Abstract
Protein thiols with regulatory functions play a critical role in maintaining the homeostasis of the redox state in mitochondria. One major host of regulatory cysteines in mitochondria is complex I, with the thiols primarily located on its 51 kDa FMN-binding subunit. In response to oxidative stress, these thiols are expected to form intra-molecular disulfide bridges as one of their oxidative post-translational modifications. Here, to test this hypothesis and gain insights into the molecular pattern of disulfide in complex I, the isolated bovine complex I was prepared. Superoxide (O2•−) is generated by complex I under the conditions of enzyme turnover. O2•−-induced intra-molecular disulfide formation at the 51 kDa subunit was determined by tandem mass spectrometry and database searching, with the latter accomplished by adaptation of the in-house developed database search engine, MassMatrix [Xu H., et. al J. Proteome Res. (2008) 7, 138–44]. LC/MS/MS analysis of tryptic/chymotryptic digests of the 51 kDa subunit from alkylated complex I revealed that four specific cysteines (C125, C142, C187, and C206) of the 51 kDa subunit were involved in the formation of mixed intra-molecular disulfide linkages. In all, three cysteine pairs were observed: C125/C142, C187/C206, and C142/C206. The formation of disulfide bond was subsequently inhibited by superoxide dismutase, indicating the involvement of O2•−. These results elucidated by mass spectrometry indicates that the residues of C125, C142, C187, and C206 are the specific regulatory cysteines of complex I, and they participate in the oxidative modification with disulfide formation under the physiological or pathophysiological conditions of oxidative stress.
INTRODUCTION
Mitochondrial complex I (EC 1.6.5.3. NADH:ubiquinone oxidoreductase, NQR) is the first energy-conserving segment of the electron transport chain (ETC) [1–3]. Purified bovine heart complex I contains up to 45 different subunits with a total molecular mass approaching 1 MDa [4]. With the use of chaotropic anions such as perchlorate, complex I can be resolved into three fractions: a flavoprotein fraction (Fp), an iron sulfur protein fraction (Ip), and a hydrophobic fraction (Hp) [see Supplemental Figure 1]. The enzyme catalyzes electron transfer from NADH to ubiquinone coupled with the translocation of four protons across the membrane. In addition to its functions of electron transfer and energy transduction, the catalysis of complex I provides the major source of oxygen free radical generation in mitochondria [5–7].
The generation of superoxide anion radical (O2•−) and the oxidants derived from it in mitochondria can act as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence. The redox pool in mitochondria is enriched in glutathione (GSH) with a high physiological concentration (in the mM range) [8]. Protein thiols are also abundant in the proteins of the mitochondrial ETC, which play a critical role in maintaining the homeostasis of the cellular redox state [9, 10]. It has been documented that complex I is the major component of the ETC to host protein thiols, which comprise structural thiols involved in the ligands of iron sulfur clusters and the reactive/regulatory thiols which are thought to have biological functions of antioxidant defense and redox signaling [11, 12]. The physiological roles of complex I-derived regulatory thiols have been implicated in the regulation of the respiration, nitric oxide utilization [13, 14], and redox status of mitochondria [8–10].
The 51-kDa subunit of bovine complex I is nuclear-encoded and constitutes the major component of the flavoprotein [Fp contains the enzymatic activity of NADH dehydrogenase (NDH) and can be isolated as a three-subunit subcomplex indicated in the Supplemental Figure 1] of complex I. This subunit contains the NADH-binding site, the primary electron acceptor FMN, and the 4F-4S cluster (N3 center). The 51-kDa subunit is also one of the major components to host the reactive/regulatory thiols of complex I [11, 12, 15, 16]. The bovine protein has 12 cysteine residues, but only 5 of them are conserved in bacterial enzyme. Four of the conserved cysteines form a typical 4Fe-4S cluster motif, C379-X2-C382-X2-C385-X39-C425 (X represents amino acid residue), near the C-terminal domain. The other conserved cysteine is located at position 206 [1, 3].
It has been demonstrated that the biological relevance of C206 of the 51 kDa subunit in the oxidative damage of complex I is to play the unique role of reactive thiol, based on the evidence of immunospin trapping with 5,5-dimethyl pyrroline N-oxide (DMPO) and mass spectrometry [15]. With a proteomic approach, C206 of the 51 kDa subunit was further determined to be involved in the redox modification of protein S-glutathiolation [16]. An X-ray crystal structure of the hydrophilic domain of respiratory complex I from bacterial enzyme of Thermus Thermophilius indicates that this conserved cysteine (Cys182 in Thermus Thermophilius) is only 6Å from the FMN, which is consistent with the role of C206 as a redox-sensitive thiol and FMN’s serving as a source of O2•− [17].
The residues of C125, C142, C187, C238, C255, C286, and C332 are the seven remaining cysteinyl residues; these are not conserved in the bacterial enzyme, but they are conserved in mammalian enzymes. Protein disulfide formation is the other important oxidative post-translational modification for the regulatory thiols of 51 kDa subunit of complex I, and could be induced by oxidative stress. To date, this issue has never been clarified, and its molecular pattern remains unclear.
Due to the nature of complexity in the structure of mitochondrial complex I, elucidation of the disulfide profile in the complex I is a challenge. Recently, mass spectrometric approach has become popular for studying the structure and post-translational modification of mammalian complex I [2, 4, 18–20]. With the development of MassMatrix program and its applications [21–23], it is feasible to solve its molecular pattern of disulfide linkage through mass spectrometric analysis. This study was undertaken to address the fundamental questions regarding the molecular pattern of disulfide bond formation in the FMN-binding subunit from the complex I under the conditions of oxidative stress. Bovine complex I was prepared under reducing conditions in the presence of dithiothretol (DTT) and subsequently subjected to O2•− generation under the conditions of enzyme turnover in the presence of NADH and Q1. Complex I was thus exposed to the physiologically relevant conditions of oxidative stress. The approaches of proteomics, mass spectrometry, and data base search with MassMtrix was then employed to identify the specific cysteine residues involved in the formation of the disulfide linkage from the 51 kDa subunit of complex I. The biological implication of this study is also discussed.
EXPERIMENTAL METHODS
Preparations of Mitochondrial Complex I
Bovine heart mitochondrial complex I was prepared according to the published method with modifications as follows [24]. Submitochondrial particles (SMP) were prepared as described and used as the starting material [25], starting with 2.5 lb of trimmed bovine hearts with the fat and connective tissues removed. The SMP preparation was suspended in 50 mM Tris-Cl buffer, pH 8.0, containing 1 mM histidine and 0.66 M sucrose (TSH), and then subjected to precipitation with KCl in the presence of deoxycholate (0.3 mg/mg protein). The supernatant thus obtained was mixed with an appropriate amount of cold water to precipitate trace amounts of cytochrome c oxidase, and then dialyzed against 10 mM Tris-Cl, pH 8.0, containing 1 mM EDTA for 6 h with one change of buffer. The dialysate was subjected to centrifugation (96,000 × g for 75 min). The pellet containing complexes I, II, and III was homogenized in TSH buffer, and then subjected to repeated ammonium acetate fractionation in the presence of deoxycholate (0.5 mg/mg protein). Complex I was finally resolved (39% saturation of ammonium sulfate) and separated using ammonium sulfate precipitation (35.9% saturation) in the presence of potassium cholate (0.4 mg/mg protein).
Analytical Methods
Optical spectra were measured on a Shimadzu 2401 UV/VIS recording spectrophotometer. The protein concentration of complex I was determined by the Lowry method using BSA as standard. The concentration of Q1 was determined by absorbance spectra from NaBH4 reduction using a millimolar extinction coefficient ε(275nm-290nm) = 12.25 mM−1cm−1 [26]. The enzyme activity of complex I was assayed by measuring rotenone-sensitive NADH oxidation by Q1. An appropriate amount of complex I was added to an assay mixture (1 ml) containing 20 mM potassium phosphate buffer, pH 8.0, 2mM NaN3, and 0.1 mM Q1, and 0.15 mM NADH as developed by Hatefi et al. [27]. The complex I activity was determined by measuring the decrease in absorbance at 340nm. The specific activity of complex I was calculated using a molar extinction coefficient ε340nm = 6.22 mM−1cm−1. The purified complex I exhibited a specific activity of ~1.0 μmol NADH oxidized min−1 mg−1.
