Neary 27 years after the first reported cases of the acquired immunodeficiency syndrome (AIDS) and 25 years after the discovery of the etiologic agent, effective control of the AIDS pandemic remains elusive. At the root of this challenge is the molecular pathogenesis of human immunodeficiency virus (HIV) type 1 (HIV-1), a virus that has evolved a number of mechanisms to elude immune control. Among the most prominent of these are the heavy glycosylation of the external glycoprotein, which protects neutralization epitopes; the virus’ direct targeting of the CD4 molecule expressed by the key T lymphocyte in immune orchestration; integration into the host-cell genome, which implies that cells that are not killed are infected permanently; and the potential of the virus to mutate and therefore evade the host immune system (mutational escape).1,2 This last mechanism results in a remarkable degree of viral diversity within HIV-1 and its rapid adaptation, in response to both immune activity and antiretroviral therapy. Over the past decade, advances in sequencing technology and expanded disease surveillance have allowed researchers to characterize the variation in HIV-1 within individual patients and around the world.
The initial view that the virus is classifiable into distinct subtypes or clades now needs to reflect the reality of a dynamic genetic evolutionary process, through which new HIV-1 strains are constantly emerging. The resultant viral diversity has implications for possible differential rates of disease progression, responses to antiretroviral therapy (including the development of resistance), and vaccine development.
ORIGIN OF HIV AND MECHANISMS OF HIV DIVERSITY
The origin of HIV-1 among nonhuman primates has been traced to a simian virus, SIVcpz, which infected several geographically isolated chimpanzee communities in southern Cameroon. This HIV-1 progenitor probably was passed from chimpanzees to human hunters through bloodborne transmission. Phylogenetic analysis of HIV-1 and related viruses from nonhuman primates suggests that three independent transmission events early in the 20th century spawned three HIV-1 groups: major (M, between 1915 and 1941), outlier (O), and nonmajor and nonoutlier (N).3,4 Although strains related to the M and N groups have been found in chimpanzees, recent evidence suggests that group O HIV-1 may have originated in gorillas, in which the closest relatives of this group have been identified.5 It is speculated that the virus then spread among humans along the Congo River into Kinshasa, Zaire, where the earliest documented case of HIV-1 infection (with group M strain) in humans has been traced to a blood sample from 1959.6
HIV has several intrinsic mechanisms that ensure rapid viral evolution. The reverse transcriptase of HIV lacks proofreading activity, the ability to confirm that the DNA transcript it makes is an accurate copy of the RNA code, and confers a mutation rate of approximately 3.4×10−5 mutations per base pair per replication cycle. Since the HIV genome is an estimated 104 base pairs in length and the baseline rate of viral production is approximately 1010 virions per day, millions of viral variants are produced within any infected person in a single day.7 HIV-1 recombination can lead to further viral diversity and occurs when one person is coinfected with two separate strains of the virus that are multiplying in the same cell (Fig. 1).8,9
Figure 1. Evolution of Diversity in HIV-1 during the Typical Viral Life Cycle and Creation of Unique Recombinant Forms in the Context of Coinfection with Two Subtypes.
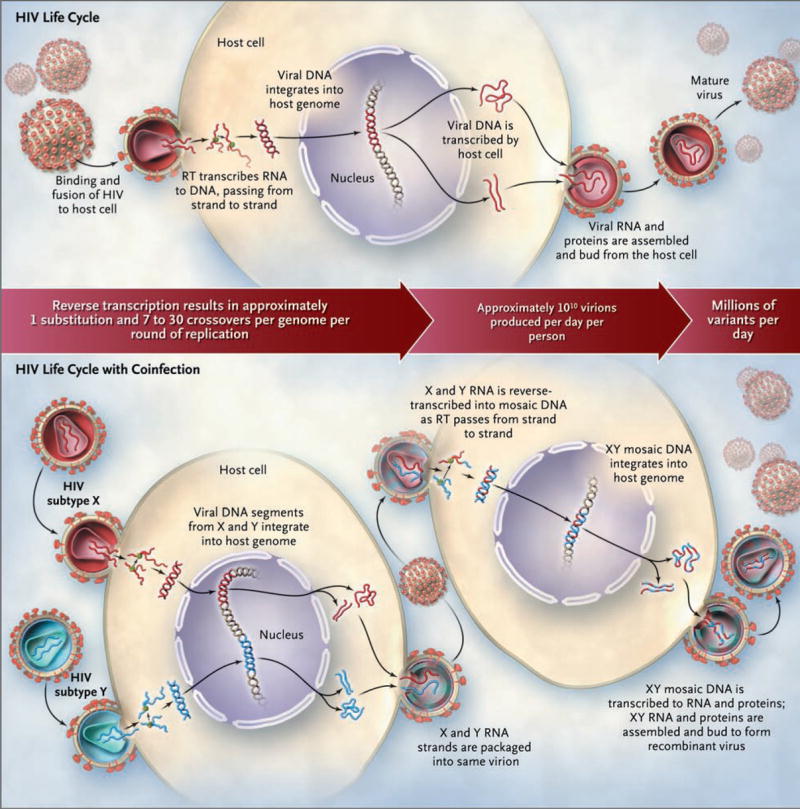
RT denotes reverse transcriptase.
CLASSIFICATION AND MOLECULAR EPIDEMIOLOGY OF HIV
Group M is the predominant circulating HIV-1 group. It has been divided into subtypes, denoted with letters, and sub-subtypes, denoted with numerals. Subtypes A1, A2, A3, A4, B, C, D, F1, F2, G, H, J, and K are currently recognized. HIV-1 subtypes, also called clades, are phylogenetically linked strains of HIV-1 that are approximately the same genetic distance from one another; in some cases, subtypes are also linked geographically or epidemiologically. Genetic variation within a subtype can be 15 to 20%, whereas variation between subtypes is usually 25 to 35%.10 Over the past decade, advances in full-genome sequencing of HIV have led to the identification of circulating and unique recombinant forms (CRFs and URFs, respectively). These are the result of recombination between subtypes within a dually infected person, from whom the recombinant forms are then passed to other people. The recombinant progeny are classified as circulating recombinant forms if they are identified in three or more people with no direct epidemiologic linkage; otherwise they are described as unique recombinant forms (Table 1).11
Table 1.
