumor suppressor proteins, p53 and Sal2, may interact with E2F. As overexpressed p53 and Sal2 strikingly decreased the level of E2F1 in contrast to stabilized E2F by Rb, the suppression of δ-catenin expression by these tumor suppressor proteins can be through different mechanisms. Interestingly, there are multiple putative p53 binding sites in the human δ-catenin promoter, raising the possibility that p53 may suppress δ-catenin transcription directly in a manner independent of E2F1.
One indication from the expression profiles of δ-catenin in 4 human prostate cell lines is that the regulation of δ-catenin expression in these cancer cells is controlled at the transcriptional level. Interestingly, the effects of E2F1 on δ-catenin expression are observed in human tumor cell lines expressing abundant levels of endogenous δ-catenin but not in tumor cell lines with undetectable levels of endogenous δ-catenin, suggesting the presence of negative δ-catenin transcription regulator(s) strong enough to suppress E2F1-induced δ-catenin expression. Alternatively, there may be additional factor(s), i.e. co-activator(s), other than E2F1 itself, which contribute to an E2F1 upregulation of δ-catenin transcription.
In prostate cancer, upregulation of E2F and δ-catenin have been reported [6; 22]. Different isoforms of E2F may exert different effects on δ-catenin expression and function. For example, Wang et al. reported that the effects of E2F1 and E2F4 on cell proliferation in vivo are similar, but their apoptotic and oncogenic properties are quite different [23]. Future experiments will be needed to clarify this issue. Interestingly, Davis et al. showed that E2F1 expression is low in benign and localized prostate cancer, modestly elevated in metastatic lymph nodes from hormone-naive patients, and significantly elevated in metastatic tissues from hormone-resistant prostate cancer patients [24]. As E2F1 can repress androgen receptor transcription, elevated E2F1 may contribute to the progression of hormone-refractory prostate cancer [24; 25]. Although δ-catenin immunoreactivity continues to increase in both intensity and extent until the prostate carcinoma reaches a Gleason score of 10 [6], we do not know the expression profiles of δ-catenin in metastatic tissues from hormone-resistant prostate cancer patients. It will be of great interests to determine in the future if the upregulation of E2F and δ-catenin and the downregulation of androgen receptor in human prostate cancer coincide and are functionally linked.
Acknowledgments
We thank Melissa Clark and Yan Zeng for technical assistance. The authors also wish to thank Werner Franke and Kwang Youl Lee for providing reagents. This study was supported in part by Korean Health 21 R&D Project #A040042, Ministry of Health & Welfare (K.K.) and by US National Institutes of Health CA111891 (Q.L.) and Department of Defense W81XWH-05-1-0028 (Q.L.).
References
- 1.Zhou J, Liyanage U, Medina M, Ho C, Simmons AD, Lovett M, Kosik KS. Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport. 1997;8:2085–90. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 2.Paffenholz R, Franke WW. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61:293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- 3.Kim K, Sirota A, Chen YH, Jones YhSB, Dudek R, Lanford GW, Thakore C, Lu Q. Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp Cell Res. 2002;275:171–84. doi: 10.1006/excr.2002.5503. [DOI] [PubMed] [Google Scholar]
- 4.Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14:1657–63. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 5.Lu Q, Paredes M, Medina M, Zhou J, Cavallo R, Peifer M, Orecchio L, Kosik KS. delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J Cell Biol. 1999;144:519–32. doi: 10.1083/jcb.144.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Q, Dobbs LJ, Gregory CW, Lanford GW, Revelo MP, Shappell S, Chen YH. Increased expression of delta-catenin/neural plakophilin-related armadillo protein is associated with the down-regulation and redistribution of E-cadherin and p120ctn in human prostate cancer. Hum Pathol. 2005;36:1037–48. doi: 10.1016/j.humpath.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Duparc RH, Boutemmine D, Champagne MP, Tetreault N, Bernier G. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300:647–55. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 8.Waterman ML. Lymphoid enhancer factor/T cell factor expression in colorectal cancer. Cancer Metastasis Rev. 2004;23:41–52. doi: 10.1023/a:1025858928620. [DOI] [PubMed] [Google Scholar]
- 9.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51. [PubMed] [Google Scholar]
- 10.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–23. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 12.Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002;1:466–76. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 13.Cao L, Faha B, Dembski M, Tsai LH, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–9. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 14.Shirodkar S, Ewen M, DeCaprio JA, Morgan J, Livingston DM, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–66. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 15.O'Connor RJ, Schaley JE, Feeney G, Hearing P. The p107 tumor suppressor induces stable E2F DNA binding to repress target promoters. Oncogene. 2001;20:1882–91. doi: 10.1038/sj.onc.1204278. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–52. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 17.Phillips SM, Morton DG, Lee SJ, Wallace DM, Neoptolemos JP. Loss of heterozygosity of the retinoblastoma and adenomatous polyposis susceptibility gene loci and in chromosomes 10p, 10q and 16q in human prostate cancer. Br J Urol. 1994;73:390–5. doi: 10.1111/j.1464-410x.1994.tb07602.x. [DOI] [PubMed] [Google Scholar]
- 18.Shan B, Durfee T, Lee WH. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc Natl Acad Sci U S A. 1996;93:679–84. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Ki H, Park HS, Kim K. Presenilin-1 D257A and D385A mutants fail to cleave Notch in their endoproteolyzed forms, but only presenilin-1 D385A mutant can restore its gamma-secretase activity with the compensatory overexpression of normal C-terminal fragment. J Biol Chem. 2005;280:22462–72. doi: 10.1074/jbc.M502769200. [DOI] [PubMed] [Google Scholar]
- 20.Li D, Tian Y, Ma Y, Benjamin T. p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP) Mol Cell Biol. 2004;24:3885–93. doi: 10.1128/MCB.24.9.3885-3893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina M, Marinescu RC, Overhauser J, Kosik KS. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63:157–64. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- 22.Waghray A, Schober M, Feroze F, Yao F, Virgin J, Chen YQ. Identification of differentially expressed genes by serial analysis of gene expression in human prostate cancer. Cancer Res. 2001;61:4283–6. [PubMed] [Google Scholar]
- 23.Wang D, Russell JL, Johnson DG. E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Mol Cell Biol. 2000;20:3417–24. doi: 10.1128/mcb.20.10.3417-3424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis JN, Wojno KJ, Daignault S, Hofer MD, Kuefer R, Rubin MA, Day ML. Elevated E2F1 inhibits transcription of the androgen receptor in metastatic hormone-resistant prostate cancer. Cancer Res. 2006;66:11897–906. doi: 10.1158/0008-5472.CAN-06-2497. [DOI] [PubMed] [Google Scholar]
- 25.Kaseb AO, Chinnakannu K, Chen D, Sivanandam A, Tejwani S, Menon M, Dou QP, Reddy GP. Androgen receptor and E2F-1 targeted thymoquinone therapy for hormone-refractory prostate cancer. Cancer Res. 2007;67:7782–8. doi: 10.1158/0008-5472.CAN-07-1483. [DOI] [PubMed] [Google Scholar]