Electron Paramagnetic Resonance Measurements
EPR measurements were performed on a Bruker EMX spectrometer operating at 9.863 GHz with 100 kHz modulation frequency at room temperature. The reaction mixture was transferred to a 50 μl capillary, which was then positioned in the high sensitivity (HS) cavity (Bruker Instrument, Billerica, MA). The sample was scanned using the following parameters: center field, 3510 G; sweep width, 140 G; power, 19.97 mW; receiver gain, 2×105; modulation amplitude, 1 G; time of conversion, 81.92 ms; time constant, 163.84 ms; number of scans, five. The spectral simulations were performed using the WinSim program developed at NIEHS by Duling [28]. The hyperfine coupling constants used to simulate the spin adduct of DEPMPO/•OOH were isomer 1: aN = 13.14 G, aHβ= 11.04 G, aHγ = 0.96 G, aP= 49.96 G (79.8 % relative concentration); isomer 2: aN = 13.18 G, aHβ = 12.59 G, aHγ = 3.46 G, aP= 48.2 G (20.2 % relative concentration).
Disulfide Formation Mediated by O2•− Generation of Complex I
Purified complex I (5 mg/ml) was reduced with DTT (1 mM) to ensure it’s in the fully cysteine sulfhydryl form. Excess DTT was removed by dialysis against TSH buffer (600× volumes) at 4 °C for 90 min. Complex I (DTT-treated, 0.2 mg/ml in PBS) was incubated with NADH (1 mM) and Q1 (0.4 mM) at room temperature for 1 h in the absence or presence of Mn-SOD (1 unit/μl). The reaction mixture was then subjected to alkylation with iodoacetamide prior to in-gel proteolytic digestion.
Alkylation of Complex I with Iodoacetamide
The sample of complex I was subjected to alkylation with iodoacetamide (ICH2CONH2) to block free thiols of protein. Complex I (0.2 mg/ml) in reaction mixture was incubated with iodoacetamide (1 mM) at room temperature. After 1 h incubation, more iodoacetamide was added to the final concentration of 1.5 mM and the mixture was incubated at 4 °C for 8 h. The protein band of the 51 kDa subunit in the SDS-PAGE of complex I overlapped with the bands from the subunits of 49 kDa (Ip) and ND5 (Hp) [16]. Therefore, to facilitate the identification of disulfide bond(s) in the 51 kDa subunit, the Fp fraction of alkylated complex I was partially purified by a procedure involving ethanol (9% v/v) extraction at 40 °C that removed the Hp fraction and most of the Ip fraction of complex I [15]. This partially purified Fp fraction containing multiple subunits was subjected to SDS-PAGE. The resulting gel band of 51 kDa was subjected to in-gel digestion with trypsin or chymotrypsin or both, and followed by LC/MS/MS analysis.
Trypsin, Chymotrypsin and Trypsin/Chymotrypsin Digestion
The protein separated by SDS-PAGE gels was digested with sequencing grade trypsin from Promega (Madison, WI) or sequencing grade chymotrypsin from Roche (Indianapolis, IN) using the Montage In-Gel Digestion Kit from Millipore (Bedford, MA) following the manufacturer’s recommended protocols. Briefly, bands were trimmed as close as possible to minimize background polyacrylamide material. Gel pieces were then washed in 50% methanol/5% acetic acid for one hour. The washing step was repeated once before gel pieces were dehydrated in acetonitrile. The gel bands were washed again with cycles of acetonitrile and ammonium bicarbonate (100mM) in 5 min increments. After the gels were dried in a speed vac, the protease was driven into the gel pieces by re-hydrating them in 50 μL of sequencing grade modified trypsin, chymotrypsin, or a combination of both at 20 μg/mL in 50 mM ammonium bicarbonate for 10 min. An aliquot of 20 μL of 50 mM ammonium bicarbonate was added to the gel bands and the mixture was incubated at room temperature overnight. The peptides were extracted from the polyacrylamide with 50% acetonitrile and 5% formic acid several times and pooled together. The extracted pools were concentrated in a speed vac to ~25 μl.
Capillary-Liquid Chromatography-Nanospray Tandem Mass Spectrometry (Nano-LC/MS/MS)
Nano LC/MS/MS was performed on a Thermo Finnigan LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system was an UltiMate™ Plus system from LC-Packings A Dionex Co (Sunnyvale, CA) with a Famos autosampler and Switchos column switcher. Solvent A was water containing 50mM acetic acid and solvent B was acetonitrile. Five microliters of each sample was first injected onto the trapping column (LC-Packings A Dionex Co, Sunnyvale, CA), and washed with 50 mM acetic acid. The injector port was switched to inject and the peptides were eluted off the trap onto the column. A 5 cm 75 μm ID ProteoPep II C18 column (New Objective, Inc. Woburn, MA) packed directly in the nanospray tip was used for chromatographic separations. Peptides were eluted directly off the column into the LTQ system using a gradient of 2–80% B over 30 minutes, with a flow rate of 300 nL/min. A total run time was 58 minutes. The MS/MS was acquired according to standard conditions established in the laboratory. Briefly, a nanospray source operated with a spray voltage of 3 KV and a capillary temperature of 200 °C is used. The scan sequence of the mass spectrometer was based on the TopTen™ method; the analysis was programmed for a full scan recorded between 350 – 2000 Da, and a MS/MS scan to generate product ion spectra in consecutive instrument scans of the ten most abundant peak in the spectrum. The CID fragmentation energy was set to 35%. To exclude multiple MS/MS, dynamic exclusion is enabled with a repeat count of 30 s, exclusion duration of 350 s and a low mass width of 0.5 and high mass width of 1.50 Da.
Sequence information from the MS/MS data was processed using the Mascot 2.0 active perl script with standard data processing parameters to form a peaklist (mgf file). Database searching was performed against NCBInr database using the MASCOT 2.0 (Matrix Science, Boston, MA) for the identification of carbamoylmethylated cysteines. The mass accuracy of the precursor ions was set to 1.5 Da to accommodate accidental selection of the 13C ion and the fragment mass accuracy was set to 0.5 Da. The number of missed cleavages permitted in the search was 2 for both tryptic and chymotryptic digestions. The considered modifications (variable) were methionine oxidation and cysteine carbamoylmethylation. The data was later searched by MassMatrix, a program designed for the identification of disulfide bonds [21]. The mass accuracy of the precursor ions was set to 2.0 Da and the fragment mass accuracy was set to 0.8 Da as default value in MassMatrix search in order to include all possible disulfide bonds linkage. Possible hits from MASCOT and MassMatrix were manually verified.
RESULTS
DEPMPO Spin Trapping of O2•− Generated by Complex I under the Conditions of Enzyme Turnover
The isolated complex I showed an electron transfer activity in catalyzing rotenone-sensitive NADH oxidation by ubiquinone (Q1) at room temperature. Due to the low oxygen tension (Po2 =1–2 mmHg) and high concentration of GSH in mitochondria [29], it is reasonable to assume that most of the protein thiols (except the structural thiols for the ligands of iron-sulfur clusters) are present as cysteine sulfhydryls in vivo. The isolated complex I was reduced with DTT (1 mM) to ensure it’s in the fully cysteine sulfhydryl form. Excess DTT was removed by dialysis against TSH buffer at 4 °C for 90 min.