Phylogenetic Classifications of HIV-1.
| Classification | Definition | Examples |
|---|---|---|
| Subtypes or clades | Genetically related HIV-1 strains that are essentially phylogenetically equidistant, generating a starlike, rather than a treelike, phylogeny | Subtypes A, B, C, D, F, G, H, J, and K are currently known; A through D are highly prevalent, others have low prevalence and limited geographic distributions |
| Sub-subtypes | Distinct lineages within a subtype; genetic distance between sub-subtypes is smaller than that between subtypes | Subtypes A and F are subdivided into sub-subtypes A1 through A4 and F1 and F2, respectively; mostly these circulate in Central and West Africa |
| Intersubtype recombinant forms | Mosaic strains with segments from two or more subtypes alternating across the genome | Common in mixed-subtype epidemics; thought to result from infection of a person with more than one HIV-1 subtype |
| Circulating recombinant forms | Specific recombinant forms that are spreading in a population; new forms are defined when three people without direct epidemiologic linkage are found to be infected; the assigned name reflects sequence of discovery and subtype composition, with “cpx” indicating forms containing three or more subtypes | Currently, 43 forms are described; CRF01_AE and CRF02_AG are found principally in Southeast Asia and West Africa, respectively; others have more limited distributions |
| Unique recombinant forms | Intersubtype recombinant forms recovered from only a single person | Hundreds of forms have been described on the basis of partial or complete genome sequences; their potential for epidemic spread is unknown |
| Geographically distinct lineages | Lineages, often country-specific, that are distinguishable phylogenetically; unlike sub-subtypes, they are not phylogenetically equidistant within subtypes | Thai B, Indian C, West vs. East African D, and Former Soviet Union A (FSU-A) |
These definitions have been evolving over the past decade. Nomenclature in the published literature varies, and certain subtypes were found to be more complex after their full genomes have been sequenced. For example, what was previously described as subtype “E,” circulating in Southeast Asia, proved to be a circulating recombinant form containing components of subtype A and was redefined as CRF01_AE in 1998.11–13
The global distribution of subtypes and circulating recombinant forms reflects the complexity of the molecular epidemiology of HIV-1 (Fig. 2). The CRF01_AE virus was first identified in Thai-land in the late 1980s.12,14 This strain and its close relatives in Central Africa have had very different fates. CRF01_AE dominates in Southeast Asia, whereas in Africa, this circulating recombinant form remains relatively rare.15,16 Two new circulating recombinant forms, combining the Thai B and Indian C strains in related but distinct mosaic structures, emerged in southern China among injection-drug users and spread, along various drug-trafficking routes, across the country.17,18 A subtype A strain of low diversity, and a new circulating recombinant form derived from it, CRF03_AB, emerged in the former Soviet Union.19,20 More recently, CRF14_BG arose among injection-drug users in Spain and Portugal and has continued to spread.21 Subtype F, rare in Central Africa, emerged in South America in the form of BF recombinant strains; the subtype F parent of these recombinants has never gained significant prevalence.22,23 Finally, the HIV-1 epidemics in Afghanistan and Iran, fueled principally by the use of injection drugs, were shown to be linked: the newly emerged CRF35_AD strain was implicated in both.24,25
Figure 2.
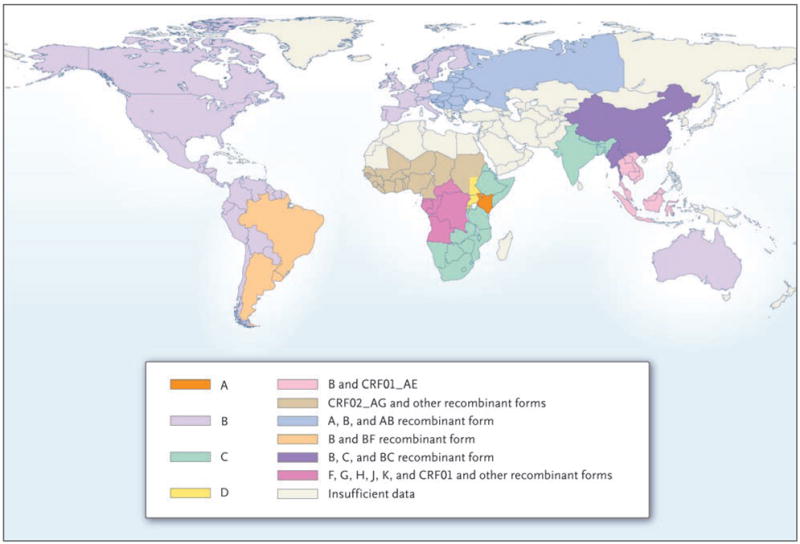
Current Global Distribution of HIV-1 Subtypes and Recombinant Forms.
CORECEPTOR USE BY HIV-1 SUBTYPES, TRANSMISSION, AND DISEASE PROGRESSION
Differential characteristics of viral subtypes and their interactions with the human host may influence HIV transmission and disease progression. The HIV strains capable of using the chemokine coreceptor CCR5 (R5 viruses) are more frequently transmitted than strains that use the CXCR4 co-receptor (X4 viruses); X4 viruses emerge later in infected patients and are associated with more rapid disease progression.26 All HIV-1 subtypes can use both coreceptors, but subtype D may be dualtropic (i.e., an R5X4 virus) most frequently.27 The percentage of X4 virus appears to be lower in subtype C than in subtype B, even when the viruses are obtained from patients with advanced AIDS.28
There are suggestions in the published literature that HIV-1 subtype or CRF may affect efficiency of transmission. Early data on mother-to-child transmission implied that subtype C was transmitted more frequently than subtype B.29 Pregnant women in Kenya infected with subtype C were more likely than those infected with subtype A or D to shed HIV-1–infected vaginal cells, implying that sexual transmission may be more likely with this subtype.30 A study in a longitudinal cohort of injection-drug users in Thailand conducted from 1995 through 1998 found an increased probability of transmission of CRF01_AE as compared with subtype B, though it was unclear whether epidemiologic, virologic, or host factors were affecting viral spread.31 An examination of subtype distribution between 1986 and 2000 in Kenya did not show an increase in the prevalence of subtype C; instead, an increase in the number of recombinant viruses was found.32
Another important question is whether subtype differences result in variable rates of disease progression. There have been several prospective, observational studies of the course of HIV-related disease in cohorts infected with various subtypes. An early study was published in 1999 by Kanki et al.,33 who had examined subtypes in 54 female sex workers in Senegal who were infected with HIV-1. Subtypes A, C, D, and G were represented, and the likelihood of AIDS developing was increased by a factor of eight among women infected with a non-A subtype as compared with those infected with subtype A. The subtypes were determined on the basis of the standard at the time, envelope-gene subtyping only; thus, further genetic complexity may have been present.
Subsequent studies have reported discordant results. In a large Swedish study of patients infected with subtype A, B, C, or D, disease progression did not differ significantly according to subtype or ethnic group.34 A second prospective multicenter study in Western and West-Central Africa did not show a significant difference in survival or clinical disease progression among people infected with CRF02_AG, as compared with those infected with other CRFs or subtypes.35 A survival study of 836 Thai heterosexual men and women infected with CRF01_AE showed a shorter time from HIV-1 infection to death than among those in Western populations.36
A cohort of 1045 Ugandans did show a faster progression to death among people infected with subtype D than among those infected with subtype A, even after controlling for CD4+ count at enrollment.37 Data from the Rakai cohort in Uganda also suggest that HIV-1 disease progresses more rapidly, and that the risk of death is greater, among persons infected with subtype D, with recombinant forms, or with multiple subtypes than among those infected with subtype A.38 A recent study of a Kenyan cohort showed that 21 patients infected with subtype D had a higher mortality rate and a faster decline in CD4+ count than those infected with subtype A or C.39 The propensity of subtype D to exhibit a greater degree of dualtropism than other subtypes27 may help to explain the observation that subtype D appears to be associated with a more rapid rate of disease progression than other HIV-1 subtypes. The notable caveat relevant to all these studies of disease progression is that confounders such as access to medical care, nutritional status, host genetic factors, and mode of viral transmission (e.g., sexual, injection-drug, or vertical) may contribute to the divergent results.
