Abstract
Sphingosine kinase-1 (SK1) is a key enzyme catalyzing the phosphorylation of sphingosine to sphingosine-1–phosphate (S1P). Recent studies suggest that SK1, and its product S1P, regulate diverse biological functions, including cell growth, differentiation, proliferation, and apoptosis. S1P may also play an important role in cardiac development and ischemic preconditioning, but the mechanism underlying these effects is not known. Using a yeast 2-hybrid screen with SK1 as bait and a cardiac cDNA library to identify novel proteins involved in regulating SK1 activity in cardiomyocytes, we identified the LIM-only factor FHL2 (SLIM3) as a SK1-interacting protein in both yeast and mammalian cells. FHL2, but not FHL1 or FHL3, interacted with SK1, and FHL2 colocalized with SK1 in the cytoplasm. The interaction sites with SK1 consisted of at least 4 LIM domains in FHL2, whereas the C-terminal portion of SK1 mediates the binding of FHL2 in SK1. Overexpression of FHL2 attenuated the activity and antiapoptotic effects of SK1. Indeed, endothelin-1, which is a potent survival factor in cardiomyocytes, inhibited FHL2-SK1 association and increased SK1 activity. These findings indicate that FHL2 is a novel inhibitor of SK1 activity in cardiomyocytes and suggest that targeting FHL2 for inhibition may prevent myocardial apoptosis through activation of SK1.
Keywords: sphingosine kinase, FHL2, cardiac myocytes, apoptosis
Sphingosine kinase (SK) is a key enzyme catalyzing the phosphorylation of sphingosine to sphingosine-1–phosphate (SPP or S1P).1 Recent studies have shown that S1P is a potent lipid messenger that plays important and diverse roles in biological processes such as cell growth, proliferation, calcium homeostasis, and survival, by directly acting on G protein–coupled S1P receptors and/or functioning as an intracellular second messenger.2 To date, 5 receptors, EDG-1/S1P1, EDG-5/S1P2, EDG-3/S1P3, EDG-6/S1P4, and EDG-8/S1P5 have been characterized to bind to both S1P and sphinganine-1–phosphate (dihydro-S1P) with high specificity.3 These receptors, via coupling to different G proteins, exert a wide array of cellular responses including vascular maturation and angiogenesis,4-6 migration,7-9 and cardiac development.7 Because S1P mediates many important biological effects, understanding how SK is activated may provide important insights into how cellular functions are regulated under pathophysiological conditions. Two different isoforms of SK have been identified, SK1 and SK2.10,11 Overexpression of SK1 promotes cell survival and protects cells from apoptotic insults, such as serum withdrawal.12,13 Indeed, various growth factors, cytokines, antigens, and G protein–coupled receptor agonists have been shown to activate SK1 and increase the intracellular levels of S1P.14-16 In contrast, overexpression of SK2 suppresses cell growth and enhances apoptosis in cultured cells.17,18 Thus, SK may function as an internal sensor that can decide the fate of cells to either undergo apoptosis or survival.
Regulation of SK1 appears to occur at both the transcriptional and posttranscriptional level.1 At the posttranscriptional level, SK1 has been shown to be regulated by phosphorylation and translocation.19-21 In addition, protein–protein interactions have also been shown to play critical roles in the regulation of SK activity. So far, at least 5 proteins, including SK-interacting protein (SKIP), platelet endothelial cell adhesion molecule (PECAM)-1, aminoacylase 1, δ-catenin/NPRAP, and tumor necrosis factor (TNF) receptor–associated factor 2 (TRAF2) have been identified to interact with SK1 and regulate its activity.1 However, the mechanisms underlying the regulation of SK1 activity by its associated proteins remain largely obscure.
Although both SK1 and SK2 have been identified in adult mouse heart, SK1 is the predominant isoform.22 Protein kinase C (PKC)-dependent activation of SK has been shown to play a critical role in mediating myocardial ischemic preconditioning.10,23 Furthermore, exogenously administered SK product S1P has been shown to protect the heart against ischemia/reperfusion injury, an effect that is independent of PKCε.23,24 To further define the roles of SK1 in cardiac myocytes, we performed a yeast 2-hybrid screening to identify SK1-interacting proteins by using a human adult heart cDNA library. We identify the four-and-a-half LIM domain 2 (FHL2 or SLIM3) as a SK1-interacting protein that negatively regulates its SK1 activity in cardiac myocytes.
Materials and Methods
Yeast Two-Hybrid Library Screening and Interaction Assays
To identify novel proteins that interact with SK1, we screened a human heart cDNA library using human SK1 as bait and the MATCHMAKER GAL4 yeast 2-hybrid system 3 (Clontech Laboratories Inc). Positive colonies were subject to multiple rounds of additional selection in the appropriate media and β-galactosidase filter assays to verify specificity. Mammalian 2-hybrid assay was performed with NIH3T3 cells in which a Gal4-dependent luciferase reporter construct was cotransfected with expression vectors encoding the Gal4DBD-SK1, the transcriptional activating protein VP16 fused to FHL1, FHL2, and FHL3. Approximately 36 hours after transfection, luciferase activity was measured and expressed as relative light units (RLU) per microgram of protein.
Primary Culture of Neonatal Rat Ventricular Myocytes
We obtained ventricles from 1-day-old Sprague–Dawley rats and isolated cardiac myocytes by digestion with trypsin-EDTA and type 2 collagenase as previously described.24 Neonatal rats were obtained from Charles River Laboratories North Wilmington, Mass. This study was reviewed and approved by the Institutional Animal Care and Use Committe at New Jersey Medical School.
Coimmunoprecipitation of SK1 and FHL2 in HEK293 Cells and Cardiomyocytes
HEK293 cells were transiently transfected with SK1 and FHL2 cDNAs using FuGENE 6 as described by the manufacturer (Roche Applied Science). Hearts from 24-week-old C57BL/6 male mice were homogenized in the buffer containing 1% Nonidet P-40, 150 mmol/L NaCl, 50 mmol/L Tris (pH 8), 100 μmol/L EDTA, and protease inhibitors. Coimmunoprecipitation of SK1 and FHL2 was performed essentially as described.25 The following antibodies were used for detection and immunoprecipitation: rabbit polyclonal c-Myc (Invitrogen), mouse monoclonal FLAG M2 (Sigma), mouse monoclonal FHL2 (MBL), and rabbit polyclonal SK1 (Abgent). Secondary antibodies were peroxidase-conjugated donkey anti-rabbit or anti-mouse (Jackson ImmunoResearch). Detection of the peroxidase was performed with ECL reagents.
