Radiation damages DNA through two distinct types of reaction pathways, direct-type and indirect-type.1 The indirect-type pathways entail reactions of water radiolysis products with DNA, for example, the formation of strand breaks by hydroxyl radical attack, which has been extensively studied.2 Direct-type effects are caused by direct ionization of DNA or by transfer of holes and electrons to DNA from the surrounding hydration waters.3-5 The direct component may constitute a significant fraction of the total radiation damage to DNA in cells where water radiolysis products are effectively scavenged by the medium.6 Much less is known about the direct-type mechanisms and related quantitative characteristics, especially in regards to strand break formation. It is known that electron attachment to DNA is not a significant source of strand breaks.7 Sugar radicals generated through one-electron oxidation of the sugar–phosphate moiety has been implicated as the main source of strand scission from direct-type damage.7
This study reports the first direct measurement of radiation–chemical yields of individual strand break products induced by the direct effect in DNA. Two DNA oligonucleotides, d(CGCG)2 (1) and d(CGCGCG)2 (2), duplexes in crystalline form, were X-irradiated at two different temperatures, 4 and 293 K. HPLC in combination with photodiode array detection was used to identify and measure the chemical yields of all cleavage fragments that retained at least one unaltered base. Product identifications are based on comparison with the retention times and optical absorption spectra of authentic compounds and are supported by MALDI-TOF measurements.8 Crystals of oligonucleotides offer the advantage of exact knowledge of the helical packing and conformation. Both 19 and 210 are in Z-conformation, and the hydration levels are relatively low (7.1 and 7.2 waters per nucleotide, respectively),4 well below the level required for detectable production of OH radicals from the waters of hydration.5 Previous work has shown that trapped hydroxyl radicals are not detected at 4 K in crystals of either 1 or 2.4 Indirect-type reactions, therefore, cannot be a significant source of strand breaks in these samples.
A typical HPLC chromatogram of irradiated 2 is shown in Figure 1 along with the reference and zero dose chromatograms. Chromatograms of the irradiated 1 differ from Figure 1 by the absence of peaks corresponding to end-phosphorylated tetramers and pentamers, and by the parent-compound retention time. From comparison of the crystal and reference spectra, all major damage products found in the crystal are represented in the reference sample and vice versa. The fragmentation products are identified as free bases, 3′-, and 5′-end phosphorylated strand fragments. The production of other degradation products not present in the reference sample is minimal. For example, there is no evidence of 3′-phosphoglycolates that are the products of the H4′-abstraction from DNA in the presence of oxygen (see ref 12 for a review).
Figure 1.
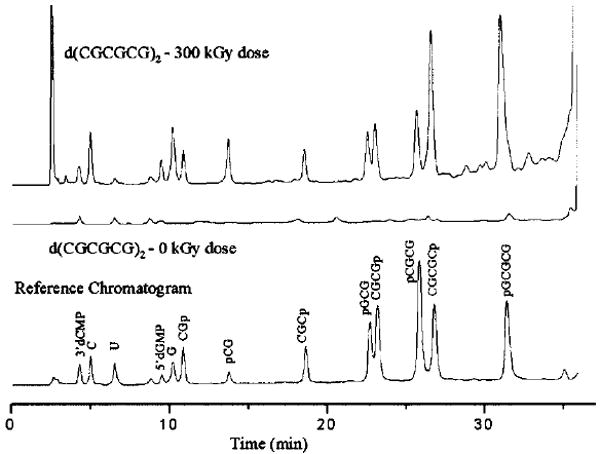
HPLC profile at 260 nm of X-irradiated11 2 (top), unirradiated 2 (middle), and the reference sample (bottom). The irradiated sample received a dose of 300 kGy at 293 K. Samples were dissolved in a 20 mM phosphate buffer and held at 70 °C for 30 min to ensure the complete conversion of sugar damage to strand breaks. Samples were then analyzed using a Phenomenex C18 column at 30 °C, washed with 25 mM ammonium acetate at 1 mL/min with a 7.5% acetonitrile gradient applied over 40 min.
The standard irradiated crystalline sample is about 200 μg, and the products each make up <1% of this initial sample. Sample sizes for 1 were larger, and the standard deviations were significantly smaller. The yield of each DNA strand fragment is given in Figure 2.
Figure 2.