Interactions between the host and HIV-1 that vary according to subtype may also be important. The known differences in HIV-1 transmission and disease progression in hosts carrying specific HLA class I types may vary according to infecting HIV-1 subtype (Table 2). In an infected person, T cells specific to HIV-1 can exhibit cross-subtype specificity and recognize viral epitopes within subtypes other than the one that generated the initial response.52,53 CD8+ T cells obtained from persons infected with subtype B recognize viral epitopes within conserved regions of the consensus sequences from genomes of subtypes A, B, and C. However, the immune response tends to be greatest against the infecting subtype, and CD8+ T-cell responses can wane over time. Despite some similarities in T-cell responses, there may be intersubtype differences in the plasma HIV-1 viral load levels after in vivo infection, though data thus far are conflicting.54,55 This is an important area of investigation because of the well-described link between viral load and transmission and the rate of disease progression.56
Table 2.
Features of the HIV-1 Pandemic, According to Subtype or Circulating Recombinant Form (CRF).*
| Subtype or CRF | Location | Global Prevalence | Tropism and Replication | Disease Progression | Response to Therapy |
|---|---|---|---|---|---|
| Subtype | |||||
| A | East and Central Africa, Central Asia, Eastern Europe | 12.3% | Mostly uses CCR5, even in late infection40 | NA | No significant difference as compared with C and D41 |
| B | Americas, Western Europe, East Asia, Oceania | 10.2% | Uses CCR5 early, with increasing use of CXCR4 in late infection28 | HLA-B7 associated with poor CTL response and increased viremia42,43; HLA-B57 associated with slow progression42; B strain in Brazil associated with slow progression44 | NA |
| C | India, Eastern and Southern Africa | 49.9% | Mostly uses CCR5, even in late infection28; increased vaginal shedding30 and mother-to-child transmission29,45 | HLA-B57 associated with slow progression42 | No significant difference as compared with A and D41; differential pathways to resistance46–49 |
| D | East Africa | 2.5% | Uses CXCR4 in early infection27 | Progression more rapid than A in Uganda, Kenya, and Tanzania37–39 | NA |
| G | West Africa | 6.3% | NA | NA | NA |
| F, H, J, and K | Various | Each <1.0% | NA | NA | NA |
| CRF | |||||
| CRF01_AE | Southeast Asia | 4.7% | May have higher initial viral load than B but subtype may be a confounder50 | Possibly accelerated progression as compared with B36 | NA |
| CRF02_AG | West Africa | 4.8% | Higher rate of replication in vitro than B51 | NA | NA |
| Other | Various | Each <0.1% | NA | NA | NA |
Location and prevalence data are from Hemelaar et al.10 Other CRFs include CRF03 through CRF43, and this category is expanding. CTL denotes cytotoxic T lymphocyte, and NA not available.
RESPONSE TO THERAPY
Does HIV-1 subtype influence the response to antiretroviral treatment? This question is urgent, since only 12% of global infections are caused by the most studied subtype, B; and 50% of prevalent HIV infections and 47% of all new HIV-1 infections are with subtype C.10 This discrepancy in the availability of clinical data for non-B subtypes is exacerbated by the fact that, until the past few years, antiretroviral treatment had been largely unavailable in many countries with non-B subtypes of HIV-1.
Initial data from treatment cohorts in Africa raise two concerns: first, that certain subtypes of HIV-1 might spread or progress more rapidly than others, making treatment decisions more challenging,37 and second, that the data on baseline antiretroviral susceptibility derived from studies of subtype B may not be applicable to non-B subtypes.57 This concern is illustrated by HIV type 2 and group O strains of HIV-1, which possess intrinsic resistance to nonnucleoside reverse-transcriptase inhibitors.58,59
Though there are potential problems with comparing responses to therapy among persons infected with group M, non-B–subtype strains — who frequently live in settings with limited resources — and those infected with subtype B, the data available thus far are encouraging. In 2002, Alexander et al.60 published data from a retrospective cohort of 485 patients receiving antiretroviral treatment in British Columbia, Canada, 4.4% of whom were infected with non-B subtypes of HIV-1. Though initial CD4+ counts were lower in the patients infected with non-B subtypes than in those infected with subtype B, the proportion of patients with an HIV-1 RNA viral load of less than 400 copies per milliliter at 18 months did not differ significantly between the two groups. A French cohort study of 416 patients, 24% of whom carried non-B subtypes of HIV-1, showed that at 3, 6, and 12 months after initiation of antiretroviral therapy, HIV-1 subtype did not affect clinical progression, CD4+ count, or viral load in response to treatment.61
In their study of patients of African origin who were infected with a non-B subtype of HIV-1 and were living in London, Frater et al.41 found no significant difference in the response to therapy among patients infected with subtype A, those infected with subtype C, and those infected with subtype D. In the Paediatric European Network for Treatment of AIDS (PENTA) 5 trial,62 there was no significant difference according to HIV-1 subtype in the virologic response to treatment or in the frequency of development of resistance among children. Overall, it appears that HIV-1 subtypes do not effect major differences in the response to antiretroviral therapy. What has emerged is a growing body of evidence that polymorphisms found in various subtypes before antiretroviral therapy is begun may affect genetic pathways of resistance.
EMERGENCE OF RESISTANCE TO ANTIRETROVIRAL THERAPY
HIV resistance to antiretroviral therapy can be divided into two categories: primary resistance, which reflects acquisition of a drug-resistant strain of HIV by a newly infected person; and secondary, or acquired, resistance, which develops after a period of HIV treatment. Not surprisingly, studies of non-B subtypes in patients who have never received antiretroviral therapy reveal that protease and reverse transcriptase sequences vary from those of the subtype B reference strains in a somewhat predictable manner.63 For example, polymorphisms in the reverse transcriptase gene do not typically occur in known sites of resistance to nucleoside reverse-transcriptase inhibitors.64,65 In contrast, data from several studies indicate that protease sequences from non-B subtypes in patients who have never received antiretroviral therapy contain amino acid substitutions associated with mutations in subtype B known to contribute to secondary resistance, including K20 → R, M36 → I, and H69 → K/Q.66,67 However, these genotypic changes do not confer consistently decreased susceptibility by themselves when viral strains are subjected to phenotypic testing (Fig. 3 and Table 3).65,66 For the fusion inhibitor enfuvirtide, substantial differences in resistance-associated mutations between B and non-B subtypes have not been found, but some polymorphisms, such as N42 → S in the heptadrepeat region 1 of glyco-protein 41, appear to be more common in non-B subtypes.79 Data on in vitro resistance against maraviroc, the recently approved entry inhibitor and CCR5 antagonist, do not suggest that there are differences among subtypes,80 but the differences in coreceptor tropism noted above lead to concern that intersubtype variation in response to therapy could exist in vivo.