Immunofluorescence Staining
HEK293 cells plated on 18-mm microcover glasses (Matsunami) were transfected with 2 μg of each FLAG-tagged SK1 and Myc-tagged FHL2 plasmids using FuGENE 6. For immunostaining, fixed HEK293 cells or freshly isolated neonatal rat cardiomyocytes were sequentially incubated with primary antibodies and appropriate fluorescent-labeled secondary antibodies. Images were visualized using an Olympus IX70 epifluorescence microscope.
SK Activity Assay
SK activity assay was performed essentially according to a previously described procedure.14,26 Briefly, cells were washed with ice-cold PBS and homogenized in ice-cold 0.1 mol/L phosphate buffer (pH 7.4) containing 20% glycerol, 1 mmol/L dithiothreitol, 1 mmol/L EDTA, 20 μmol/L ZnCl2, 1 mmol/L Na3VO4, 15 mmol/L NaF, 10 μg/mL leupeptin, 10 μg/mL aprotinin, 1 mmol/L phenylmethylsulfonyl fluoride, and 0.5 mmol/L 4-deoxypyridoxine. After ultracentrifugation at 100 000g for 30 minutes, lysates were assayed for SK1 activity in the reaction buffer favored SK1 activity (50 mmol/L HEPES [pH 7.4], 0.5% Triton X-100, 15 mmol/L MgCl2, 10% glycerol, 10 mmol/L NaF, and 1.5 mmol/L semicarbazide), based on the SK-catalyzed transfer of the γ-phosphate group of ATP (using a mixture of cold ATP and [γ32P]ATP [1 μCi/ sample]) to a specific substrate, and the products were separated by thin layer chromatography on Silica Gel G60 (Whatman) using chloroform/methanol/acetic acid/water (90:90:15:6). The radioactive spots corresponding to sphingosine phosphate were scraped and counted in a scintillation counter. In the SK1 reaction buffer used which contained 0.5% Triton X-100, SK2 activity has been shown to be inhibited by ≈95%.26
Gene Silencing with Small Interfering RNA
FHL2 small interfering RNA (siRNA) was designed based on sequence specific for rat FHL2 cDNA (5′-GCAAGGACTTGTCCTACAA-3′). Antisense and sense siRNA oligonucleotides with dTdT 3′ overhang were synthesized and transfected to the cells with Trans-Messenger Reagent according to the instructions of the manufacturer (Qiagen). The control cells were transfected with a control siRNA duplex (sense, UUCUCCGAACGUGUCACGUdTdT; antisense, ACGUGACACGUUCGGAGAAdTdT), this control siRNA has no known target in mammalian genomes. All siRNAs were purchased from Qiagen. For the knock-in studies, the FHL2 gene was mutated (152GCAAGGACTTGTCCTACAA170 to 152GCAAAGATCTGTCTTATAA170) to prevent destruction of exogenous mRNA by the corresponding siRNA, leaving the amino acid sequence unchanged. Mutations were made using the Quick Change Site-Directed Mutagenesis kit (Stratagene) according to the protocol of the manufacturer. Adenovirus-harboring siRNA resistant FHL2 mutant or DN-SK1(G82D) was made using AdMax (Microbix).
Assays for Apoptosis
Histone-associated DNA fragments were quantitated by the Cell Death Detection ELISA (Roche).27
Statistical Analyses
Data are given as mean±SEM. Statistical analyses were performed using ANOVA and post hoc tests by the Tukey method. Significant differences were taken at P<0.05.
Results
Interaction of FHL2 With SK1
Although SK1 has already been shown to interact with several proteins,1 our goal was to identify novel proteins that interact with SK1 in the heart. Accordingly, a yeast 2-hybrid screening was performed with human SK1 as bait in conjunction with a human heart cDNA library. After screening 2.4×106 clones, a total of 15 positive clones were identified, 3 of which encoded a full-length cDNA of the cardiac-restricted LIM-only factor, FHL2. To confirm the specificity of interaction between SK1 and FHL2, constructs containing the Gal4-activating domain fused with full-length FHL2 (pGAD-FHL2) and either the Gal4-binding domain fused with SK1 (pGBD-SPHK1) or the Gal4-binding domain alone (pGBD) were coexpressed in yeast AH109. Coexpression of pGAD-FHL2 and pGBD-SK1 resulted in a 5.1-fold increase in β-galactosidase activity compared with coexpression of pGBD and pGAD-FHL2 (Figure 1A).
Figure 1.
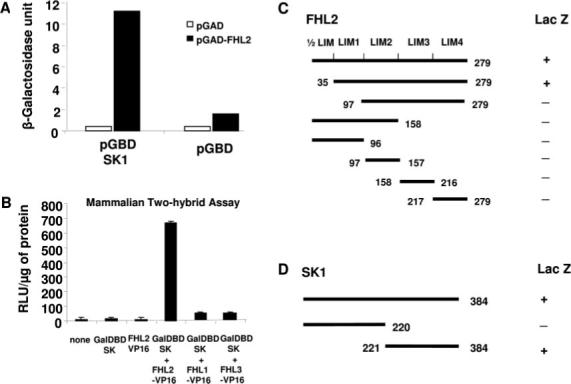
Two-hybrid screening identifies FHL2 as a SK1-interacting protein. A, Yeast 2-hybrid assay with strain AH109, which contains an interaction-dependent β-galactosidase reporter. The bait consisted of the Gal4 DBD fused to SK1, and the identified prey contained the Gal4 activation domain (AD) fused to FHL2. B, Mammalian 2-hybrid assay performed with NIH3T3 cells in which a Gal4-dependent luciferase reporter construct was cotransfected with expression vectors encoding the Gal4DBD-SK1, the transcriptional activating protein VP16 fused to FHL2, FHL1, and FHL3 (n=4). Luciferase activity was measured as relative light units (RLU) per microgram of protein. C, Analysis of the SK1-binding domains in FHL2. D, Analysis of the FHL2-binding domain in SK1. The relative strength of the interactions of the proteins was measured by a quantitative β-galactosidase assay (ONPG assay).