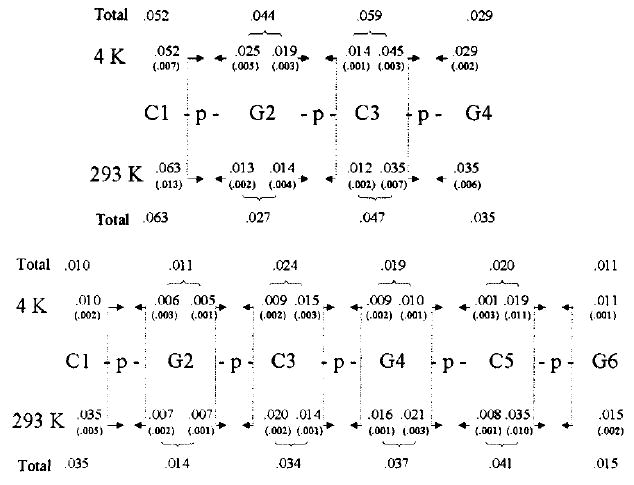
Yields (in μmol/J) of products in 1 and 2 after irradiation at (above) and 293 K (below). Yields were determined from the slopes of the least-squares lines fit to the dose response data. The errors, given in parentheses, are the standard deviations derived from the linear fits to these data. The oligomer sequences are given using standard notation, reading from left-to-right 5′-to-3′. Arrows emanate from the side of the phosphodiester bond that is cleaved. At the base of an arrow, the yield is given for the oligomer that the arrow points toward. For example, irradiation of 1 at 4 K gives the following products and yields in μmol/J: pG2pC3pG4 is 0.052, C1p is 0.025, pC3pG4 is 0.019, etc. The totals refer to the sum of strand breaks due to cleavage of both 3′- and 5′-phosphodiester bonds originating from damage at each residue. A total of 5 and 4 samples were used to determine yields of 1 at 4 and 293 K respectively, and 9 and 14 samples for 2 at 4 and 293 K, respectively.
In summary, this study finds that (1) the major products of strand breakage from the direct-type effect are free bases and oligodeoxynucleotides with 3′-, and 5′-phosphate end groups and (2) the yield of immediate strand breaks is between 0.05 and 0.15 μmol/J.
Strand Break Products
It is known that the sugar radicals produced by hydrogen abstraction from any site of the deoxyribose moiety lead to primary products consisting of free bases and shorter oligomers with 3′- or 5′-phosphorylated ends.12 It was concluded in our previous study7 that irradiation of crystalline DNA produces a population of sugar radicals, the distribution of which is different than that generated by the indirect effect. In the current study, we find that the DNA 4 K fragmentation induced by direct-type effects supports the proposal that direct ionization of DNA gives rise to damaged sugar, presumably sugar-centered radicals, that are precursors to strand breaks. A major fraction of the radical cations formed by ionization of the sugar–phosphate backbone either do not undergo hole transfer to the base stack of DNA, or if transfer does occur, it occurs after reactions that leave the sugar moiety damaged.
Sugar radicals in directly ionized DNA are less prone to dose saturation than base-centered radicals.13 Resistance to saturation has also been observed for unaltered base release from X-irradiated DNA crystals, supporting the conclusion that sugar radicals are precursors to unaltered base release in crystalline DNA.7 Here, too, we find resistance to dose saturation for the radiation products in both crystals, up to at least 100 kGy. This adds to the growing evidence that an appreciable fraction of direct-type damage in DNA consists of sugars radicals that are precursors to strand breaks.14
Strand Break Yields
There are three approaches to calculating the yield of strand breaks from our data. Given that no other cleavage products are observed except the bases, oligomers with 3′-phosphate ends, and oligomers with 5′-phosphate ends, the yields of strand breaks can be calculated by summation over the individual yields within each of these three groups. However, each of these approaches has drawbacks. Base release does not necessarily translate into an immediate strand break; therefore, assuming one strand break per base release may overestimate the yield of immediate strand breaks. A drawback in summing over oligomers with 3′-phosphate ends is that no 3′-phosphate can be generated from the sugar radicals localized on the 5′-terminus (this sugar is not phosphorylated in the 5′ position). Similarly, no 5′-phosphate could originate from the sugar radicals generated at the terminal 3′-end nucleotide. Therefore, a summation over the yields of either 3′- or 5′-phosphate ends will underestimate the yield of strand breaks.
To avoid these problems, we sum over products that are representative of each cleavage site. For 1, this means that the summation includes the individual yields of Cp, CGp, CGCp, and pGCG, and alternatively, pG, pCG, pGCG, and CGCp. These two sums are then averaged. Strand break yields, G(ssb), calculated in this manner, are shown in the Table 1 along with the measured yields of free base release, G(Cfree+Gfree). The yields are in good agreement with that of 0.06 μmol/J measured for release of unaltered bases in a similar hexamer crystal, a CACGC: GTGCGC duplex.7
Table 1.