Figure 3. Predominant Amino Acid (AA) Changes Conferred by Polymorphisms in HIV-1 Protease, According to Subtype.
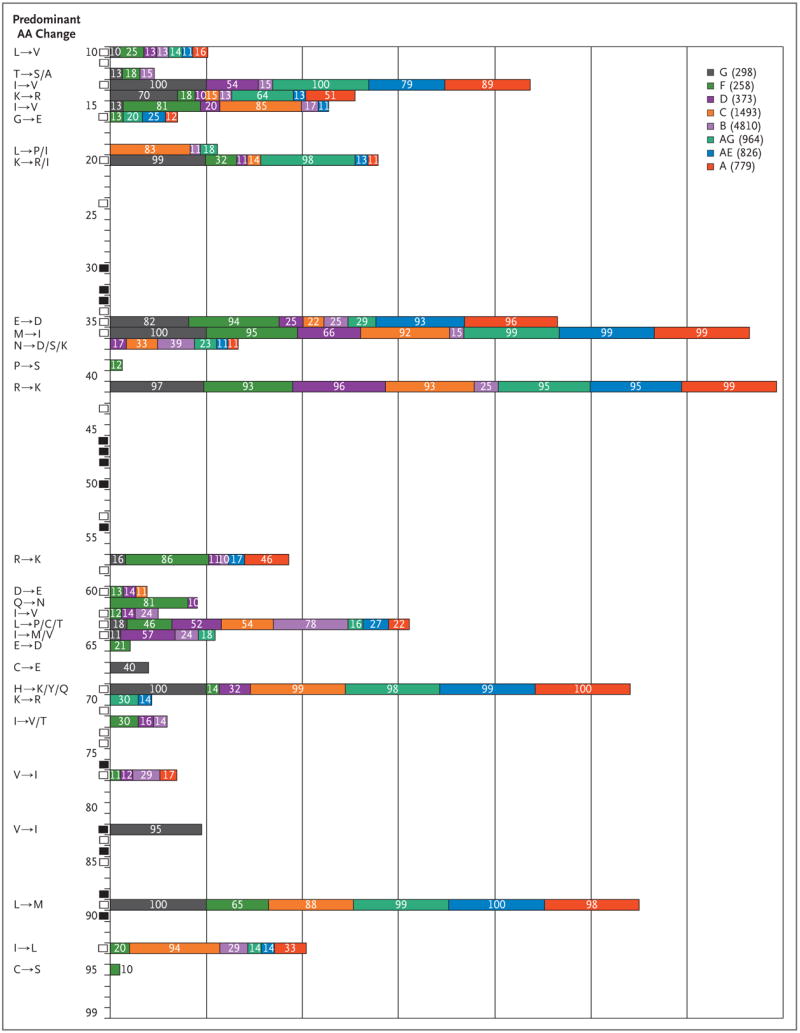
AA sequences for HIV-1 protease were compared with a consensus subtype B sequence. Polymorphism data were obtained in May 2007 with the use of the HIVseq program from the Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu/pages/algs/HIVseq.html). Polymorphisms in AAs 10 through 99 of the HIV-1 protease are shown for a given subtype if 10% or more of the sequences in the database were polymorphic at that site; the percentage of sequences that were polymorphic appear within the colored bars. The subtype is shown in the key, followed by the number of sequences in the analysis in parentheses. Black rectangles indicate the sites of major protease resistance mutations, and white rectangles indicate the sites of minor protease resistance mutations, according to the International AIDS Society–USA in 2007. The predominant AA changes at each site are shown on the left-hand side. Polymorphisms in HIV-1 reverse transcriptase AAs 40 through 240 are shown in the Supplementary Appendix, available with the full text of this article at www.nejm.org.
Table 3.
Details of Selected Studies of Mutations Conferring Primary Resistance to Antiretroviral Therapy.*
| Study | Location and Year | No. of Subjects | Prevalence of Subtypes or CRFs | Prevalence of Known Resistance Mutations |
|---|---|---|---|---|
| Nkengafac et al.68 | Cameroon, 2005–2006 | 180 | B (42%), G (29%), other (29%) | 8% |
| Palma et al.69 | Portugal, 2003 | 180 | B (42%), G (29%), other (29%) | 8% |
| Tee et al.70 | Malaysia, 2003–2004 | 100 | CRF01_AE (65%), AE/B (22%), B (12%) | 1% |
| Paraskevis et al.71 | Greece, 2002–2003 | 101 | B (48%), A (33%) | 9% |
| Ly et al.72 | Cambodia, 2003–2004 | 144 | CRF01_AE (97%) | 5% |
| Descamps et al.73 | France, 2001–2002 | 662 | B (71%), non-B (29%) | 12% in patients with acute infection, 2% in those with chronic B infection, lower in those with non-B infection |
| Wensing et al.74 | Europe, 1996–2002 | 2208 | B (70%), C (10%) | 13% in patients infected with B, 5% in those infected with non-B |
| Vazquez de Parga et al.75 | Former Soviet Union, 1997–2004 | 278 | A (80%) | 13% for reverse transcrip tase mutations, 4% for protease mutations |
| Roudinskii et al.76 | Former Soviet Union, 1995–2003 | 119 | A (97%) | None detected |
| Grossman et al.49 | Israel, 1999–2003 | 117 | C (75%), B (25%) | 50% in patients infected with C, 10% in those infected with B |
| Deshpande et al.77 | India, 2003 | 128 | C (96%) | <2% |
| Maljkovic et al.78 | Sweden, 1998–2001 | 100 | B (55%), C (29%) | 9% |
Studies that included at least 100 isolates were selected during a literature review in May 2007. Minor protease mutations were not included in the calculation of the prevalence of known resistance mutations. CRF denotes circulating recombinant form.
Studies of resistance patterns that emerge in non-B subtypes in patients receiving antiretroviral therapy indicate that polymorphisms present in these subtypes before therapy may provide a background for the emergence of subtype-specific pathways to secondary resistance. Mutations leading to resistance appear to be similar among subtypes, but certain mutations seem to occur more frequently in non-B subtypes in particular. In a study of patients in Botswana infected with subtype C, in those without a response to a didanosine- or stavudine-based regimen, virus containing K65 → R mutations developed within 8 months, more rapidly than is seen in patients infected with subtype B.41 When exposed to tenofovir in culture, subtype C developed K65 → R mutations more rapidly than other subtypes,47 but data from clinical trials remain inconclusive.48 Subtype C viruses also develop resistance against nonnucleoside reverse-transcriptase inhibitors through either the K103 → N or V106 → M mutations, whereas subtype B viruses rarely develop V106 → M mutations.49 Nelfinavir resistance appears to occur primarily through L90 → M mutations in subtypes G and C and other non-B subtypes, whereas subtype B acquires either D30 → N or L90 → M nelfinavir-resistance mutations.81,82 Overall, it appears that most antiretroviral resistance in non-B subtypes is accounted for within the current resistance databases.83 Further studies of treated cohorts infected with non-B HIV-1 are needed to determine whether other subtype-specific pathways to resistance exist (Table 3).