To determine whether this interaction also occurs in mammalian cells, FHL2 and SK1 cDNAs were cloned into mammalian expression vectors to permit a 2-hybrid analysis in NIH3T3 cells using the Gal4-dependent luciferase reporter construct. Transfection of either the Gal4 DBD-SK1 fusion vector or the VP16 activation domain-FHL2 fusion vector alone did not activate Gal4-dependent luciferase reporter (Figure 1B). However, cotransfection of the vectors encoding Gal4 DBD-SK1 with FHL2-VP16 resulted in a substantial increase in Gal4-dependent transcriptional activity, indicating interaction of SK1 and FHL2 in mammalian cells. In addition to FHL2, we also investigated whether FHL1 or FHL3 interacted with SK1. Constructs encoding either the FHL1-VP16 fusion or FHL3-VP16 fusion were used with SK1. In contrast to FHL2, neither FHL1 nor FHL3 interacted with SK1, suggesting selective interaction of SK1 with FHL2 (Figure 1B).
We further characterized the interaction between SK1 and FHL2 to map the essential binding domains of SK1 and FHL2. The relative strength of the interactions of the proteins in yeast was determined by a quantitative β-galactosidase assay using O-nitrophenyl β-d-galactopyranoside. We found that full-length FHL2 and constructs containing LIM1–4 retained β-galactosidase activities (Figure 1C). Constructs with 3 or fewer LIM domains had no activity, suggesting that the last 4 LIM domain of FHL2 is involved in SK1 binding to FHL2. In addition, the C-terminal portion of SK1 mediated the FHL2 binding on SK1 (Figure 1D).
Colocalization of FHL2 with SK1
To confirm that FHL2 interacts with SK1, coimmunoprecipitation experiments were performed in transfected HEK293 cells. Immunoprecipitation of FLAG-tagged SK1 led to coimmunoprecipitation of Myc-tagged FHL2 when both proteins were cotransfected (Figure 2A). As a control, the anti-FLAG antibody did not immunoprecipitate Myc-tagged FHL2 in the absence of FLAG-SK1. Similarly, immunoprecipitation of Myc-tagged FHL2 resulted in coimmunoprecipitation of FLAG-tagged SK1, whereas the anti-Myc antibody did not immunoprecipitate FLAG-SK1 in the absence of Myc-FHL2. Neither FHL1 nor FHL3 interacted with SK1 in the immunoprecipitation experiments (Figure 2B). SK2, which shares approximately 80% similarity with SK1 in the C-terminal portion,11 also interacted with FHL2 (Figure 2C). Together, these findings indicate that FHL2 and SK1 exist in the same complex.
Figure 2.
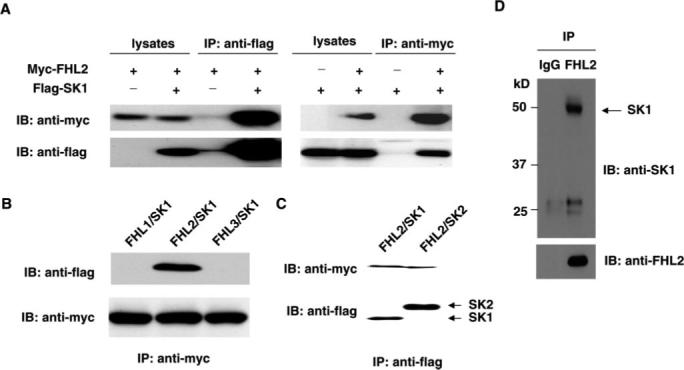
Physical interaction between SK1 and FHL2 by immunoprecipitation analysis. A, Expression vector in combination of either empty vector, pMT2-Myc-FHL2, or pFLAG-SK1 were cotransfected into HEK293 cells. Extracted proteins were precipitated by either anti-FLAG antibody (left) or anti-Myc antibody (right) and then separated by 10% SDS-PAGE. The transferred membrane was immunoblotted with either horseradish peroxidase (HRP)-conjugated anti-Myc or HRP-conjugated anti-Flag antibody. B, Expression vector in combination of either pMT2-FHL1, pMT2-Myc-FHL2, pMT2-FHL3, or pFlag-SK1 were cotransfected into HEK283 cells. Extracted proteins were precipitated by either anti-Myc antibody and then separated by 10% SDS-PAGE. The transferred membrane was immuno-blotted with either HRP-conjugated anti-Myc or HRP-conjugated anti-FLAG antibody. C, Expression vector in combination of either pFlag-SK1 or pFlag-SK2 with pMT2-Myc-FHL2 were cotransfected into HEK293 cells. Extracted proteins were precipitated by either anti-Flag antibody and then separated by 12% SDS-PAGE. The transferred membrane was immunoblotted with either HRP conjugated anti-myc or HRP-conjugated anti-FLAG antibody. D, Cell lysates obtained from adult mouse heart were immunoprecipitated with anti-FHL2 antibody and then separated by 12% SDS-PAGE. Transferred membrane was immunoblotted with either anti-SK1 or FHL2 antibody.
To determine whether there is interaction of endogenous FHL2 and SK1 in cardiomyocytes, we performed immunoprecipitation with anti-FHL2 antibody using lysates obtained from murine heart. SK1 coprecipitated with the anti-FHL2 antibody but not with the nonimmune control (Figure 2D). These findings indicate that endogenous levels of FHL2 can interact with SK1 in cardiomyocytes. To determine the intracellular localization of this interaction, we performed immunofluorescence staining in both HEK293 cells cotransfected with FLAG-SK1 and Myc-FHL2 cDNAs and neonatal rat cardiac myocytes. Immunofluorescent microscopy showed that FHL2 and SK1 are colocalized in the cytoplasm (Figure 3).
Figure 3.
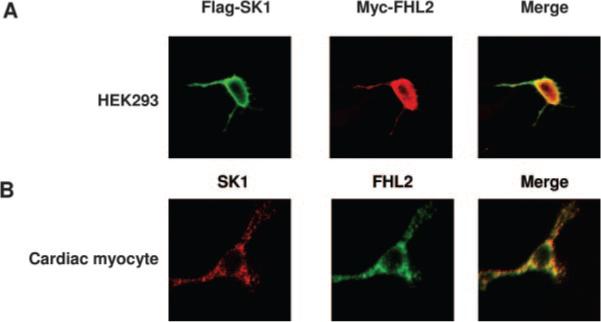
Subcellular localization of SK1 and FHL2 in mammalian cells. A, HEK293 cells were transiently transfected with FLAG-SK1 and Myc-FHL2, stained with anti-FLAG monoclonal antibody and rabbit polyclonal anti-Myc antibody, and processed for confocal imaging. B, Fixed neonatal rat cardiac myocytes were stained with anti-FHL2 monoclonal antibody and rabbit polyclonal anti-SK1 antibody and processed for confocal imaging. The merged image shows clear colocalization of these 2 proteins in cytoplasm.