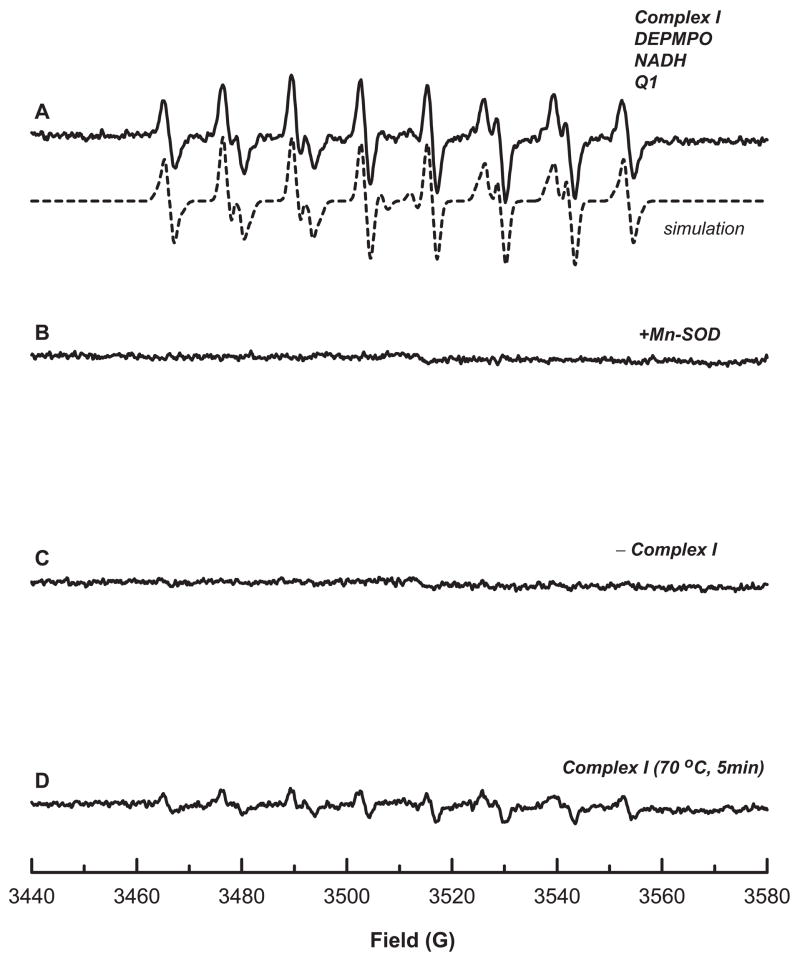
Mitochondrial complex I-mediated O2•− production can be induced under the conditions of enzyme turnover in the presence of NADH and ubiquinone-1 (Q1). The generated O2•− can be detected and measured by EPR spin-trapping. Complex I in the fully cysteine sulfhydryl form (0.2 mg/ml) was incubated with the nitrone spin trap, 5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide (DEPMPO, 20 mM) in PBS. The reaction was initiated by the addition of NADH (1 mM) and Q1 (0.4 mM). A multi-line EPR spectrum was acquired that was characteristic of DEPMPO/•OOH (Figure 1A, solid line) based on hyperfine coupling constants [isomer 1: aN = 13.14 G, aHβ = 11.04 G, aHγ = 0.96 G, aP= 49.96 G (80% relative concentration); isomer 2: aN = 13.18 G, aHβ = 12.59 G, aHγ = 3.46 G, aP= 48.2 G (20% relative concentration)] obtained from the computer simulation (Figure 1A, dashed line) [15]. That the DEPMPO/•OOH adduct arose from the trapping of O2•− was confirmed by the addition of Mn-containing superoxide dismutase (Mn-SOD, 1 unit/μl) to the reaction system (Figure 1B); upon its addition the adduct formation was completely prevented. In the absence of complex I, no DEPMPO/•OOH was detected (Figure 1C). When complex I was replaced with heat-denatured (70 °C for 5 min) complex I, the formation of DEPMPO/•OOH was inhibited (Figure 1D). These results indicate the enzymatic dependence of the DEPMPO adduct formation.
Figure 1. EPR spin trapping of O2•− generated from complex I in the presence of DEPMPO under the conditions of enzyme turnover.
A, the computer simulation (dashed line) superimposed on the experimental spectrum (solid line) obtained using complex I (0.2 mg/ml, reduced by DTT), DEMPO (20 mM), DTPA (1 mM), NADH (1 mM), and Q1 (0.4 mM) in PBS. The experimental spectrum was recorded after signal averaging 5 scans at room temperature. The absolute basal activity of complex I-mediated O2•− generation is 0.87 nmol O2•− production/min/mg NQR using 4-hydroxy-2,2,6,6-tetramethylpiperidinyloxy (TEMPOL) as a standard for spin quantitation. B, the same as A, except that Mn-SOD (1 U/μl) was added to the mixture before the reaction was initiated by NADH. C, the same as A, except that the enzyme was heated at 70 °C for 5 min prior to EPR measurement. D, the same as A, except that the enzyme was omitted from the system.
Mediation of Disulfide Formation in the 51 kDa Subunit of Complex I by Oxidative Stress
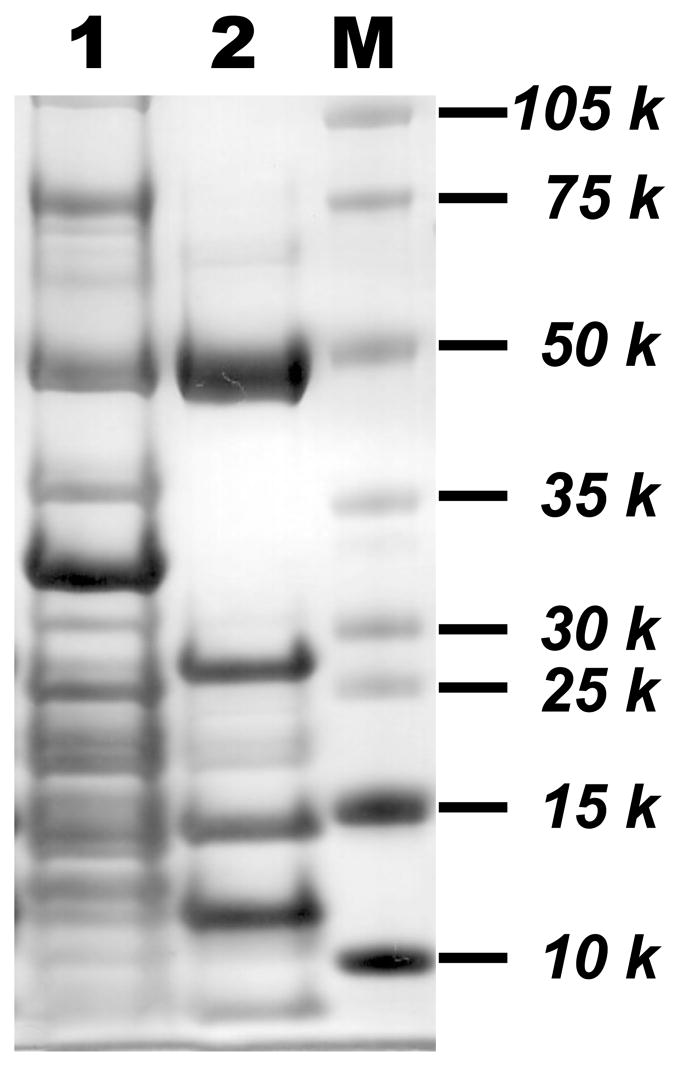
In the absence of spin trap, O2•− mediated by compelx I was expected to induce oxidative modification with disulfide bond formation at the 51 kDa subunit since FMN-binding Fp is involved in the electron leakage for O2•− production [15]. To test this hypothesis, oxidative attack of complex I was initiated by incubation of enzyme (0.2 mg/ml in PBS) with NADH (1 mM) and Q1 (0.4 mM) at room temperature for 1 h. The amount of O2•− generation was ~52 nmol/mg complex I (or 0.87 nmol/min/mg complex I) during the enzyme turnover. The reaction mixture was subjected to alkylation with iodoacetamide. In order to investigate the disulfide formation in the 51 kDa subunit, iodoacetamide, an alkylation reagent that specifically reacts with free cysteines present in the complex I, was used to block free cysteines through carbamoylmethylation (addition of −CH2CONH2) as described in EXPERIMENTAL METHODS. The alkylation with iodoacetamide potentially prevents the disulfide formation caused by disproportionation such as thiol-disulfide exchange. The alkylated 51 kDa subunit was then separated from other components in complex I (lane 1 of Figure 2) using 9% ethanol extraction at 40 °C and SDS-PAGE gel indicated in the Figure 2 (lane 2), and the protein band of 51 kDa was subjected to in-gel digestion with trypsin and chymotrypsin or a combination of trypsin/chymotrypsin under non-reducing conditions. The sequence of the digested peptide was analyzed by LC/MS/MS and the results were searched with Mascot (Matrix Science, Boston, MA) and MassMatrix [21] to identify disulfide formation.
Figure 2. Preparation of the crude NADH dehydrogenase (NDH, Fp subcomplex) from carbamoylmethylated complex I.
Complex I was alkylated with iodoacetamide according to the procedure described in Experimental Methods. 100 % ethanol was added to a final concentration of 9% (v/v), and the mixture was incubated at 40 °C for 10 min. The sample was then chilled in an ice-salt water bath for 10 min and subjected to centrifugation at 25,000 rpm for 30 min. The supernatant containing crude NDH was collected and concentrated with Centricon 30. Carbamoylmethylated complex I and crude NDH were subjected to SDS-PAGE using 4–12% Bis-Tris polyacrylamide gel. Lane 1, Carbamoylmethylated complex I (30 μg); lane 2, crude NDH preparation (15 μg) from carbamoylmethylated complex I. M represents a molecular weight marker.
