Short abstract
Comparative genomic analysis of the Impact locus, which is imprinted in Glires but not in other mammals, reveals features required for genomic imprinting.
Abstract
Background
Imprinted genes are exclusively expressed from one of the two parental alleles in a parent-of-origin-specific manner. In mammals, nearly 100 genes are documented to be imprinted. To understand the mechanism behind this gene regulation and to identify novel imprinted genes, common features of DNA sequences have been analyzed; however, the general features required for genomic imprinting have not yet been identified, possibly due to variability in underlying molecular mechanisms from locus to locus.
Results
We performed a thorough comparative genomic analysis of a single locus, Impact, which is imprinted only in Glires (rodents and lagomorphs). The fact that Glires and primates diverged from each other as recent as 70 million years ago makes comparisons between imprinted and non-imprinted orthologues relatively reliable. In species from the Glires clade, Impact bears a differentially methylated region, whereby the maternal allele is hypermethylated. Analysis of this region demonstrated that imprinting was not associated with the presence of direct tandem repeats nor with CpG dinucleotide density. In contrast, a CpG periodicity of 8 bp was observed in this region in species of the Glires clade compared to those of carnivores, artiodactyls, and primates.
Conclusions
We show that tandem repeats are dispensable, establishment of the differentially methylated region does not rely on G+C content and CpG density, and the CpG periodicity of 8 bp is meaningful to the imprinting. This interval has recently been reported to be optimal for de novo methylation by the Dnmt3a-Dnmt3L complex, suggesting its importance in the establishment of imprinting in Impact and other genes.
Background
Genomic imprinting is an epigenetic modification that leads to monoallelic gene expression in a parent-of-origin-specific manner. In mammals, approximately 100 'imprinted' genes are subject to this regulation [1]. Identification of a specific sequence that is recognized as the target for epigenetic marking is the foremost problem in this field. Researchers have compared genomic sequences of human and mouse imprinted and non-imprinted genes in order to identify motifs that are characteristic of, or responsible for, genomic imprinting [2-5]. Especially, finding target sequences for de novo DNA methylation during gametogenesis would further our understanding of the molecular mechanisms of imprinting, as well as development, tissue-specific gene regulation, and the etiology of various cancers. However, genomic features unique to imprinted genes, which could lead to their discovery, have not been described, with one exception [6]. It has been suggested that the absence of such features is due to variability in the molecular mechanisms of imprinting [7,8].
Therefore, instead of identifying common features, we limited our study to one imprinted gene, Impact, but performed comparative genomics among thirty eutherian species. The Impact gene was first identified in mouse as a novel imprinted gene by a systematic screening method using mRNA display PCR [9]. Its protein product is suggested to have a role in response to amino acid starvation [10,11]. This gene exhibits species-specific imprinting; it is imprinted in species of the Glires clade (rodents and lagomorphs), but not in other mammals such as primates and artiodactyls (even-toed ungulates) [12]. Since the Glires clade diverged from primates approximately 70 million years ago [13], the acquisition of the imprinting in these species is quite recent compared to other imprinted genes, most of which are imprinted in both mouse and human. This makes the comparative analysis between imprinted and non-imprinted orthologues more straightforward. By contrast, if we studied, for example, the Igf2 gene by the same strategy, we would have to compare two clades, for example, eutherians and monotremes, which diverged about 200 million years ago [14]. Generally, such sequences are too divergent to allow DNA motifs to be found by sequence alignment. The recent evolution of Impact as an imprinted gene provides a unique opportunity to perform this kind of comparative genomics.
In species of the Glires clade, Impact bears a differentially methylated region (DMR) in its first intron that is de novo methylated during oogenesis, but not in spermatogenesis, and maintained in all types of somatic cells to adulthood [15]. Hence, this region is a so-called primary DMR, which is the key cis-regulatory element directing the correct establishment and maintenance of genomic imprinting. In our previous analysis of the Impact DMR in species of the Glires clade, the sequences of mouse, rat, and rabbit were determined. The DMR in these species is characterized by a CpG island, and the DMR in rodents contains characteristic tandem repeats in the CpG island [12]. Because the mechanism by which the de novo DNA methylation machinery recognizes the DMRs is not yet known, we have tried in the present study to search for the target sequences of the allele-specific methylation by sequencing the genomic region of various Glires animals, including beaver, porcupine, chipmunk, and prairie dog. Fortunately, the first intron could readily be amplified by PCR using primers located in the first and second exons. Including data from our previous study [12], 27 out of 30 eutherian species were successfully sequenced.
More than a decade ago, direct tandem repeats were suggested to be related to genomic imprinting [16]; however, the numbers of identified imprinted genes and available mouse and human genomic sequences were considerably limited at that time. Later, Impact was identified, and it was reported that imprinted mouse Impact bears these characteristic repeats whereas the non-imprinted human orthologue lacks any apparent repeats [17]. It was subsequently reported that the repeat is absent in the imprinted rabbit Impact gene [12]. Since tandem repeats are abundant and widespread throughout mammalian genomes [18], it is therefore difficult to associate these with the imprinting status of specific genes. One strategy to address this is to increase the number of species studied at a given locus. A recent study determining the extent and boundaries of all known primary DMRs enabled the analysis of their specific nucleotide sequences and content [19]. Some characteristic features were described; however, the number of primary DMRs in mouse is limited to only 15 to date. Our study provides additional data that are needed to characterize such intriguing regions.