Interaction Domains of FHL2 and SK1
To further confirm the interaction domains of FHL2 and SK1 obtained in yeast 2-hybrid assays, we performed immunoprecipitation in HEK293 cells cotransfected with different deletion mutants of FHL2 and SK1. We first investigated the binding domains of SK1 in FHL2. We generated a series of FHL2 deletion mutants subcloned into pCS26MT vector with 6×Myc tag and transfected these mutants into HEK293 cells along with FLAG-SK1 (Figure 3). Lysates from transfected HEK293 cells were immunoprecipitated with anti-Myc antibody and analyzed by Western blot analysis using anti-FLAG and anti-Myc antibodies. We found that FLAG-SK1 only bound to FHL2 mutant bearing 4 LIM domains but not to FHL2 mutants with 3 or fewer LIM domains (Figure 4B). As a negative control, the anti-Myc antibody did not immunoprecipitate FLAG-SK1 in cells cotransfected with FLAG-SK1 and empty vector. These results further indicate that the C-terminal 4 LIM domains of FHL2 are necessary for interaction with SK1.
Figure 4.
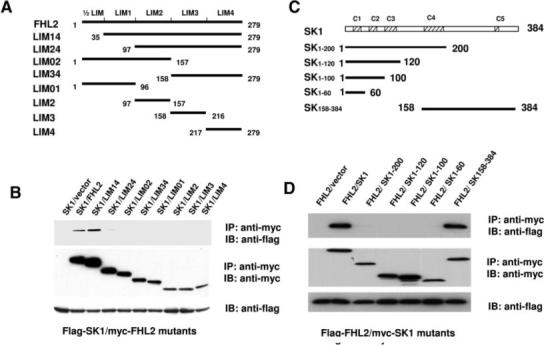
Identification of SK1 and FHL2 interaction sites. A, Schematic representation of FHL2 deletion mutants. B, FLAG-SK1 expression vector in combination of either empty vector or expression vectors of Myc-FHL2 mutants were cotransfected into HEK293 cells. Extracted proteins were precipitated by anti-Myc antibody and then separated by 15% SDS-PAGE. The transferred membrane was immunoblotted with either horseradish peroxidase (HRP)-conjugated anti-FLAG or HRP-conjugated anti-Myc antibody. Lysates were also immunoblotted with anti-FLAG antibody to show the expression levels of FLAG-SK1in HEK293 cells. C, Schematic representation of SK1 deletion mutants. D, FLAG-FHL2 expression vector in combination of either empty vector or expression vectors of Myc-SK1 mutants were cotransfected into HEK293 cells. Extracted proteins were precipitated by anti-Myc antibody and then separated by 15% SDS-PAGE. The transferred membrane was immunoblotted with either HRP-conjugated anti-FLAG or HRP-conjugated anti-Myc antibody. Lysates were also immunoblotted with anti-FLAG antibody to show the expression levels of FLAG-FHL2 in HEK293 cells.
Alignment of the conserved subdomains of SKs from various species revealed that SK1 contains 5 highly conserved domains, namely C1 through C5.10 To map the FHL2-binding domain in SK1, Myc-tagged SK1 mutants were cotransfected into HEK293 cells with Flag-tagged FHL2. Lysates from transfected HEK293 cells were immunoprecipitated with anti-Myc antibody and analyzed by Western blot analysis using anti-FLAG and anti-Myc-antibodies. As shown in Figure 3D, FHL2 interacted only with SK1 C-terminal portion consisting of C4 and C5 domains but not with other deletion mutants including SK1 to 200-containing C4 domain. Together, these results further suggest that SK1 C-terminal portion containing C5 domain mediates its interaction with FHL2.
Inhibition of SK1 Activity by FHL2
To determine whether the interaction of FHL2 with SK1 has functional consequences in terms of affecting SK1 activity, we cotransfected SK1 and FHL2 in HEK293 cells and determined SK1 expression and activity. Transfection of FHL2 did not affect SK1 expression (Figure 5A) but markedly decreased SK1 activity (Figure 5A). In contrast, the FHL2 mutant consisting of LIM2 to -4 that did not interact with SK1, had no effects on SK1 activity (Figure 5A). Because SK1 has been characterized to suppress cell apoptosis,12,13 we investigated whether FHL2 can affect apoptosis via inhibitory effects on SK1 in stably transfected NIH3T3 cells. Transfection of FHL2 attenuated the antiapoptotic effects of SK1 as determined by the extent of DNA fragmentation, whereas FHL2 mutant LIM24 had no effects (Figure 5B). These findings suggest that the interaction of FHL2 with SK1 may be functionally important in terms of regulating SK1-mediated cell survival.
Figure 5.
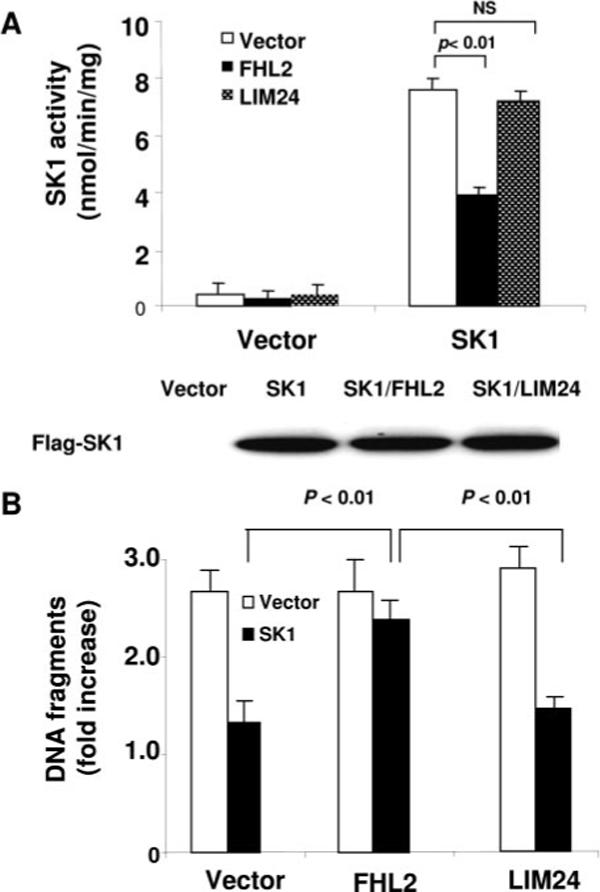
Effects of FHL2 on SK1 activity and function. A, HEK293 cells were cotransfected with FLAG-SK1 and vector, Myc-FHL2, or Myc-LIM24. After 48 hours, cells were lysed and SK1 activity was measured. The data are representative of 3 independent experiments. B, Antiapoptotic effects of SK1. NIH 3T3 cells stably transfected with vector (open bars) or SK1 (filled bars) were transiently transfected with either FHL2 or LIM24 or empty vector, then serum starved. After 24 hours, cell apoptosis was quantitated by the Cell Death Detection ELISA (Roche). The experimental data are normalized by those obtained in HEK293 cells cultured in 10% serum. The data are representative of 3 independent experiments.