IMPLICATIONS FOR VACCINE DEVELOPMENT
Ultimate control of the HIV-1 pandemic is dependent on the development of an effective, preventive vaccine (see the recent review by Johnston and Fauci84). Among the many challenges to achieving this goal, one of the greatest is HIV-1 diversity — reflected by the presence of HIV-1 subtypes, circulating recombinant forms, and continuous viral evolution within populations and individual hosts. Hosts infected with HIV-1 have cellular and humoral immune responses to their infecting strains, but there is evidence of mutational escape by viruses from responses by CD8+ cytotoxic T cells and neutralizing antibodies over time.85,86 Although cross-reactive responses to other viral subtypes have been shown,53,87 the strength and breadth of these responses are typically limited.52 Several researchers have developed multisubtype consensus sequences or expression cassettes and have coadministered vaccines and selected cytokines in attempts to increase the breadth and strength of the cyto-toxic T-cell response.88,89 Critical to the development of a successful HIV-1 vaccine will be our ability to decipher the genetic diversity of the virus, elicit broadly neutralizing antibodies, and generate strong CD4+ and CD8+ T-cell responses.90,91
Development of a vaccine that induces neutralizing antibodies that bind to the trimeric envelope on the surface of the virus remains a great challenge. In two large, phase 3 trials of a monomeric form of the external glycoprotein 120, conducted in the United States92 and Thailand,93 the protein failed to protect healthy subjects from HIV infection. Some current approaches to the design of a neutralizing immunogen are to mimic glyco-protein 120–glycoprotein 41 envelope trimers on the virion surface, to produce envelope molecules with enhanced expression of neutralizing epitopes and thereby improve their relative immunogenicity, and to remove or mask the variable regions and expose conserved epitopes to focus the immune response.84,91
There has been progress as well as challenges in the development of vaccines that induce T-cell–mediated immune responses that may not prevent HIV-1 infection but may modulate the viral load and subsequent disease progression in people who do become infected.94 The diversity of T-cell epitopes makes it unlikely that the use of one natural isolate for a vaccine will protect against other viral subtypes or variants within the same subtype. Many strategies are being pursued to confront this problem, including the use of consensus sequences, the deployment of a combination of immunogens from different subtypes, the creation of mosaic immunogens assembled through computational optimization from pieces of natural sequences, and the construction of multisubtype immunogens derived from conserved regions of the HIV-1 consensus proteome.95
Two advanced approaches to the development of T-cell vaccines, both of which use a recombinant adenovirus type 5 vector, illustrate different strategies to address the challenge of HIV-1 subtype diversity. The first, developed by Merck Research Laboratories, involved the immunization of HIV-1–seronegative persons with a recombinant adenovirus type 5 vector containing gag, pol, and nef genes from subtype B. An unexpected development for the HIV vaccine field occurred in September 2007: a data and safety monitoring board review for the phase 2B, test-of-concept, efficacy trial of this product in the Americas (the Step study [HIV Vaccine Trials Network study 502, Merck protocol 023]) recommended that vaccinations in this trial be stopped, since statistical criteria for futility had been met. Vaccinations were also discontinued in a sister, phase 2B study, with partial enrollment, of the same product in South Africa (HIV Vaccine Trials Network study 503). Data released subsequently also raised the issue of whether there was an increased risk of HIV-1 acquisition conferred by the vaccine in persons with preexisting immunity to adenovirus type 5.96,97
The second approach is illustrated by the preventive vaccine regimen being developed by the Vaccine Research Center of the National Institute of Allergy and Infectious Diseases. This regimen involves a DNA prime, recombinant adenovirus type 5 boost with gag, pol, and nef genes derived from subtype B and envelope genes from subtypes A, B, and C; thus, it is a multigene, multisubtype vaccine.98,99 The regimen is currently in phase 1–2 testing, and phase 2B, preliminary efficacy testing is under consideration. Ultimately, a fully effective, preventive vaccine regimen will probably need to induce strong, cross-subtype HIV-specific T-cell immunity as well as broadly reactive, neutralizing antibody activity to overcome the challenge of HIV diversity.
CONCLUSIONS
With the continuing spread of HIV, the world faces a pandemic of unprecedented genetic and geographic complexity. Five subtypes and two circulating recombinant forms have each established a global prevalence greater than 2.5%, a level that virtually ensures their continued presence in the decades to come. Factors that influence the spread of particular subtypes or circulating recombinant forms in different geographic regions are incompletely understood. Mutation and recombination, both essential features of the HIV replication cycle, are major forces driving diversity. Only through a deeper understanding of this diversity and its implications for HIV prevention, vaccine development, and antiretroviral therapy will we be able to end the pandemic.
Acknowledgments
Supported by grants from the National Institutes of Health (training grant T32 A149821-06, to Dr. Taylor; and Clinical Trials Unit Grant UO1 AI069470-01, to Drs. Sobieszczyk and Hammer).
Footnotes
Dr. Hammer reports receiving consulting fees from Merck, Progenics, and Wyeth and serving on a data and safety monitoring board for a clinical trial sponsored by Bristol-Myers Squibb. No other potential conflict of interest relevant to this article was reported.