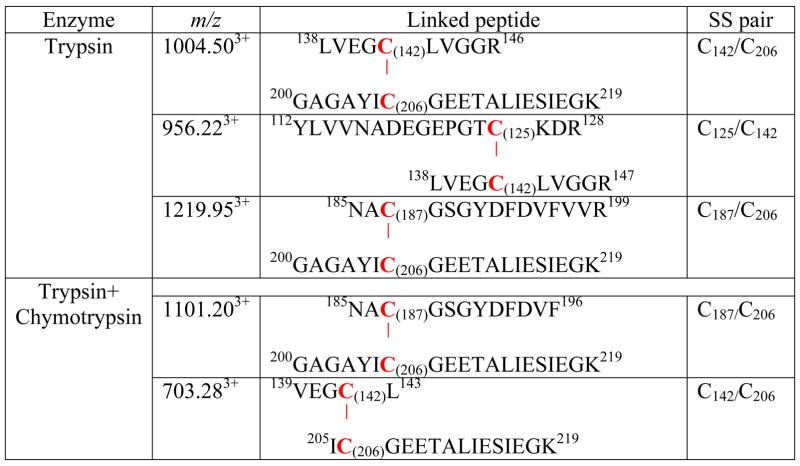
As shown in Figure 3, 12 of 12 cysteine residues were identified with more than 90% of the protein sequence coverage by MS/MS. The MS/MS data were further searched with MassMatrix to identify any disulfide bonds present in this protein. MassMatrix is a database search engine available in house for identification of peptide sequences from tandem mass spectrometry data [21]. The program was adapted such that it would generate tandem mass spectrometry spectra for all putative peptides containing intra-molecular disulfide bonds. The program was then able to score matches between the tandem mass spectra and the disulfide linked peptides. This approach required no derivatization of the sample provided efforts were undertaken to reduce disulfide exchange. The search results were all manually examined. The results showed that 4 cysteines, C125, C142, C187 and C206, were involved in the formation of disulfide bonds, but in a random pattern. Overall, three pairs of disulfide bonds were observed in tryptic digestion: C125/C142, C142/C206 and C187/C206 (Figures 4–6, vide infra) and two pairs of disulfide bonds were observed in tryptic/chymotryptic digestion: C142/C206 and C187/C206 (vide infra). The results are summarized in the Table I.
Figure 3. Amino acid sequence of the precursor of the complex I-51 kDa FMN-binding subunit.
The regions labeled with bold represent the amino acid residues identified with LC/MS/MS. The underlined regions are the sequence motif of 4Fe-4S binding (aa 379–425). The cysteines involved in the 4Fe-4S binding are C379, C382, C385, and C425. The cysteinyl residues involved in disulfide linkage are highlighted with gray and they are C125, C142, C187, and C206. The region labeled with a dotted underline is the N-terminal extension (aa 1–20), which acts as an import sequence and does not exist in the mature protein.
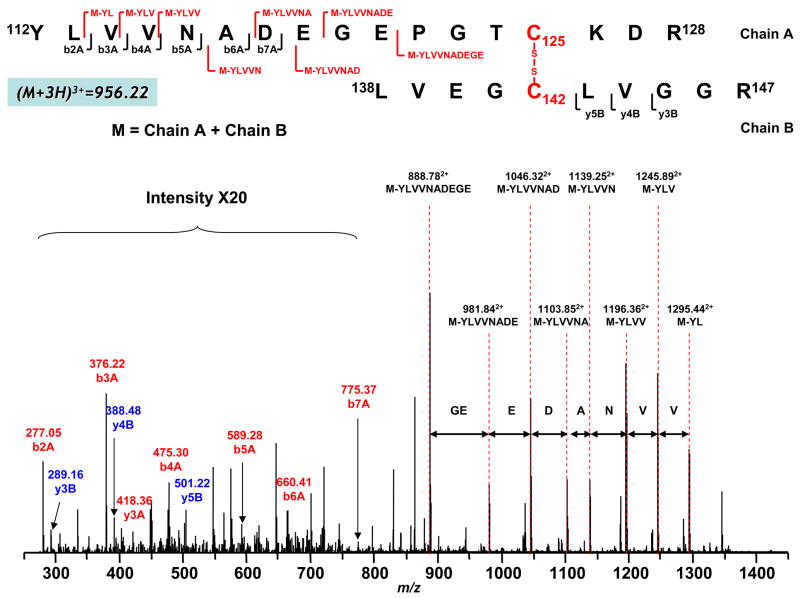
Figure 4. Disulfide bond linkage between C125 and C142 as determined by MS/MS spectrum of tryptic digests of the 51 kDa subunit of complex I.
Complex I was exposed to the conditions of superoxide production, and then alkylated by iodoacetamide as described in “Materials and Methods”. Crude NADH dehydrogenase (NDH) was prepared from complex I as described in the published procedure (16). The obtained crude NDH was subjected to SDS-PAGE under non-reducing conditions. The gel band of the 51 kDa polypeptide was subjected to in-gel digestion prior to LC/MS/MS as described in “Materials and Methods”. Chain A and Chain B represent peptides containing aa 112–128 and aa 138–147 respectively. A disulfide bond linking Chain A with Chain B is indicated in red. The sequence-specific ions are labeled as bnA (n=2–7) for Chain A, and ynB (n=3–5) for Chain B on the spectrum. M indicates Chain A plus Chain B.
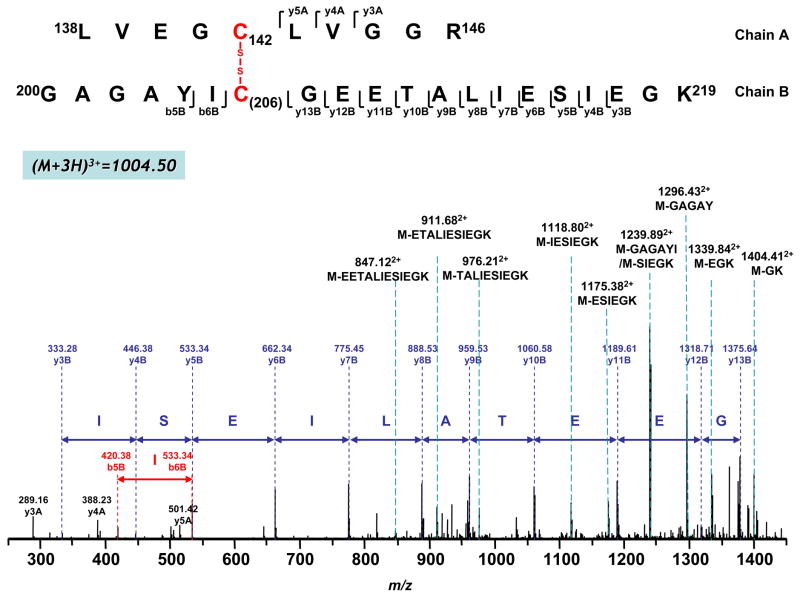
Figure 6. Disulfide bond linkage between C142 and C206 as determined by MS/MS spectrum of doubly protonated molecular ion obtained from tryptic digests of the 51 kDa subunit of complex I.
Chain A and Chain B represent peptides containing aa 138–147 and aa 200–219 respectively. A disulfide bond linking Chain A with Chain B is indicated in red. The sequence-specific ions are labeled as ynA (n=3–5) for Chain A, and ynB (n=3–13), bnB (n=5–6) for Chain B on the spectrum.
Table I.
Detection of oxygen free radical(s)-mediated disulfide bond formation in the 51 kDa polypeptide of complex I by LC/MS/MS.
|
Identification of Disulfide Bonds in the 51 kDa subunit of complex I by LC/MS/MS
(I) Involvement of C125 and C142 in the disulfide bond formation
When MS/MS was performed on the triply charged [(M+3H)3+] peak at m/z 956.223+, b ions (b2A-b7A) from peptide chain 112–128 (chain A) and y ions (y3B-y5B) from peptide chain 138–146 (chain B) were observed as indicated in the Figure 4. In addition, a series of ions with loss of part of the sequence was also observed, including loss of YL (M-YL, m/z 1295.442+), YLV (M-YLV, m/z 1245.892+), YLVV (M-YLVV, m/z 1196.362+), YLVVN (M-YLVVN, m/z 1139.252+), YLVVNA (M-YLVVNA, m/z 1103.852+), YLVVNAD (M-YLVVNAD, m/z 1046.322+), YLVVNADE (M-YLVVNADE, m/z 981.842+) and YLVVNADEGE (M-YLVVNADEGE, m/z 888.782+) from chain A (aa 112–128); these ions subsequently disappeared under reducing conditions in the presence of DTT or β-mercaptoethanol (β-ME). Therefore, they were identified to be peptide fragments containing the disulfide linkage between C126 and C142, and the species with m/z 956.223+ consisted of two fragments, peptide 112–128 and peptide 138–146, connected through the disulfide bond between C125 and C142. Detailed MS/MS assignment is also listed in the Supplemental Table 1.