Calculated Yields G (in μMol/J) of Immediate Strand Breaks Compared to the Measured Yields of Free Base in Crystalline 1 and 2
| oligomer | temp (K) | G(ssb) | G(Cfree+Gfree) |
|---|---|---|---|
| 1 | 4 | 0.13 ± 0.02 | 0.12 ± 0.01 |
| 1 | 293 | 0.14 ± 0.03 | 0.13 ± 0.03 |
| 2 | 4 | 0.06 ± 0.02 | 0.03 ± 0.01 |
| 2 | 293 | 0.11 ± 0.02 | 0.07 ± 0.01 |
Implications for DNA in Vivo
The strand break yields for 1 and 2 suggest that in DNA, strand break yields from direct-type damage should be comparable to the yields from indirect-type damage in vivo. For indirect effects in oxygenated solution, reported yields range from 0.0001 to 0.1 μmol/J, depending on conditions.2,15 While a major fraction of the ionizing energy is deposited in water, histoproteins and other cellular components protect DNA from indirect-type damage by competing for hydroxyl radicals generated in the water but do not protect against direct-type damage. The yield of direct-type strand breaks is substantial (~0.1 μmol/J), and that of the radicals trapped by the bases is even higher (~0.6 μmol/J).16 Both of these lesions are likely to be important components of multiply damaged sites.17
Other Observations
The phosphodiester bond on the 3′-side of cytosine residues appears to be 1.5–2.5× more prone to cleavage than on the 3′-side of guanine residues, and generally there is a 1.8× greater yield of oligomer products with 5′-phosphorylated ends than with 3′-ends (see Figure 2). These preliminary observations need to be verified in longer oligonucleotides and different sequences, and this is currently underway.
Acknowledgments
This study was supported by PHS Grant 2-R01-CA32546, awarded by the National Cancer Institute. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor and Francis; New York: 1987. [Google Scholar]
- 2.(a) Milligan J, Ng J, Wu C, Aguilera J, Fahey R, Ward J. Radiat Res. 1995;143:273–280. [PubMed] [Google Scholar]; (b) Ito T, Baker SC, Stickley CD, Peak JG, Peak MJ. Int J Radiat Biol. 1993;63:289–296. doi: 10.1080/09553009314550391. [DOI] [PubMed] [Google Scholar]; (c) Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiat Res. 1992;129:333–44. [PubMed] [Google Scholar]; (d) Sies H, Schuttz WA, Steenken S. J Photochem Photobiol. 1996;32:97–102. doi: 10.1016/1011-1344(95)07192-x. [DOI] [PubMed] [Google Scholar]
- 3.Mroczka NE, Mercer KR, Bernhard WA. Radiat Res. 1997;147:560–568. [PubMed] [Google Scholar]
- 4.Debije MG, Strickler MD, Bernhard WA. Radiat Res. 2000;154:163–170. doi: 10.1667/0033-7587(2000)154[0163:oteoha]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Vere T, Becker D, Sevilla MD. Radiat Res. 1996;145:673–680. [PubMed] [Google Scholar]
- 6.Krisch RE, Flick MB, Trumbore CN. Radiat Res. 1991;126:251–259. [PubMed] [Google Scholar]
- 7.Razskazovskiy Y, Debije MG, Bernhard WA. Radiat Res. 2000;153:436–441. doi: 10.1667/0033-7587(2000)153[0436:drdtcd]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MALDI-TOF was used to determine the molecular mass of the fragments with m/z > 500 from 1 assigned to CGp, pCG, CGCp, and pGCG (m/z, product/reference: 637.3/637.1, 637.3/637.0, 926.1/926.5, and 966.5/966.0, respectively). Identification of the products from 2 by MALDI-TOF was not possible due to insufficient quantities of the material.
- 9.Gessner RV, Frederick CA, Quigley GJ, Rich A, Wang AH-J. J Biol Chem. 1989;264:7921–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- 10.Crawford JL, Kolpak FJ, Wang A, Quigley G, van Boom J, van der Marel G, Rich A. Proc Natl Acad Sci USA. 1980;77:4016–4020. doi: 10.1073/pnas.77.7.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer KR, Bernhard WA. J Magn Reson. 1987;74:66–71. [Google Scholar]
- 12.Pogozelski WK, Tullius TD. Chem ReV. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 13.Becker D, Razskazovskii Y, Callaghan MU, Sevilla MD. Radiat Res. 1996;146:361–368. [PubMed] [Google Scholar]
- 14.Debije MG, Bernhard WA. Radiat Res. 2001 In Press. [Google Scholar]
- 15.Lücke-Huhle C, Braun A, Hagen U. Z Naturforsch. 1970;25b:1264–1268. doi: 10.1515/znb-1970-1111. [DOI] [PubMed] [Google Scholar]
- 16.(a) Debije MG, Milano MT, Bernhard WA. Angew Chem Int Ed. 1999;38:2752–2756. [PMC free article] [PubMed] [Google Scholar]; (b) Debije MG, Bernhard WA. Radiat Res. 1999;152:583–589. [PMC free article] [PubMed] [Google Scholar]; (c) Debije MG, Bernhard WA. J Phys Chem B. 2000;104:7845–7851. [Google Scholar]
- 17.Ward JF. Radiat Res. 1981;86:185–195. [PubMed] [Google Scholar]