References
- 1.Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–98. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frost SD, Wrin T, Smith DM, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A. 2005;102:18514–9. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keele BF, Van Heuverswyn F, Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–6. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korber B, Muldoon M, Theiler J, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–96. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 5.Van Heuverswyn F, Li Y, Neel C, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 6.Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–7. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 7.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–6. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 8.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374:124–6. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 9.Blackard JT, Cohen DE, Mayer KH. Human immunodeficiency virus superinfection and recombination: current state of knowledge and potential clinical consequences. Clin Infect Dis. 2002;34:1108–14. doi: 10.1086/339547. [DOI] [PubMed] [Google Scholar]
- 10.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 11.Leitner T, Foley B, Hahn B, et al. HIV sequence compendium 2005. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 2005. [Google Scholar]
- 12.Carr JK, Salminen MO, Koch C, et al. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–43. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao F, Robertson DL, Morrison SG, et al. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–29. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCutchan FE, Hegerich PA, Brennan TP, et al. Genetic variants of HIV-1 in Thailand. AIDS Res Hum Retroviruses. 1992;8:1887–95. doi: 10.1089/aid.1992.8.1887. [DOI] [PubMed] [Google Scholar]
- 15.Tovanabutra S, Beyrer C, Sakkhachornphop S, et al. The changing molecular epidemiology of HIV type 1 among northern Thai drug users, 1999 to 2002. AIDS Res Hum Retroviruses. 2004;20:465–75. doi: 10.1089/088922204323087705. [DOI] [PubMed] [Google Scholar]
- 16.Kijak GH, Tovanabutra S, Sanders-Buell E, et al. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v. 2. Virology. 2007;358:178–91. doi: 10.1016/j.virol.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Piyasirisilp S, McCutchan FE, Carr JK, et al. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J Virol. 2000;74:11286–95. doi: 10.1128/jvi.74.23.11286-11295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCutchan FE, Carr JK, Murphy D, et al. Precise mapping of recombination breakpoints suggests a common parent of two BC recombinant HIV type 1 strains circulating in China. AIDS Res Hum Retroviruses. 2002;18:1135–40. doi: 10.1089/088922202320567879. [DOI] [PubMed] [Google Scholar]
- 19.Nabatov AA, Kravchenko ON, Lyulchuk MG, Shcherbinskaya AM, Lukashov VV. Simultaneous introduction of HIV type 1 subtype A and B viruses into injecting drug users in southern Ukraine at the beginning of the epidemic in the former Soviet Union. AIDS Res Hum Retroviruses. 2002;18:891–5. doi: 10.1089/08892220260190380. [DOI] [PubMed] [Google Scholar]
- 20.Saad MD, Aliev Q, Botros BA, et al. Genetic forms of HIV type 1 in the former Soviet Union dominate the epidemic in Azerbaijan. AIDS Res Hum Retroviruses. 2006;22:796–800. doi: 10.1089/aid.2006.22.796. [DOI] [PubMed] [Google Scholar]
- 21.Delgado E, Thomson MM, Villahermosa ML, et al. Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J Acquir Immune Defic Syndr. 2002;29:536–43. doi: 10.1097/00126334-200204150-00016. [DOI] [PubMed] [Google Scholar]
- 22.Thomson MM, Delgado E, Herrero I, et al. Diversity of mosaic structures and common ancestry of human immunodeficiency virus type 1 BF intersubtype recombinant viruses from Argentina revealed by analysis of near full-length genome sequences. J Gen Virol. 2002;83:107–19. doi: 10.1099/0022-1317-83-1-107. [DOI] [PubMed] [Google Scholar]
- 23.Thomson MM, Sierra M, Tanuri A, et al. Analysis of near full-length genome sequences of HIV type 1 BF intersubtype recombinant viruses from Brazil reveals their independent origins and their lack of relationship to CRF12_BF. AIDS Res Hum Retroviruses. 2004;20:1126–33. doi: 10.1089/aid.2004.20.1126. [DOI] [PubMed] [Google Scholar]
- 24.Naderi HR, Tagliamonte M, Tornesello M, et al. Molecular and phylogenetic analysis of HIV-1 variants circulating among injecting drug users in Mashhad-Iran. Infect Agent Cancer. 2006;1:4. doi: 10.1186/1750-9378-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders-Buell E, Saad MD, Abed AM, et al. A nascent HIV type 1 epidemic among injecting drug users in Kabul, Afghanistan is dominated by complex AD recombinant strain, CRF35_AD. AIDS Res Hum Retroviruses. 2007;23:834–9. doi: 10.1089/aid.2006.0299. [Erratum, AIDS Res Hum Retroviruses 2007;23:953–4.] [DOI] [PubMed] [Google Scholar]
- 26.Berger EA, Doms RW, Fenyö EM, et al. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 27.Huang W, Eshleman SH, Toma J, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81:7885–93. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cilliers T, Nhlapo J, Coetzer M, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol. 2003;77:4449–56. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renjifo B, Gilbert P, Chaplin B, et al. Preferential in-utero transmission of HIV-1 subtype C as compared to HIV-1 subtype A or D. AIDS. 2004;18:1629–36. doi: 10.1097/01.aids.0000131392.68597.34. [DOI] [PubMed] [Google Scholar]
- 30.John-Stewart GC, Nduati RW, Rousseau CM, et al. Subtype C is associated with increased vaginal shedding of HIV-1. J Infect Dis. 2005;192:492–6. doi: 10.1086/431514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudgens MG, Longini IM, Jr, Vanichseni S, et al. Subtype-specific transmission probabilities for human immunodeficiency virus type 1 among injecting drug users in Bangkok, Thailand. Am J Epidemiol. 2002;155:159–68. doi: 10.1093/aje/155.2.159. [DOI] [PubMed] [Google Scholar]
- 32.Rainwater S, DeVange S, Sagar M, et al. No evidence for rapid subtype C spread within an epidemic in which multiple subtypes and intersubtype recombinants circulate. AIDS Res Hum Retroviruses. 2005;21:1060–5. doi: 10.1089/aid.2005.21.1060. [DOI] [PubMed] [Google Scholar]
- 33.Kanki PJ, Hamel DJ, Sankalé JL, et al. Human immunodeficiency virus type 1 subtypes differ in disease progression. J Infect Dis. 1999;179:68–73. doi: 10.1086/314557. [DOI] [PubMed] [Google Scholar]
- 34.Alaeus A, Lidman K, Björkman A, Giesecke J, Albert J. Similar rate of disease progression among individuals infected with HIV-1 genetic subtypes A-D. AIDS. 1999;13:901–7. doi: 10.1097/00002030-199905280-00005. [DOI] [PubMed] [Google Scholar]
- 35.Laurent C, Bourgeois A, Faye MA, et al. No difference in clinical progression between patients infected with the predominant human immunodeficiency virus type 1 circulating recombinant form (CRF) 02_AG strain and patients not infected with CRF02_AG, in Western and West-Central Africa: a four-year prospective multicenter study. J Infect Dis. 2002;186:486–92. doi: 10.1086/341833. [DOI] [PubMed] [Google Scholar]