(II) Involvement of C187 and C206 in the disulfide bond formation
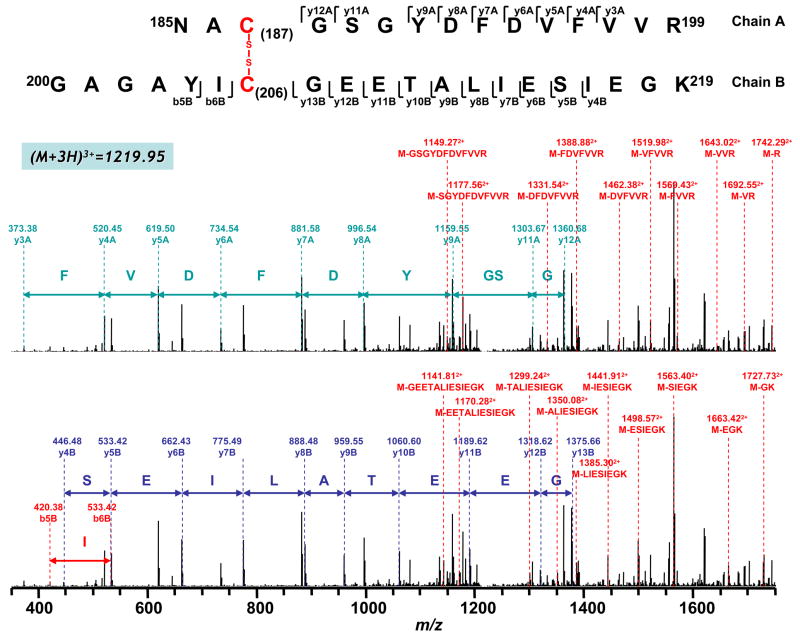
Figure 5 shows the MS/MS spectrum indicating disulfide bond linkage between peptide 185–199 (Chain A) and 200–219 (Chain B) through C187 and C206. As illustrated in Figure 5, MS/MS was performed on a triply charged peak with an m/z of 1219.953+. Several b ions (b5B, b6B) and y ions (y4B-y13B) from peptide chain 200–219 (Chain B) were observed, as well as y ions (y3A-y9A, y11A, y12A) from peptide chain 185–199 (Chain A). Fragments containing the partial sequence of the whole branched peptide (M=Chain A + Chain B) were also observed as the cleavage occurred at the C-terminus from both chains, including the cleavage at R through GYDFDVFVVR from peptide chain 200–219 (Figure 5, shown in the upper panel) and GK through GEETALIESIEGK from peptide chain 185–199 (the same spectrum of Figure 5 shown in the lower panel). The molecular ions of these fragments subsequently disappeared in the presence of thiol-containing reducing agents such as DTT or β-ME, thus supporting a disulfide linkage between C187 and C206. Detailed MS/MS assignment is also listed in the Supplemental Table 2.
Figure 5. Disulfide bond linkage between C187 and C206 as determined by MS/MS spectrum of tryptic digests of the 51 kDa subunit of complex I.
Chain A and Chain B represent peptides containing aa 185–199 and aa 200–219 respectively. A disulfide bond linking Chain A with Chain B is indicated in red. The sequence-specific ions are labeled as ynA (n=3–9 and 11–12) for Chain A and ynB (n=4–13), bnB (n=5–6) for Chain B on the spectrum. M indicates Chain A plus Chain B. Note that the same spectrum is shown in upper and lower panels.
The molecular pattern of disulfide linkage between C187 and C206 was further evidenced by the detection of a doubly protonated ion (M+2H)2+ with m/z 1101.202+ obtained from trypsin/chymotrypsin combined digests (Table I). MS/MS was performed on this doubly charged peak. Several y ions (y3B-y13B) and two b ions (b5B and b6B) from chain B (aa 200–219) were observed (indicated in the Supplemental Figure 2). In addition, a series of ions with loss of part of the sequence from chain A (aa 185–196) was also observed, including loss of F (M-F, m/z 1569.382+), VF (M-VF, m/z 1520.202+), DVF (M-DVF, m/z 1462.302+), FDVF (M-FDVF, m/z 1388.592+), DFDVF (M-DFDVF, m/z 1331.072+), GYDFDVF (M-GYDFDVF, m/z 1221.352+), and GSGYDFDVF (M-GSGYDFDVF, m/z 1149.112+) (indicated in the Supplemental Figure 2). These ions were not detected by MS under reducing conditions.
(III) Involvement of C142 and C206 in the disulfide bond formation
As shown in Figure 6, when MS/MS was performed on a triply charged species at an m/z of 1004.503+, MS/MS fragments were observed at 289.16, 388.23 and 501.42, which can be assigned as y3A, y4A and y5A from peptide 138–147 (chain A). Similarly, a second series of fragment ions was assigned as y3B-y13B from peptide 200–219 (chain B). In addition, a series of ions with loss of part of the sequence from chain B (aa 200–219) was also observed, including loss of GK (M-GK, m/z 1404.412+), EGK (M-EGK, m/z 1339.842+), SIEGK (M-SIEGK, m/z 1239.892+), ESIEGK (M-ESIEGK, m/z 1175.382+), IESIEGK (M-IESIEGK, m/z 1118.802+), TALIESIEGK (M-TALIESIEGK, m/z 976.212+), ETALIESIEGK (M-ETALIESIEGK, m/z 911.682+), EETALIESIEGK (M-EETALIESIEGK, m/z 847.122+), GAGAY (M-GAGAY, m/z 1296.432+) and GAGAYI (M-GAGAYI, m/z 1239.892+) (indicated in the Figure 6). This information, combined with the determined molecular weight, suggested that this species consisted of peptide fragments 138–147 and 200–219 linked together through a disulfide bond between C142 and C206. Detailed MS/MS assignment is also listed in the Supplemental Table 3.
The molecular pattern of disulfide bond formation involved in the C142 and C206 was further evidenced by detecting a triply protonated ion with m/z 703.283+ obtained from proteolytic digestion with trypsin/chymotrypsin combination. MS/MS was performed on this triply charged peak. Several y ions (y2B-y9B) from chain B (aa 205–219) were observed (indicated in the Supplemental Figure 3). In addition, a series of ions with loss of part of the sequence from chain B was also observed, including loss of K [M-K, m/z 980.402+; M=chain A (aa 139–143) + chain B (aa 205–219) ], GK (M-GK, m/z 951.902+), IEGK (M-IEGK, m/z 830.982+), SIEGK (M-SIEGK, m/z 787.382+), ESIEGK (M-ESIEGK, m/z 722.692+), IESIEGK (M-IESIEGK, m/z 666.342+), and LIESIEGK (M-LIESIEGK, m/z 609.602+) indicated in the Supplemental Figure 3. Likewise, these ions were not detected by MS under reducing conditions in the presence of β-ME.
Identification of carbamoylmethylated cysteine of complex I-51 kDa by LC/MS/MS
The sequences of peptides with carbamoylmethylated cysteines were further verified manually. As indicated in Table II, all cysteines of the alkylated 51 kDa subunit were observed to be carbamoylmethylated, including the residues (C379, C382, C385, and C425) involved in the ligands of the 4Fe-4S cluster N3 center. Presumably, the structure of the 4Fe-4S coordinates may suffer partial destruction under the conditions of oxidative stress and extensive alkylation (incubation with 1.5 mM iodoacetamide for 8 h).
Table II.
Detected fragments containing carbamoylmethylated cysteines from the 51 kDa subunit of complex I subjected to oxidative attack by O2•−.