Regulation of SK1 and FHL2 Interaction by Endothelin-1
Endothelin (ET)-1 has been shown to protect cardiomyocytes from undergoing apoptosis.28-30 Because SK1 and its product S1P are cardioprotective during ischemic injury,24,31 we investigated whether SK1 can mediate some of the cardioprotective effects of ET-1. We found that treatment of neonatal rat cardiomyocytes with ET-1 (100 nmol/L, 30 minutes) decreased FHL2 interaction with SK1 (Figure 6A) and increased SK1 activity by 85% (P<0.01) (Figure 6B). To determine whether the increase in SK1 activity by ET-1 could contribute to its antiapoptotic effects in cardiomyocytes, we investigated the effects of ET-1 on H2O2-induced cardiomyocyte apoptosis in the presence or absence of the SK1 competitive inhibitor N′,N′-dimethylsphingosine (DMS). Treatment of cardiomyocytes with ET-1 inhibited H2O2-induced apoptosis, which was blocked in the presence of DMS (5 μmol/L), whereas DMS alone slightly but not significantly increased cellular apoptosis (Figure 6C). Furthermore, adenovirus expressing dominant negative SK1(G82D) (Ad-DNSK1)32 also diminished ET-1–mediated antiapoptotic effects in cardiomyocytes (Figure 6D). Collectively, these findings indicate that some of the cardioprotective effects of ET-1 on myocyte apoptosis may be mediated by the upregulation of SK1 activity.
Figure 6.
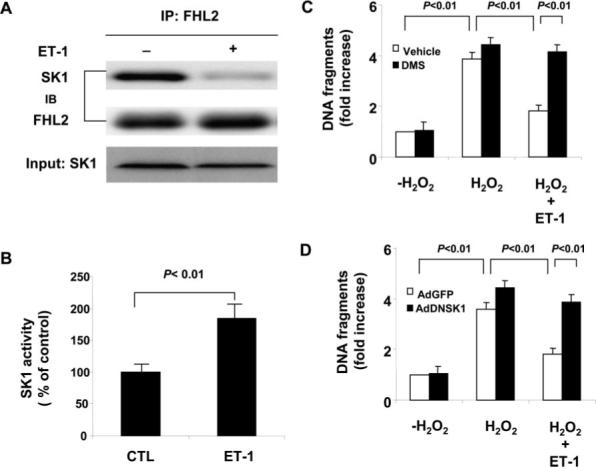
Role of SK1 in ET-1 induced antiapoptosis in cardiac myocytes. A, Effect of ET-1 treatment on the interaction of SK1 and FHL2. Myocytes cultured in serum-free condition were treated with either vehicle or ET-1 (100 nmol/L) for 30 minutes and cell lysates were then immunoprecipitated with anti-FHL2 antibody. Transferred membrane was immunoblotted with either anti-SK1 or FHL2 antibody. B, Effect of ET-1 on SK1 activity. Cardiac myocytes cultured in serum-free condition were treated with ET-1 (100 nmol/L) and SK1 activity measured. The data are representative of 3 independent experiments. C, Effect of ET-1 (100 nmol/L) on H2O2 (100 μmol/L) induced cardiac myocytes apoptosis. Myocytes were treated as indicated for 48 hours. Cell apoptosis was quantitated by the Cell Death Detection ELISA (Roche). The experimental data are normalized by those obtained in control myocytes without both H2O2 and DMS (5 μmol/L) treatments. The data are representative of 3 independent experiments. D, Effect of dominant negative SK1 (DNSK1) on ET-1(100 nmol/L) mediated antiapoptotic effects against H2O2 (100 μmol/L) treatment. Myocytes were transduced with either Ad-GFP (control) or Ad-DNSK1. Twenty-four hours after transduction, myocytes were treated as indicated for 48 hours. Cell apoptosis was quantitated by the Cell Death Detection ELISA (Roche). The experimental data are normalized by those obtained in control myocytes without H2O2 treatment. The data are representative of 3 independent experiments.
Knockdown of FHL2 Protects Cardiomyocytes From Apoptosis
To further investigate the role of FHL2 in the regulation of SK1 in cardiomyocytes, we used siRNA to knock down the expression of FHL2. Transfection of FHL2 siRNA reduced FHL2 expression but not SK1 expression in cardiomyocytes (Figure 7A). In addition, immunoprecipitation with anti-SK1 antibody demonstrated a reduced amount of FHL2 that is associated with SK1 (Figure 7A). Knockdown of FHL2 expression was found to markedly increase SK1 activity and protect cardiomyocytes from H2O2-induced apoptosis compared with the cells treated with control siRNA (Figure 6B). Similarly, the SK1 inhibitor, DMS (5 μmol/L), blocked the antiapoptotic effects of FHL2 knockdown (Figure 6B). To confirm that the observed RNA interference effect is gene specific, cardiomyocytes were transduced with adenovirus harboring siRNA resistant FHL2 mutant FHL2(re). Forty-eight hours after infection, FHL2 expression was analyzed by immunoblotting. The level of FHL2 in FHL2 siRNA-transfected cells was comparable with that of endogenous FHL2 in control siRNA-transfected cells (Figure 7C). Indeed, the ectopic expression of FHL2(re) reversed the antiapoptotic effects of FHL2 knockdown in cardiomyocytes (Figure 7D). Taken together, these results indicate that FHL2 functions as a negative regulator of SK1 in cardiomyocytes.
Figure 7.