- 36.Costello C, Nelson KE, Suriyanon V, et al. HIV-1 subtype E progression among northern Thai couples: traditional and non-traditional predictors of survival. Int J Epidemiol. 2005;34:577–84. doi: 10.1093/ije/dyi023. [DOI] [PubMed] [Google Scholar]
- 37.Kaleebu P, French N, Mahe C, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–50. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- 38.Kiwanuka N, Laeyendecker O, Robb M, et al. Effect of human immunodeficiency virus type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, wtih incident HIV-1 infection. J Infect Dis. 2008;197:707–13. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 39.Baeten JM, Chohan B, Lavreys L, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195:1177–80. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 40.Laeyendecker O, Li X, Arroyo M, et al. The effect of HIV subtype on rapid disease progression in Rakai, Uganda. Presented at the 13th Conference on Retroviruses and Opportunistic Infections; Denver. February 5–8, 2006; abstract. [Google Scholar]
- 41.Frater AJ, Beardall A, Ariyoshi K, et al. Impact of baseline polymorphisms in RT and protease on outcome of highly active antiretroviral therapy in HIV-1-infected African patients. AIDS. 2001;15:1493–502. doi: 10.1097/00002030-200108170-00006. [DOI] [PubMed] [Google Scholar]
- 42.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 43.Tang J, Tang S, Lobashevsky E, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–84. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Brito A, Komninakis SC, Novoa P, et al. Women infected with HIV type 1 Brazilian variant, subtype B (B′-GWGR motif) have slower progression to AIDS, compared with patients infected with subtype B (B-GPGR motif) Clin Infect Dis. 2006;43:1476–81. doi: 10.1086/508875. [DOI] [PubMed] [Google Scholar]
- 45.Renjifo B, Fawzi W, Mwakagile D, et al. Differences in perinatal transmission among human immunodeficiency virus type 1 genotypes. J Hum Virol. 2001;4:16–25. [PubMed] [Google Scholar]
- 46.Doualla-Bell F, Avalos A, Brenner B, et al. High prevalence of the K65R mutation in human immunodeficiency virus type 1 subtype C isolates from infected patients in Botswana treated with didanosine-based regimens. Antimicrob Agents Chemother. 2006;50:4182–5. doi: 10.1128/AAC.00714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenner BG, Oliveira M, Doualla-Bell F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20(9):F9–F13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- 48.Miller MD, Margot N, McColl D, Cheng AK. K65R development among subtype C HIV-1-infected patients in tenofovir DF clinical trials. AIDS. 2007;21:265–6. doi: 10.1097/QAD.0b013e32801199ee. [DOI] [PubMed] [Google Scholar]
- 49.Grossman Z, Istomin V, Averbuch D, et al. Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS. 2004;18:909–15. doi: 10.1097/00002030-200404090-00008. [DOI] [PubMed] [Google Scholar]
- 50.Hu DJ, Vanichseni S, Mastro TD, et al. Viral load differences in early infection with two HIV-1 subtypes. AIDS. 2001;15:683–91. doi: 10.1097/00002030-200104130-00003. [DOI] [PubMed] [Google Scholar]
- 51.Njai HF, Gali Y, Vanham G, et al. The predominance of human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness. Retrovirology. 2006;3:40. doi: 10.1186/1742-4690-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKinnon LR, Ball TB, Kimani J, et al. Cross-clade CD8(+) T-cell responses with a preference for the predominant circulating clade. J Acquir Immune Defic Syndr. 2005;40:245–9. doi: 10.1097/01.qai.0000184858.16447.04. [DOI] [PubMed] [Google Scholar]
- 53.Brown SA, Slobod KS, Surman S, Zirkel A, Zhan X, Hurwitz JL. Individual HIV type 1 envelope-specific T cell responses and epitopes do not segregate by virus subtype. AIDS Res Hum Retroviruses. 2006;22:188–94. doi: 10.1089/aid.2006.22.188. [DOI] [PubMed] [Google Scholar]
- 54.Hu DJ, Subbarao S, Vanichseni S, et al. Higher viral loads and other risk factors associated with HIV-1 seroconversion during a period of high incidence among injection drug users in Bangkok. J Acquir Immune Defic Syndr. 2002;30:240–7. doi: 10.1097/00042560-200206010-00013. [DOI] [PubMed] [Google Scholar]
- 55.Gray CM, Williamson C, Bredell H, et al. Viral dynamics and CD4+ T cell counts in subtype C human immunodeficiency virus type 1-infected individuals from southern Africa. AIDS Res Hum Retroviruses. 2005;21:285–91. doi: 10.1089/aid.2005.21.285. [DOI] [PubMed] [Google Scholar]
- 56.Quinn TC, Wawer MJ, Sewankambo N, et al. Viral load and heterosexual transmission of human immunodeficiency virus type 1. N Engl J Med. 2000;342:921–9. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 57.Snoeck J, Kantor R, Shafer RW, et al. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob Agents Chemother. 2006;50:694–701. doi: 10.1128/AAC.50.2.694-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Descamps D, Collin G, Letourneur F, et al. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J Virol. 1997;71:8893–8. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuaillon E, Gueudin M, Lemée V, et al. Phenotypic susceptibility to nonnucleo-side inhibitors of virion-associated reverse transcriptase from different HIV types and groups. J Acquir Immune Defic Syndr. 2004;37:1543–9. doi: 10.1097/00126334-200412150-00001. [DOI] [PubMed] [Google Scholar]
- 60.Alexander CS, Montessori V, Wynhoven B, et al. Prevalence and response to antiretroviral therapy of non-B subtypes of HIV in antiretroviral-naive individuals in British Columbia. Antivir Ther. 2002;7:31–5. [PubMed] [Google Scholar]
- 61.Bocket L, Cheret A, Deuffic-Burban S, et al. Impact of human immunodeficiency virus type 1 subtype on first-line antiretroviral therapy effectiveness. Antivir Ther. 2005;10:247–54. [PubMed] [Google Scholar]
- 62.Pillay D, Walker AS, Gibb DM, et al. Impact of human immunodeficiency virus type 1 subtypes on virologic response and emergence of drug resistance among children in the Paediatric European Network for Treatment of AIDS (PENTA) 5 trial. J Infect Dis. 2002;186:617–25. doi: 10.1086/342680. [DOI] [PubMed] [Google Scholar]
- 63.Kantor R, Katzenstein D. Drug resistance in non-subtype B HIV-1. J Clin Virol. 2004;29:152–9. doi: 10.1016/S1386-6532(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 64.Bellocchi MC, Forbici F, Palombi L, et al. Subtype analysis and mutations to antiviral drugs in HIV-1-infected patients from Mozambique before initiation of antiretroviral therapy: results from the DREAM programme. J Med Virol. 2005;76:452–8. doi: 10.1002/jmv.20382. [DOI] [PubMed] [Google Scholar]
- 65.Deroo S, Robert I, Fontaine E, et al. HIV-1 subtypes in Luxembourg, 1983–2000. AIDS. 2002;16:2461–7. doi: 10.1097/00002030-200212060-00012. [DOI] [PubMed] [Google Scholar]
- 66.Holguin A, Paxinos E, Hertogs K, Womac C, Soriano V. Impact of frequent natural polymorphisms at the protease gene on the in vitro susceptibility to protease inhibitors in HIV-1 non-B subtypes. J Clin Virol. 2004;31:215–20. doi: 10.1016/j.jcv.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Grossman Z, Vardinon N, Chemtob D, et al. Genotypic variation of HIV-1 reverse transcriptase and protease: comparative analysis of clade C and clade B. AIDS. 2001;15:1453–60. doi: 10.1097/00002030-200108170-00001. [Erratum, AIDS 2001;15: 2209.] [DOI] [PubMed] [Google Scholar]