| Detected m/z | Enzyme | Fragment | Cys Detected |
|---|---|---|---|
| 826.532+ | Trypsin | 112YLVVNADEGEPGTCK126 | C125 |
| 826.652+ | Tryp/Chymo | 112YLVVNADEGEPGTCK126 | C125 |
| 962.172+/641.713+ | Trypsin | 112YLVVNADEGEPGTCKDR128 | C125 |
| 745.122+ | Tryp/Chymo | 113YLVVNADEGEPGTCK126 | C125 |
| 587.743+ | Tryp/Chymo | 113LVVNADEGEPGTCKDR128 | C125 |
| 530.582+ | Tryspin | 138LVEGCLVGGR147 | C142 |
| 530.532+ | Tryp/Chymo | 138LVEGCLVGGR147 | C142 |
| 1010.382+ | Chymo | 178EAGLIGKNACGSGYDFDVF196 | C187 |
| 1351.331+ | Tryp/Chymo | 185NACGSGYDFDVF196 | C187 |
| 853.602+ | Trypsin | 185NACGSGYDFDVFVVR199 | C187 |
| 1034.702+/690.143+ | Trypsin | 200GAGAYICGEETALIESIEGK219 | C206 |
| 1034.802+ | Tryp/Chymo | 200GAGAYICGEETALIESIEGK219 | C206 |
| 824.762+ | Tryp/Chymo | 205ICGEETALIESIEGK219 | C206 |
| 1133.872+ | Trypsin | 225LKPPFPADVGVFGCPTTVANVETVAVSPTICR256 | C238 and C255 |
| 1066.782+ | Tryp/Chymo | 237GCPTTVANVETVAVSPTICR256 | C238 and C255 |
| 1307.582+/872.073+ | Trypsin | 275LFNISGHVNNPCTVEEEMSVPLKE297 | C286 |
| 1121.682+ | Chymo | 277NISGHVNNPCTVEEEM(Ox)SVPL296 | C286 |
| 1121.692+ | Tryp/Chymo | 277NISGHVNNPCTVEEEM(Ox)SVPL296 | C286 |
| 1185.772+/791.023+ | Tryp/Chymo | 277NISGHVNNPCTVEEEM(Ox)SVPLK297 | C286 |
| 573.442+ | Chymo | 327IPKSVCETVL336 | C332 |
| 928.042+ | Chymo | 327IPKSVCETVLM(Ox)DFDAL342 | C332 |
| 1086.083+ | Trypsin | 330SVCETVLMDFDALIQAQTGLG TAAVIVM(Ox)DR359 | C332 |
| 465.043+ | Trypsin | 376HESCGQCTPCR386 | C379, C382 and C385 |
| 1096.872+ | Trypsin | 418QIEGHTICALGDGAAWPVQGL438 | C425 |
| 1231.872+/821.143+ | Trypsin | 418QIEGHTICALGDGAAWPVQGLIR440 | C425 |
| 822.043+ | Tryp/Chymo | 418QIEGHTICALGDGAAWPVQGLIR440 | C425 |
It is worth noting that alkylation of three 4Fe-4S ligands, C379, C382, C385, was not observed in the MS spectra under modest conditions (incubation with 1.0 mM iodoacetamide for 1 h). Presumably these three structural thiols involved in 4Fe-4S binding are buried inside the complex I and structurally protected, which only can be alkylated under vigorous conditions.
Iodine, a potential oxidative agent, could be produced through drastic conditions of iodoacetamide alkylation. However, the detection of O2•−-induced specific disulfide formation was not affected by iodoacetamide pre-treatment. That is, we have detected the same molecular pattern of disulfide linkage without alkylation of complex I, thus eliminating the possibility of artifactual disulfide bond formation caused by iodine.
It should be noted that the residues (C125, C142, C187, and C206) involved in disulfide linkage were also observed to be carbamoylmethylated (Table II), suggesting that both cysteine sulfhydryl (or free thiol form) and cysteine disulfide (induced by O2•−) are present in residues C125, C142, C187, and C206 of the 51 kDa subunit of complex I. Therefore, these four specific cysteines only partially participated in the disulfide linkage and the detected disulfide formation induced by oxidative stress was not uniform.
Involvement of O2•− in the disulfide bond formation
When Mn-SOD (1 unit/μl) was included in the reaction system containing complex I/NADH/Q1, no disulfide linkage was detected in the 51 kDa subnit of complex I by LC/MS/MS, suggesting the involvement of oxidative attack by O2•−/or O2•−-derived oxidants.
In the control experiment, complex I (DTT-treated) was incubated at room temperature for 1 h in the absence of NADH and Q1 prior to alkylation with iodoacetamide. LC/MS/MS analysis of tryptic/or chymotryptic peptides of 51 Da revealed no disulfide formation. These results illustrate that the formation of the 51 kDa-derived disulfide bond is physiologically relevant and is involved in the biological event of oxidative stress.
DISCUSSION
Advantage of the Mass Spectrometry Coupled with MassMatrix Program in the Detection of Protein Disulfide Linkages
The investigation of disulfide bonds is critical for understanding the biological properties and function of a protein but has not always been an easy task for researchers. X-ray crystallography and NMR are excellent tools for studying the 3D structure of proteins, thus providing information regarding disulfide linkages [30–33]. However, large amounts of well purified protein with good solubility or the proper size crystal are required for these two methods respectively [34]. Furthermore, some proteins must interact with other proteins in order to accomplish their biological function. It is usually difficult to separate one protein from other associated components in a solution phase. Some proteins containing regulatory thiols can only form disulfide linkages under conditions of oxidative stress (the 51 kDa FMN-binding subunit of complex I in this study). The disulfide bonds thus formed are not necessarily uniform, and are unlikely to be well characterized by X-ray crystallography.
Mass spectrometry has been widely used to identify disulfide bonds [35–44]. Proteins can be investigated either in solution phase [34, 37, 42] or from a gel [40, 45]. Since protease is used to decrease the size of the protein/peptide in MS analysis, large-sized proteins can also be analyzed. It has been demonstrated that disulfide bridge patterns can be identified by partial reduction followed by MS analysis [42, 43, 46, 47]. However, when a protein is highly disulfide-bond-bridged, this approach requires multiple reduction and separation steps for the complete mapping of protein disulfide bonds. In addition, this method fails when multiple disulfide bonds show similar reduction kinetics [46].
Tandem MS followed by proteolytic digestion under non-reducing conditions is another approach to characterizing the disulfide pattern of a given protein [34, 37, 38, 40, 45, 48–51]. With appropriate protease(s) employed, the pattern of disulfide linkages can be identified in a single run. However, most of the data are manually analyzed, which is time-consuming and labor-intensive. This factor makes data analysis extremely difficult, especially for proteins with unknown disulfide linkages such as the 51 kDa FMN-binding subunit of complex I in this study. Also, it tends to miss some information when the sample contains large number of cysteines with multiple mixed intra-molecular disulfide bonds in the protein.
In this study, the modified database search program MassMatrix was applied to provide the candidate list of disulfide linked peptides, which greatly simplified data analysis and reduced the rate of false negative matches [21]. With the aid of MassMatrix, we were able to completely and accurately identify oxygen free radical-induced disulfide bonds in the FMN-binding subunit of complex I. Four out of 12 cysteine residues were identified as involved in the formation of three disulfide bonds: C125/C142, C187/C206, and C142/C206 in the complex I under the conditions of oxidative stress. None of the identified cysteines is associated with the cysteinyl ligands of the 4Fe-4S cluster (N3 center).
Biological Implications
Mitochondrial thiols are composed of protein thiols and the GSH/GSSG pool. The proteins of the mitochondrial electron transport chain are rich in protein thiols [9, 10]. An analysis of rat liver mitochondria has shown them to contain 65–70 nmol of thiols per mg of protein. Excluding the structural thiols involved in the ligands of metal centers, ~20–25 nmol of thiols per mg of protein are exposed. The content of GSH/GSSG for isolated liver mitochondria is 3–5 nmol per mg of protein [9, 10]. Therefore, protein thiols play a critical role in controlling the redox state of mitochondria.
Mitochondrial complex I is the major component of the ETC to host protein thiols, which comprise structural thiols involved in the ligands of iron sulfur clusters and the reactive/regulatory thiols which are thought to have biological functions of antioxidant defense and redox signaling [11, 12]. The physiological roles of complex I-derived regulatory thiols have been associated with the regulation of respiration, nitric oxide utilization [13, 14], redox status of mitochondria [8–10], and redox modifications such as S-glutathiolation [16] and protein thiyl radical formation [15].
The current studies identified four cysteine residues, C125, C142, C187, and C206 that are involved in the mixed intra-molecular disulfide linkages induced by oxidative stress. However, the formed intra-molecular disulfide is not uniform since the evidence obtained from alkylation with iodoacetamide clearly indicated that free thiols of residues C125, C142, C187, and C206 are also present in the complex I that was exposed to oxidative challenge (Figure 1 and Table I). We did not detect any disulfide bridge involvement for the other four cysteine residues, C238, C255, C286, and C332, based on the data analysis obtained from LC/MS/MS and MassMatrix database searching. Therefore, it can be suggested that the residues of C125, C142, C187, and C206 function as the regulatory thiols in the FMN-binding subunit of complex I. They may contribute to the homeostasis of thiol redox in mitochondria. In previous studies, we have identified C206 to be the reactive cysteine susceptible to oxidative attack of oxygen free radical(s), forming a specific protein thiyl radical [15]. In a separate study, C206 was also identified as involved in the redox event of protein S-glutathiolation when complex I was exposed to oxidized glutathione, GSSG [16]. In the present studies, C206 and C187/or C142 were formed disulfide linkage induced by oxidative attack (Figures 5–7). Two additional cysteine residues, C125 and C142, were further identified as participants in the redox event of disulfide formation due to oxidative stress (Figure 4).
The milieu of the mitochondrial matrix is almost anoxic (oxygen tension, Po2 =1–2 mmHg) in the presence of the GSH/GSSG pool under normal physiological conditions [29]. Analysis of redox compartmentation indicates that the relative redox states from most reductive to most oxidative are as follows: mitochondria>nuclei>endoplamic reticulum>extracellular space [29]. Thus, it is expected that low oxygen tension in the mitochondrial environment should facilitate the free thiol state for most regulatory cysteines; and that mitochondrial thiols are the targets of oxygen free radical(s) and are vulnerable to oxidative stress. Presumably, the formation of disulfide linkages is a mechanism of antioxidant defense adopted by mitochondrial proteins in response to oxidative stress. This conclusion is strongly supported by the data from EPR spin trapping (Figure 1) and analysis of disulfide formation by exposure of complex I to oxidative stress (Figures 4–7).