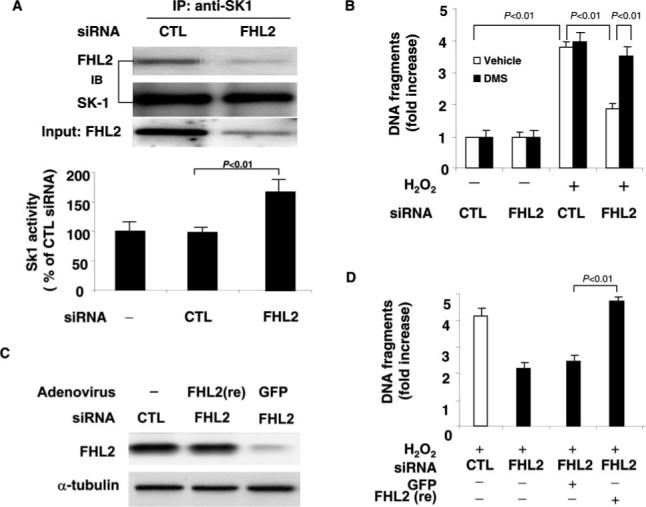
Knockdown of FHL2 by siRNA upregulates SK activity. A, Extracted proteins from control or FHL2 siRNA transfected myocytes were subjected to immunoprecipitation with anti-SK1 antibody. Transferred membrane was immunoblotted with either anti-SK1 or FHL2 antibody. Lysates were also immunoblotted with anti-FHL2 antibody to show the expression levels of FHL2 in control or FHL2 siRNA-transfected myocytes. SK1 activity was also measured in cell lysates obtained from control or FHL2 siRNA-transfected myocytes. The data are representative of 3 independent experiments. B, Myocytes were transfected with either control or FHL2 siRNA and treated with or without H2O2 (100 μmol) for 48 hours. Cell apoptosis was quantitated by the Cell Death Detection ELISA (Roche). The experimental data are normalized by those obtained in control myocytes without both H2O2 and DMS (5 μmol/L) treatment. The data are representative of 3 independent experiments. C, FHL2 siRNA-transfected myocytes were transduced with either Ad-GFP or adenovirus harboring siRNA-resistant FHL2, Ad-FHL2(re). Forty-eight hours after transduction, cell lysates were immunoblotted with anti-FHL2 monoclonal antibody. D, FHL2 siRNA-transfected myocytes were transduced with either AdGFP (control) or Ad-FHL2(re). Twenty-four hours after transduction, myocytes were treated with H2O2 (100 μmol) for 48 hours. Cell apoptosis was quantitated by the Cell Death Detection ELISA (Roche). The data are representative of 3 independent experiments.
Discussion
Using the yeast 2-hybrid system and SK1 as bait, we isolated several FHL2 cDNAs from a human heart cDNA library. The interaction of SK1 with FHL2 was further supported by coimmunoprecipitation studies showing that in both cotransfected HEK293 cells and cardiomyocytes, FHL2, but not FHL1 or FHL3, interacted with SK1. The interaction required the 4 LIM domains of FHL2 as well as C-terminal domain of SK1 and occurred with endogenous levels of FHL2 and SK1. Indeed, FHL2 and SK1 were found to colocalize in the cytoplasm. The functional consequence of this interaction was demonstrated by the ability of FHL2 to inhibit SK1 activity and SK1-mediated antiapoptotic effects. Indeed, factors such as ET-1, which promote cardiomyocyte survival, increases SK1 activity via inhibiting FHL2-SK1 interaction. These results suggest that the regulation of SK1 activity by FHL2 may be an important determinant of cell survival.
We chose to screen for SK1-interacting proteins in cardiomyocytes based on the established importance of SK1 in heart development7 and on the observations that SK1 and its product S1P have cardioprotective effects, particularly after ischemic injury.23,24,31 To identify the cellular proteins that interacted with SK1 in cardiomyocytes, we performed a yeast 2-hybrid screen using a human heart cDNA library. Three independent clones selected under high stringency conditions corresponded to a four-and-a-half LIM domain gene 2, FHL2. FHL2 expression was originally thought to be cardiac specific.33 However, recent studies suggest that FHL2 is also expressed in ovary, placenta, uterus, mammary gland, and adrenal gland.33 Furthermore, although FHL1 and FHL3 are also expressed in cardiac tissues, only FHL2 was able to interact with SK1.
The mechanism by which FHL2 inhibits SK1 is not known, but involves FHL2-SK1 interaction. As FHL2 is an adaptor protein without known enzymatic activity, it might regulate SK1 activity either by altering its conformation or by restricting its translocation from cytosol to the plasma membrane. Indeed, it has been suggested that the molecular mechanism of SK1 activation correlated with its phosphorylation at Ser225, which is necessary for translocation of the enzyme from the cytosol to the plasma membrane to gain access to its substrate sphingosine at the membrane.21 In this regard, our results suggest that FHL2 might confine the localization of SK1 to the cytoplasm and restrict the translocation of SK1 to the membrane. However, further studies are needed to elucidate the precise mechanisms by which FHL2 regulates SK1 activity.
ET-1 is a potent survival factor against apoptosis in various cell types.28-30 This antiapoptotic effect is mediated mainly through Gq protein–coupled ET-1 type A receptor.28 Activation of Gq increases intracellular calcium levels and subsequently activates the calcineurin pathway.29,34 In our study, we showed that ET-1 increases SK1 activity and that SK1 mediates the antiapoptotic effects of ET-1 in cardiomyocytes. Both FHL2 and DN-SK1, which inhibit SK1 activity, blocked the antiapoptotic effects of ET-1 in response to H2O2. In this regard, our studies provide a novel mechanism by which ET-1 exerts some of its cardioprotective effects through the regulation of FHL2-SK1 interaction. However, the cell signaling pathway mediating the regulation of FHL2-SK1 interaction still remains elusive. SK1 has been shown to be regulated by phosphorylation and translocation.19-21 Recently, the activation of PKC has been reported to enhance SK activity leading to elevated levels of S1P.20,23,35,36 For example, phosphorylation of SK1 by PKC was shown to activate SK in response to VEGF stimulation in endothelial cells.36 PKC-dependent activation of SK1 leads to SK1 translocation from cytosol to plasma membrane in HEK293 cells.20 In addition, phosphorylation of SK1 at Ser225 by extracellular signal-regulated kinase (ERK) 1/2 causes not only an increase in enzymatic activity but is also necessary for translocation of the enzyme from the cytosol to the plasma membrane.21 Interestingly, ET-1 has been shown to activate PKC and mitogen-activated protein kinases in cardiac myocytes.37 However, it remains to be determined whether these signaling proteins are involved in promoting FHL2-SK1 interaction in response to ET-1.