- 68.Nkengafac A, Tina S, Sua F, Mason T, Auyuketta N, Oben S. Molecular epidemiology and prevalence of drug resistance-associated mutations in newly-diagnosed HIV-1 patients in Cameroon. Antivir Ther. 2007;12(Suppl 1):S50. abstract. [Google Scholar]
- 69.Palma AC, Araujo F, Duque V, Borges F, Paixão MT, Camacho R. Molecular epidemiology and prevalence of drug resistance-associated mutations in newly diagnosed HIV-1 patients in Portugal. Infect Genet Evol. 2007;7:391–8. doi: 10.1016/j.meegid.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 70.Tee KK, Kamarulzaman A, Ng KP. Short communication: low prevalence of genotypic drug resistance mutations among antiretroviral-naive HIV type 1 patients in Malaysia. AIDS Res Hum Retroviruses. 2006;22:121–4. doi: 10.1089/aid.2006.22.121. [DOI] [PubMed] [Google Scholar]
- 71.Paraskevis D, Magiorkinis E, Katsoulidou A, et al. Prevalence of resistance-associated mutations in newly diagnosed HIV-1 patients in Greece. Virus Res. 2005;112:115–22. doi: 10.1016/j.virusres.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 72.Ly N, Recordon-Pinson P, Phoung V, et al. Characterization of mutations in HIV type 1 isolates from 144 Cambodian recently infected patients and pregnant women naive to antiretroviral drugs. AIDS Res Hum Retroviruses. 2005;21:971–6. doi: 10.1089/aid.2005.21.971. [DOI] [PubMed] [Google Scholar]
- 73.Descamps D, Chaix ML, André P, et al. French national sentinel survey of antiretroviral drug resistance in patients with HIV-1 primary infection and in antiretroviral-naive chronically infected patients in 2001–2002. J Acquir Immune Defic Syndr. 2005;38:545–52. doi: 10.1097/01.qai.0000155201.51232.2e. [DOI] [PubMed] [Google Scholar]
- 74.Wensing AM, van de Vijver DA, Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis. 2005;192:958–66. doi: 10.1086/432916. [Erratum, J Infect Dis 2005;192:1501.] [DOI] [PubMed] [Google Scholar]
- 75.Vázquez de Parga E, Rakhmanova A, Pérez-Alvarez L, et al. Analysis of drug resistance-associated mutations in treatment-naive individuals infected with different genetic forms of HIV-1 circulating in countries of the former Soviet Union. J Med Virol. 2005;77:337–44. doi: 10.1002/jmv.20461. [DOI] [PubMed] [Google Scholar]
- 76.Roudinskii NI, Sukhanova AL, Kazennova EV, et al. Diversity of human immunodeficiency virus type 1 subtype A and CRF03_AB protease in Eastern Europe: selection of the V77I variant and its rapid spread in injecting drug user populations. J Virol. 2004;78:11276–87. doi: 10.1128/JVI.78.20.11276-11287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deshpande A, Recordon-Pinson P, Deshmukh R, et al. Molecular characterization of HIV type 1 isolates from untreated patients of Mumbai (Bombay), India, and detection of rare resistance mutations. AIDS Res Hum Retroviruses. 2004;20:1032–5. doi: 10.1089/aid.2004.20.1032. [DOI] [PubMed] [Google Scholar]
- 78.Maljkovic I, Wilbe K, Sölver E, Alaeus A, Leitner T. Limited transmission of drug-resistant HIV type 1 in 100 Swedish newly detected and drug-naive patients infected with subtypes A, B, C, D, G, U, and CRF01_AE. AIDS Res Hum Retroviruses. 2003;19:989–97. doi: 10.1089/088922203322588341. [DOI] [PubMed] [Google Scholar]
- 79.Aghokeng AF, Ewane L, Awazi B, et al. Enfuvirtide binding domain is highly conserved in non-B HIV type 1 strains from Cameroon, West Central Africa. AIDS Res Hum Retroviruses. 2005;21:430–3. doi: 10.1089/aid.2005.21.430. [DOI] [PubMed] [Google Scholar]
- 80.Westby M, Smith-Burchnell C, Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81:2359–71. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soares EA, Santos RP, Pellegrini JA, Sprinz E, Tanuri A, Soares MA. Epidemiologic and molecular characterization of human immunodeficiency virus type 1 in southern Brazil. J Acquir Immune Defic Syndr. 2003;34:520–6. doi: 10.1097/00126334-200312150-00012. [DOI] [PubMed] [Google Scholar]
- 82.Grossman Z, Paxinos EE, Averbuch D, et al. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob Agents Chemother. 2004;48:2159–65. doi: 10.1128/AAC.48.6.2159-2165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2(4):e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnston MI, Fauci AS. An HIV vaccine — evolving concepts. N Engl J Med. 2007;356:2073–81. doi: 10.1056/NEJMra066267. [DOI] [PubMed] [Google Scholar]
- 85.Goulder PJ, Brander C, Tang Y, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–8. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 86.Kwong PD, Doyle ML, Casper DJ, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 87.Thakar MR, Bhonge LS, Lakhashe SK, et al. Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef. J Infect Dis. 2005;192:749–59. doi: 10.1086/432547. [DOI] [PubMed] [Google Scholar]
- 88.Weaver EA, Lu Z, Camacho ZT, et al. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group M consensus env immunogen. J Virol. 2006;80:6745–56. doi: 10.1128/JVI.02484-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barouch DH, Santra S, Schmitz JE, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–92. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 90.Girard MP, Osmanov SK, Kieny MP. A review of vaccine research and development: the human immunodeficiency virus (HIV) Vaccine. 2006;24:4062–81. doi: 10.1016/j.vaccine.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 91.Douek DC, Kwong PD, Nabel GJ. The rational design of an AIDS vaccine. Cell. 2006;124:677–81. doi: 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 92.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 93.Pitisuttithum P, Gilbert P, Gurwith M, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 94.Duerr A, Wasserheit JN, Corey L. HIV vaccines: new frontiers in vaccine development. Clin Infect Dis. 2006;43:500–11. doi: 10.1086/505979. [DOI] [PubMed] [Google Scholar]
- 95.Létourneau S, Im EJ, Mashishi T, et al. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS ONE. 2007;2(10):e984. doi: 10.1371/journal.pone.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robertson M, Mehotra D, Fitzgerald D, et al. Efficacy results from the STEP study (Merck V520 Protocol 023/HVTN 502): a phase II test-of-concept trial of the MRKAd5 HIV-1 Gag/Pol/Nef trivalent vaccine. Presented at the 15th Conference on Retroviruses and Opportunistic Infections; Boston. February 3–6, 2008; abstract. [Google Scholar]
- 97.Cohen J. AIDS research: did Merck’s failed HIV vaccine cause harm? Science. 2007;318:1048–9. doi: 10.1126/science.318.5853.1048. [DOI] [PubMed] [Google Scholar]
- 98.Catanzaro AT, Koup RA, Roederer M, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006;194:1638–49. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Graham BS, Koup RA, Roederer M, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J Infect Dis. 2006;194:1650–60. doi: 10.1086/509259. [DOI] [PMC free article] [PubMed] [Google Scholar]