The homology model (aa: 38–445 in bovine protein and shown in the Supplemental Figure 4) is built via MODELLER v8 [52], using the structure of the hydrophilic domain of respiratory complex I of Thermus Thermophilius (PDB ID 2FUG) as a template with a sequence identity of 46%. This model is not likely to logically explain the detected disulfide pattern (Figs 4–6) based on the crystal structure since the measured distances of the pairs including C206/C187 (34.7 Å), C206/C142 (19.5 Å), and C125/C142 (20.5 Å) are longer than a reasonable distance (< 6.4Å [53]) for disulfide formation. Therefore, it is suggested that protein may suffer an oxygen free radical-induced conformational change in solution to facilitate disulfide formation. Specifically, cysteine-related reversible oxidative alternation such as protein thiyl radical formation or cysteine sufenic/sufinic acids may contribute to this change. The presumed oxidative alteration does not include irreversible cysteine sulfonation which would not lead to disulfide formation. It should not be ruled out that other oxidative modifications, such as methionine oxidation, may induce conformational change prior to the formation of the disulfide bonds. If this was the case, one would expect that methionine oxidation should be extensive enough to bring specific cysteines into close enough contact for disulfide bond formation.
CONCLUSIONS
The use of LC/MS/MS coupled with MassMatrix program facilitates solving the molecular pattern of disulfide formation induced by oxidant stress. Specifically, we have successfully employed this technique to a complicated enzymatic system. Importantly, the approach can be potentially applied to other mitochondrial proteins containing redox thiols in vitro and in vivo. Furthermore, the intra-molecular disulfide pattern elucidated here provides a useful concept for understanding the fundamental question of how complex I utilize its specific regulatory thiols to address situations of oxidative stress caused by oxygen free radical attack. Clearly, the major role of specific disulfide formation is to regulate the redox signal of oxygen free radical or its derived oxidant. Recognition of this molecular event is important in understanding the fundamental basis of regulatory cysteines in complex I.
Supplementary Material
Acknowledgments
The authors thank Mr. Alexander Yeh (Davis Heart and Lung Research Institute) for assistance in the preparation of complex I, Dr. Chenglong Li (Division of Medicinal Chemistry and Pharmacognosy, College of Pharmacy) for building homology model of complex I-51 kDa, and Mr. Richard Sessler from the Mass Spectrometry and Proteomics Facility for assistance with the mass spectrometric experiments. This work was supported by RO1 grant from National Institutes of Health [(HL083237, to Y-R C)]. MAF was supported by NIH CA107106 and CA101956, Leukemica and Lymphoma Society (SCOR 7080-06) and the V Foundation/American Association for Cancer Research Translational Cancer Research Grant.
Footnotes
Abbreviations: NQR, NADH ubiquinone reductase, or mitochondrial complex I; NDH, NADH dehydrogenase, or the flavin subcomplex of complex I, or Fp fraction of complex I; FMN, flavin mononucleotide; Ip, iron-sulfur protein of complex I; Hp, hydrophobic protein fraction of complex I; O2•−, superoxide anion radical; DTT, dithiothretol; ETC, electron transport chain; SMP, submitochondrial particles; SDS-PAGE, SDS polyacrylamide gel electrophoresis; MS, mass spectrometry; MS/MS, tandem mass spectrometry; EPR, electron paramagnetic resonance.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The Nuclear Encoded Subunits of Complex I from Bovine Heart Mitochondria. Biochim Biophys Acta. 2003;1604:135–50. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 2.Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the Subunit Composition of Complex I from Bovine Heart Mitochondria. Mol Cell Proteomics. 2003;2:117–26. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Walker JE. The NADH:Ubiquinone Oxidoreductase (Complex I) of Respiratory Chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 4.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine Complex I is a Complex of 45 Different Subunits. J Biol Chem. 2006;281:32724–7. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 5.Galkin A, Brandt U. Superoxide Radical Formation by Pure Complex I (NADH:Ubiquinone Oxidoreductase) from Yarrowia Lipolytica. J Biol Chem. 2005;280:30129–35. doi: 10.1074/jbc.M504709200. [DOI] [PubMed] [Google Scholar]
- 6.Kudin AP, Bimpong-Buta NY, Vielhaber S, Elger CE, Kunz WS. Characterization of Superoxide-Producing Sites in Isolated Brain Mitochondria. J Biol Chem. 2004;279:4127–35. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 7.Turrens JF, Boveris A. Generation of Superoxide Anion by the NADH Dehydrogenase of Bovine Heart Mitochondria. Biochem J. 1980;191:421–7. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa NJ, Dahm CC, Hurrell F, Taylor ER, Murphy MP. Interactions of Mitochondrial Thiols with Nitric Oxide. Antioxid Redox Signal. 2003;5:291–305. doi: 10.1089/152308603322110878. [DOI] [PubMed] [Google Scholar]
- 9.Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP, Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Disulphide Formation on Mitochondrial Protein Thiols Glutathionylation of Mitochondrial Proteins. Biochem Soc Trans. 2005;33:1390–3. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- 10.Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of Mitochondrial Proteins. Antioxid Redox Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- 11.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 Catalyzes the Reversible Oxidation and Glutathionylation of Mitochondrial Membrane Thiol Proteins: Implications for Mitochondrial Redox Regulation and Antioxidant DEFENSE. J Biol Chem. 2004;279:47939–51. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 12.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible Glutathionylation of Complex I Increases Mitochondrial Superoxide Formation. J Biol Chem. 2003;278:19603–10. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 13.Beltran B, Orsi A, Clementi E, Moncada S. Oxidative Stress and S-Nitrosylation of Proteins in Cells. Br J Pharmacol. 2000;129:953–60. doi: 10.1038/sj.bjp.0703147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent Inhibition of Cell Respiration by Nitric Oxide: Crucial Role of S-Nitrosylation of Mitochondrial Complex I and Protective Action of Glutathione. Proc Natl Acad Sci U S A. 1998;95:7631–6. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide Generation from Mitochondrial NADH Dehydrogenase Induces Self-inactivation with Specific Protein Radical Formation. J Biol Chem. 2005;280:37339–48. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 16.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, Chen YR. Site-Specific S-Glutathiolation of Mitochondrial NADH Ubiquinone Reductase. Biochemistry. 2007;46:5754–65. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sazanov LA, Hinchliffe P. Structure of the Hydrophilic Domain of Respiratory Complex I from Thermus thermophilus. Science. 2006;311:1430–6. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 18.Fearnley IM, Carroll J, Walker JE. Proteomic Analysis of the Subunit Composition of Complex I (NADH:Ubiquinone Oxidoreductase) from Bovine Heart Mitochondria. Methods Mol Biol. 2007;357:103–25. doi: 10.1385/1-59745-214-9:103. [DOI] [PubMed] [Google Scholar]