FHL2 gene–targeted mice have been recently generated and are viable,38,39 most likely because of redundancy with multiple other LIM-only family members that are also expressed in the heart, such as FHL1 and FHL3.40 However, following β-adrenergic stimulation, FHL2-null mice developed greater cardiac hypertrophy.39 Interestingly, SK product S1P has recently been shown to be a potent stimulator of cardiac hypertrophy in rat neonatal cardiomyocytes.41 It is likely, therefore, that FHL2 functions as a negative regulator of SK1 in the heart, as it would explain why FHL2-null mice, which would be expected to have higher SK1 activity, would have a greater tendency to develop cardiac hypertrophy. Furthermore, it would be interesting to investigate whether ischemia-induced heart injury is attenuated in FHL2-null mice as a means of more specifically assessing the functional significance of FHL2-SK1 interaction in vivo.
In summary, we have shown that SK1 interacts with FHL2 in cardiomyocytes. This interaction is relatively specific, because neither FHL1 nor FHL3 interacts with SK1; it is functional, as it leads to decrease SK1 activity; and it is physiological, as it mediates the antiapoptotic effects of ET-1 on cardiomyocytes. These findings suggest that FHL2 may be an important therapeutic target for promoting myocardial survival in ischemia–reperfusion injury and heart failure.
Sources of Funding
This work was supported by American Heart Association Scientist Development Grant 0630047N (to J.S.), the National Natural Science Foundation of China (grants 30472024 and 30371648 to J.S.), and the NIH (grants HL52233, HL70274, and DK62729 to J.K.L.).
Footnotes
Disclosures
None.
References
- 1.Taha TA, Hannun YA, Obeid LM. Sphingosine kinase: biochemical and cellular regulation and role in disease. J Biochem Mol Biol. 2006;39:113–131. doi: 10.5483/bmbrep.2006.39.2.113. [DOI] [PubMed] [Google Scholar]
- 2.Spiegel S, Milstien S. Sphingosine-1–phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 3.Hla T, Lee MJ, Ancellin N, Paik JH, Kluk MJ. Lysophospholipids–receptor revelations. Science. 2001;294:1875–1878. doi: 10.1126/science.1065323. [DOI] [PubMed] [Google Scholar]
- 4.Kono M, Mi Y, Liu Y, Sasaki T, Allende ML, Wu YP, Yamashita T, Proia RL. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J Biol Chem. 2004;279:29367–29373. doi: 10.1074/jbc.M403937200. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Wada R, Yamashita T, Mi Y, Deng CX, Hobson JP, Rosenfeldt HM, Nava VE, Chae SS, Lee MJ, Liu CH, Hla T, Spiegel S, Proia RL. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J Clin Invest. 2000;106:951–961. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.English D, Garcia JG, Brindley DN. Platelet-released phospholipids link haemostasis and angiogenesis. Cardiovasc Res. 2001;49:588–599. doi: 10.1016/s0008-6363(00)00230-3. [DOI] [PubMed] [Google Scholar]
- 7.Ishii I, Ye X, Friedman B, Kawamura S, Contos JJ, Kingsbury MA, Yang AH, Zhang G, Brown JH, Chun J. Marked perinatal lethality and cellular signaling deficits in mice null for the two sphingosine 1-phosphate (S1P) receptors, S1P(2)/LP(B2)/EDG-5 and S1P(3)/LP(B3)/EDG-3. J Biol Chem. 2002;277:25152–25159. doi: 10.1074/jbc.M200137200. [DOI] [PubMed] [Google Scholar]
- 8.Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, Thornton R, Shei GJ, Card D, Keohane C, Rosenbach M, Hale J, Lynch CL, Rupprecht K, Parsons W, Rosen H. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 9.Lee MJ, Thangada S, Paik JH, Sapkota GP, Ancellin N, Chae SS, Wu M, Morales-Ruiz M, Sessa WC, Alessi DR, Hla T. Akt-mediated phosphorylation of the G protein-coupled receptor EDG-1 is required for endothelial cell chemotaxis. Mol Cell. 2001;8:693–704. doi: 10.1016/s1097-2765(01)00324-0. [DOI] [PubMed] [Google Scholar]
- 10.Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, Milstien S, Kohama T, Spiegel S. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem. 2000;275:19513–19520. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 12.Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, Spiegel S. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol. 1999;147:545–558. doi: 10.1083/jcb.147.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivera A, Rosenfeldt HM, Bektas M, Wang F, Ishii I, Chun J, Milstien S, Spiegel S. Sphingosine kinase type 1 induces G12/13-mediated stress fiber formation, yet promotes growth and survival independent of G protein-coupled receptors. J Biol Chem. 2003;278:46452–46460. doi: 10.1074/jbc.M308749200. [DOI] [PubMed] [Google Scholar]
- 14.Olivera A, Spiegel S. Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature. 1993;365:557–560. doi: 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- 15.Xia P, Gamble JR, Rye KA, Wang L, Hii CS, Cockerill P, Khew-Goodall Y, Bert AG, Barter PJ, Vadas MA. Tumor necrosis factor-alpha induces adhesion molecule expression through the sphingosine kinase pathway. Proc Natl Acad Sci U S A. 1998;95:14196–14201. doi: 10.1073/pnas.95.24.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alemany R, Meyer zu Heringdorf D, van Koppen CJ, Jakobs KH. Formyl peptide receptor signaling in HL-60 cells through sphingosine kinase. J Biol Chem. 1999;274:3994–3999. doi: 10.1074/jbc.274.7.3994. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Toman RE, Goparaju SK, Maceyka M, Nava VE, Sankala H, Payne SG, Bektas M, Ishii I, Chun J, Milstien S, Spiegel S. Sphingosine kinase type 2 is a putative BH3-only protein that induces apoptosis. J Biol Chem. 2003;278:40330–40336. doi: 10.1074/jbc.M304455200. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi N, Okada T, Hayashi S, Fujita T, Jahangeer S, Nakamura S. Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J Biol Chem. 2003;278:46832–46839. doi: 10.1074/jbc.M306577200. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim FB, Pang SJ, Melendez AJ. Anaphylatoxin signaling in human neutrophils. A key role for sphingosine kinase. J Biol Chem. 2004;279:44802–44811. doi: 10.1074/jbc.M403977200. [DOI] [PubMed] [Google Scholar]
- 20.Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA). J Biol Chem. 2002;277:35257–35262. doi: 10.1074/jbc.M203033200. [DOI] [PubMed] [Google Scholar]
- 21.Pitson SM, Moretti PA, Zebol JR, Lynn HE, Xia P, Vadas MA, Wattenberg BW. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 2003;22:5491–5500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuda Y, Kihara A, Igarashi Y. Distribution of sphingosine kinase activity in mouse tissues: contribution of SPHK1. Biochem Biophys Res Commun. 2003;309:155–160. doi: 10.1016/s0006-291x(03)01551-1. [DOI] [PubMed] [Google Scholar]
- 23.Jin ZQ, Goetzl EJ, Karliner JS. Sphingosine kinase activation mediates ischemic preconditioning in murine heart. Circulation. 2004;110:1980–1989. doi: 10.1161/01.CIR.0000143632.06471.93. [DOI] [PubMed] [Google Scholar]
- 24.Karliner JS, Honbo N, Summers K, Gray MO, Goetzl EJ. The lysophospholipids sphingosine-1-phosphate and lysophosphatidic acid enhance survival during hypoxia in neonatal rat cardiac myocytes. J Mol Cell Cardiol. 2001;33:1713–1717. doi: 10.1006/jmcc.2001.1429. [DOI] [PubMed] [Google Scholar]
- 25.Sun J, Liao JK. Functional interaction of endothelial nitric oxide synthase with a voltage-dependent anion channel. Proc Natl Acad Sci U S A. 2002;99:13108–13113. doi: 10.1073/pnas.202260999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billich A, Bornancin F, Devay P, Mechtcheriakova D, Urtz N, Baumruker T. Phosphorylation of the immunomodulatory drug FTY720 by sphingosine kinases. J Biol Chem. 2003;278:47408–47415. doi: 10.1074/jbc.M307687200. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto S, Yang G, Zablocki D, Liu J, Hong C, Kim SJ, Soler S, Odashima M, Thaisz J, Yehia G, Molina CA, Yatani A, Vatner DE, Vatner SF, Sadoshima J. Activation of Mst1 causes dilated cardiomyopathy by stimulating apoptosis without compensatory ventricular myocyte hypertrophy. J Clin Invest. 2003;111:1463–1474. doi: 10.1172/JCI17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki M, Hasegawa K, Iwai-Kanai E, Fujita M, Sawamura T, Kakita T, Wada H, Morimoto T, Sasayama S. Endothelin-1 as a protective factor against beta-adrenergic agonist-induced apoptosis in cardiac myocytes. J Am Coll Cardiol. 2000;36:1411–1418. doi: 10.1016/s0735-1097(00)00822-6. [DOI] [PubMed] [Google Scholar]
- 29.Kakita T, Hasegawa K, Iwai-Kanai E, Adachi S, Morimoto T, Wada H, Kawamura T, Yanazume T, Sasayama S. Calcineurin pathway is required for endothelin-1-mediated protection against oxidant stress-induced apoptosis in cardiac myocytes. Circ Res. 2001;88:1239–1246. doi: 10.1161/hh1201.091794. [DOI] [PubMed] [Google Scholar]
- 30.Ogata Y, Takahashi M, Ueno S, Takeuchi K, Okada T, Mano H, Ookawara S, Ozawa K, Berk BC, Ikeda U, Shimada K, Kobayashi E. Antiapoptotic effect of endothelin-1 in rat cardiomyocytes in vitro. Hypertension. 2003;41:1156–1163. doi: 10.1161/01.HYP.0000064342.30653.24. [DOI] [PubMed] [Google Scholar]
- 31.Jin ZQ, Zhou HZ, Zhu P, Honbo N, Mochly-Rosen D, Messing RO, Goetzl EJ, Karliner JS, Gray MO. Cardioprotection mediated by sphingosine-1-phosphate and ganglioside GM-1 in wild-type and PKC epsilon knockout mouse hearts. Am J Physiol Heart Circ Physiol. 2002;282:H1970–H1977. doi: 10.1152/ajpheart.01029.2001. [DOI] [PubMed] [Google Scholar]
- 32.Pitson SM, Moretti PA, Zebol JR, Xia P, Gamble JR, Vadas MA, D'Andrea RJ, Wattenberg BW. Expression of a catalytically inactive sphingosine kinase mutant blocks agonist-induced sphingosine kinase activation. A dominant-negative sphingosine kinase. J Biol Chem. 2000;275:33945–33950. doi: 10.1074/jbc.M006176200. [DOI] [PubMed] [Google Scholar]
- 33.Johannessen M, Moller S, Hansen T, Moens U, Van Ghelue M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci. 2006;63:268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pu WT, Ma Q, Izumo S. NFAT transcription factors are critical survival factors that inhibit cardiomyocyte apoptosis during phenylephrine stimulation in vitro. Circ Res. 2003;92:725–731. doi: 10.1161/01.RES.0000069211.82346.46. [DOI] [PubMed] [Google Scholar]
- 35.Buehrer BM, Bardes ES, Bell RM. Protein kinase C-dependent regulation of human erythroleukemia (HEL) cell sphingosine kinase activity. Biochim Biophys Acta. 1996;1303:233–242. doi: 10.1016/0005-2760(96)00092-6. [DOI] [PubMed] [Google Scholar]
- 36.Shu X, Wu W, Mosteller RD, Broek D. Sphingosine kinase mediates vascular endothelial growth factor-induced activation of ras and mitogen-activated protein kinases. Mol Cell Biol. 2002;22:7758–7768. doi: 10.1128/MCB.22.22.7758-7768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogoyevitch MA, Glennon PE, Andersson MB, Clerk A, Lazou A, Marshall CJ, Parker PJ, Sugden PH. Endothelin-1 and fibroblast growth factors stimulate the mitogen-activated protein kinase signaling cascade in cardiac myocytes. The potential role of the cascade in the integration of two signaling pathways leading to myocyte hypertrophy. J Biol Chem. 1994;269:1110–1119. [PubMed] [Google Scholar]
- 38.Chu PH, Bardwell WM, Gu Y, Ross J, Jr, Chen J. FHL2 (SLIM3) is not essential for cardiac development and function. Mol Cell Biol. 2000;20:7460–7462. doi: 10.1128/mcb.20.20.7460-7462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong Y, Shelton JM, Rothermel B, Li X, Richardson JA, Bassel-Duby R, Williams RS. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to beta-adrenergic stimulation. Circulation. 2001;103:2731–2738. doi: 10.1161/01.cir.103.22.2731. [DOI] [PubMed] [Google Scholar]
- 40.Chu PH, Ruiz-Lozano P, Zhou Q, Cai C, Chen J. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech Dev. 2000;95:259–265. doi: 10.1016/s0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- 41.Robert P, Tsui P, Laville MP, Livi GP, Sarau HM, Bril A, Berrebi-Bertrand I. EDG1 receptor stimulation leads to cardiac hypertrophy in rat neonatal myocytes. J Mol Cell Cardiol. 2001;33:1589–1606. doi: 10.1006/jmcc.2001.1433. [DOI] [PubMed] [Google Scholar]