- 19.Carroll J, Fearnley IM, Walker JE. Definition of the Mitochondrial Proteome by Measurement of Molecular Masses of Membrane Proteins. Proc Natl Acad Sci U S A. 2006;103:16170–5. doi: 10.1073/pnas.0607719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen R, Fearnley IM, Peak-Chew SY, Walker JE. The Phosphorylation of Subunits of Complex I from Bovine Heart Mitochondria. J Biol Chem. 2004;279:26036–45. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Freitas MA. A Mass Accuracy Sensitive Probability Based Scoring Algorithm for Database Searching of Tandem Mass Spectrometry Data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Zhang L, Freitas MA. Identification and Characterization of Disulfide Bonds in Proteins and Peptides from Tandem MS Data by Use of the MassMatrix MS/MS Search Engine. J Proteome Res. 2008;7:138–44. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Freitas MA. Monte Carlo Simulation-Based Algorithms for Analysis of Shotgun Proteomic Data. J Proteome Res. 2008;7:2605–15. doi: 10.1021/pr800002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galante YM, Hatefi Y. Purification and Molecular and Enzymic Properties of Mitochondrial NADH Dehydrogenase. Arch Biochem Biophys. 1979;192:559–68. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- 25.Vinogradov AD, King TE. The Keilin-Hartree Heart Muscle Preparation. Methods Enzymol. 1979;55:118–27. doi: 10.1016/0076-6879(79)55017-4. [DOI] [PubMed] [Google Scholar]
- 26.Redfearn ER, Whittaker PA. The Determination of the Oxidation-Reduction States of Ubiquinone (Coenzyme Q) in Rat-Liver Mitochondria. Biochim Biophys Acta. 1966;118:413–8. doi: 10.1016/s0926-6593(66)80050-4. [DOI] [PubMed] [Google Scholar]
- 27.Hatefi Y. Preparation and Properties of NADH: Ubiquinone Oxidoreductase (Complex I), EC 1.6.5.3. Methods Enzymol. 1978;53:11–4. doi: 10.1016/s0076-6879(78)53006-1. [DOI] [PubMed] [Google Scholar]
- 28.Duling DR. Simulation of Mmultiple Isotropic Spin-Trap EPR Spectra. J Magn Reson B. 1994;104:105–10. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 29.Hansen JM, Go YM, Jones DP. Nuclear and Mitochondrial Compartmentation of Oxidative Stress and Redox Signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–34. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 30.Collins ES, Wirmer J, Hirai K, Tachibana H, Segawa S, Dobson CM, Schwalbe H. Characterisation of Disulfide-Bond Dynamics in Non-Native States of Lysozyme and its Disulfide Deletion Mutants by NMR. Chembiochem. 2005;6:1619–27. doi: 10.1002/cbic.200500196. [DOI] [PubMed] [Google Scholar]
- 31.Haebel PW, Wichman S, Goldstone D, Metcalf P. Crystallization and Initial Crystallographic Analysis of the Disulfide bond Isomerase DsbC in Complex with the Alpha Domain of the Electron Transporter DsbD. J Struct Biol. 2001;136:162–6. doi: 10.1006/jsbi.2001.4430. [DOI] [PubMed] [Google Scholar]
- 32.Meier S, Haussinger D, Pokidysheva E, Bachinger HP, Grzesiek S. Determination of a High-precision NMR Structure of the Minicollagen Cysteine Rich Domain from Hydra and Characterization of Its Disulfide Bond Formation. FEBS Lett. 2004;569:112–6. doi: 10.1016/j.febslet.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 33.von Ossowski L, Tossavainen H, von Ossowski I, Cai C, Aitio O, Fredriksson K, Permi P, Annila A, Keinanen K. Peptide Binding and NMR Analysis of the Interaction Between SAP97 PDZ2 and GluR-A: Potential Involvement of a Disulfide Bond. Biochemistry. 2006;45:5567–75. doi: 10.1021/bi0511989. [DOI] [PubMed] [Google Scholar]
- 34.Yen TY, Joshi RK, Yan H, Seto NO, Palcic MM, Macher BA. Characterization of Cysteine Residues and Disulfide Bonds in Proteins by Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry. J Mass Spectrom. 2000;35:990–1002. doi: 10.1002/1096-9888(200008)35:8<990::AID-JMS27>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 35.Barbirz S, Jakob U, Glocker MO. Mass Spectrometry Unravels Disulfide Bond Formation as the Mechanism that Activates a Molecular Chaperone. J Biol Chem. 2000;275:18759–66. doi: 10.1074/jbc.M001089200. [DOI] [PubMed] [Google Scholar]
- 36.Fagerquist CK. Collision-Activated Cleavage of a Peptide/Antibiotic Disulfide Linkage: Possible Evidence for Intramolecular Disulfide Bond Rearrangement upon Collisional Activation. Rapid Commun Mass Spectrom. 2004;18:685–700. doi: 10.1002/rcm.1390. [DOI] [PubMed] [Google Scholar]
- 37.John H, Forssmann WG. Determination of the Disulfide Bond Pattern of the Endogenous and Recombinant Angiogenesis Inhibitor Endostatin by Mass Spectrometry. Rapid Commun Mass Spectrom. 2001;15:1222–8. doi: 10.1002/rcm.367. [DOI] [PubMed] [Google Scholar]
- 38.Krokhin OV, Cheng K, Sousa SL, Ens W, Standing KG, Wilkins JA. Mass Spectrometric Based Mapping of the Disulfide Bonding Patterns of Integrin Alpha Chains. Biochemistry. 2003;42:12950–9. doi: 10.1021/bi034726u. [DOI] [PubMed] [Google Scholar]
- 39.Mundt AA, Cuillel M, Forest E, Dupont Y. Peptide Mapping and Disulfide Bond Analysis of the Cytoplasmic Region of an Intrinsic Membrane Protein by Mass Spectrometry. Anal Biochem. 2001;299:147–57. doi: 10.1006/abio.2001.5416. [DOI] [PubMed] [Google Scholar]
- 40.Tie JK, Mutucumarana VP, Straight DL, Carrick KL, Pope RM, Stafford DW. Determination of Disulfide Bond Assignment of Human Vitamin K-Dependent Gamma-glutamyl Carboxylase by Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry. J Biol Chem. 2003;278:45468–75. doi: 10.1074/jbc.M309164200. [DOI] [PubMed] [Google Scholar]
- 41.Wolf SM, Ferrari RP, Traversa S, Biemann K. Determination of the Carbohydrate Composition and the Disulfide Bond Linkages of Bovine Lactoperoxidase by Mass Spectrometry. J Mass Spectrom. 2000;35:210–7. doi: 10.1002/(SICI)1096-9888(200002)35:2<210::AID-JMS931>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 42.Yen TY, Yan H, Macher BA. Characterizing Closely Spaced, Complex Disulfide Bond Patterns in Peptides and Proteins by Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry. J Mass Spectrom. 2002;37:15–30. doi: 10.1002/jms.241. [DOI] [PubMed] [Google Scholar]
- 43.Zeng R, Xu Q, Shao XX, Wang KY, Xia QC. Determination of the Disulfide Bond Pattern of a Novel C-Type Lectin from Snake Venom by Mass Spectrometry. Rapid Commun Mass Spectrom. 2001;15:2213–20. doi: 10.1002/rcm.500. [DOI] [PubMed] [Google Scholar]
- 44.Zubarev RA. Electron-Capture Dissociation Tandem Mass Spectrometry. Curr Opin Biotechnol. 2004;15:12–6. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Debski J, Wyslouch-Cieszynska A, Dadlez M, Grzelak K, Kludkiewicz B, Kolodziejczyk R, Lalik A, Ozyhar A, Kochman M. Positions of Disulfide Bonds and N-Glycosylation Site in Juvenile Hormone Binding Protein. Arch Biochem Biophys. 2004;421:260–6. doi: 10.1016/j.abb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 46.Jones MD, Hunt J, Liu JL, Patterson SD, Kohno T, Lu HS. Determination of Tumor Necrosis Factor Binding Protein Disulfide Structure: Deviation of the Fourth Domain Structure from the TNFR/NGFR Family Cysteine-Rich Region Signature. Biochemistry. 1997;36:14914–23. doi: 10.1021/bi971696k. [DOI] [PubMed] [Google Scholar]
- 47.Jones MD, Patterson SD, Lu HS. Determination of Disulfide Bonds in Highly Bridged Disulfide-Linked Peptides by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry with Postsource Decay. Anal Chem. 1998;70:136–43. doi: 10.1021/ac9707693. [DOI] [PubMed] [Google Scholar]
- 48.Cole AR, Hall NE, Treutlein HR, Eddes JS, Reid GE, Moritz RL, Simpson RJ. Disulfide Bond Structure and N-Glycosylation Sites of the Extracellular Domain of the Human Interleukin-6 Receptor. J Biol Chem. 1999;274:7207–15. doi: 10.1074/jbc.274.11.7207. [DOI] [PubMed] [Google Scholar]
- 49.Merewether LA, Le J, Jones MD, Lee R, Shimamoto G, Lu HS. Development of Disulfide Peptide Mapping and Determination of Disulfide Structure of Recombinant Human Osteoprotegerin Chimera Produced in Escherichia Coli. Arch Biochem Biophys. 2000;375:101–10. doi: 10.1006/abbi.1999.1636. [DOI] [PubMed] [Google Scholar]
- 50.Pitt JJ, Da Silva E, Gorman JJ. Determination of the Disulfide Bond Arrangement of Newcastle Disease Virus Hhemagglutinin Neuraminidase. Correlation with a Beta-Sheet Propeller Structural Fold Predicted for Paramyxoviridae Attachment Proteins. J Biol Chem. 2000;275:6469–78. doi: 10.1074/jbc.275.9.6469. [DOI] [PubMed] [Google Scholar]
- 51.Wallis TP, Huang CY, Nimkar SB, Young PR, Gorman JJ. Determination of the Disulfide Bond Arrangement of Dengue Virus NS1 Pprotein. J Biol Chem. 2004;279:20729–41. doi: 10.1074/jbc.M312907200. [DOI] [PubMed] [Google Scholar]
- 52.Sali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 53.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. Engineered Disulfide Bonds Support the Functional Rotation Mechanism of Multidrug Efflux Pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.