In support of the fast molecular clock of rodent genomes [20], we observe that the determined genomic sequences are considerably diverged only among rodents, but not in lagomorphs. While the data challenge the proposed role of tandem repeats and CpG content in genomic imprinting, they suggest the importance of latent CpG dinucleotide periodicity in the establishment of the Impact DMR.
Results
We previously developed a simple PCR-based strategy to determine the nucleic acid sequence of the first intron of Impact and reported the sequences of 14 eutherian species [12]. In this method, primers were designed for highly conserved regions in exons 1 and 2 for forward and reverse primers, respectively. Two forward and two reverse degenerate primers were prepared to perform nested PCR for the divergent sequences. In the present study, we used the same method to determine the corresponding sequences in two lagomorphs and 17 rodents (Table 1). All but three were successfully amplified. For these species (field mouse, agouti, and paca), specific PCR products could not be obtained even after nested PCR. This is probably due to unexpectedly divergent sequences at the exonic priming sites or excessive elongation of the intron in these animals (see Discussion).
Table 1.
Lagomorphs and rodents used in this study
| Species | Taxonomy ID* | Common name† | Accession number |
| Oryctolagus cuniculus | 9986 | Rabbit | EF470590 |
| Sylvilagus floridanus | 9988 | Cottontail | EF470591 |
| Peromyscus maniculatus | 10042 | Deer mouse | EF470592 |
| Mus musculus | 10090 | Mouse | EF470593 |
| Rattus norvegicus | 10116 | Rat | EF470594 |
| Erethizon dorsatum | 34844 | Porcupine | EF470595 |
| Hylomyscus alleni | 34858 | Wood mouse | EF470596 |
| Apodemus agrarius | 39030 | Field mouse | - |
| Dasyprocta leporina | 42152 | Agouti | - |
| Cynomys ludovicianus | 45480 | Prairie dog | EF470597 |
| Castor canadensis | 51338 | Beaver | EF470598 |
| Rhizomys pruinosus | 53275 | Bamboo rat | EF470599 |
| Tamias sibiricus | 64680 | Chipmunk | EF470600 |
| Eothenomys melanogaster | 82468 | Vole | EF470601 |
| Dicrostonyx groenlandicus | 85953 | Lemming | EF470602 |
| Agouti paca | 108852 | Paca | - |
| Reithrodontomys gracilis | 243215 | Harvest mouse | EF470603 |
| Tscherskia triton | 329627 | Hamster | EF470604 |
| Rheomys thomasi | 451894 | Water mouse | EF470605 |
*Assigned by NCBI Taxonomy. †Common names used in this paper. Some of these are short forms of GenBank common names, such as American beaver. See NCBI Taxonomy.
Following treatment with exonuclease I and shrimp alkaline phosphatase, nested PCR products were directly sequenced by the primer-walking method. The identities of these amplicons as the Impact gene were confirmed by the 30-nucleotide sequences at the beginning of exon 2. This short region was also amplified along with the first intron for this purpose. Almost all encode an amino acid sequence identical to NEEIEAMAAI seen in human IMPACT. Exceptions were mouse, wood mouse, bamboo rat, and porcupine, which code for SEEIEAMAAI, SEEIEAMAAI, NEEIEAMASI, and NEEIEALSAI, respectively. It has been surmised that Impact does not have paralogues in any vertebrate genome due to dosage sensitivity [21]. Accordingly, a PCR product amplified from a single locus was obtained in each species. We also confirmed that all of the intronic sequences meet the GT-AG rule, also known as Chambon's rule, and that they have a branch site proximal to the splice acceptor (not shown).
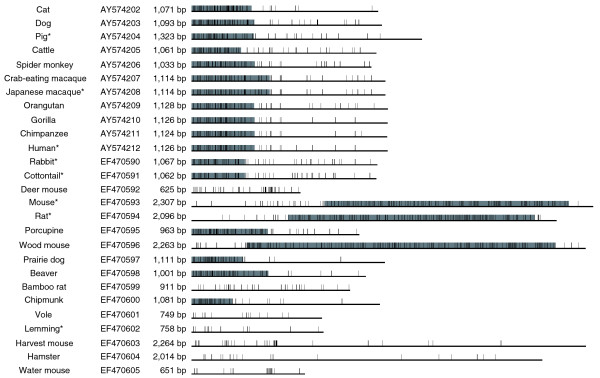
In the previous study using rodents, lagomorphs, artiodactyls, carnivores, and primates, the sequences were readily classified into two groups (Figure 1). The first group has a longer intron (approximately 2 kb), the 3' portion of which constitutes a CpG island with a characteristic tandem reiterated structure [17]. The second group has a shorter intron (approximately 1 kb), the 5' portion of which constitutes a short CpG island without any apparent repeats. Regardless of the imprinting status of the Impact gene, only mouse and rat sequences fall in the former group. Despite the fact that rabbit Impact is imprinted, it was unexpectedly categorized in the latter group. Additionally, a sequence derived from the whole genome shotgun sequencing of the rabbit was obtained [GenBank:AAGW01108706], which covers this region and confirms the absence of tandem repeats, even in the expanded flanking regions included in this sequence. In mouse, the two genes flanking Impact are not imprinted and no additional imprinted genes have been found on chromosome 18 where it is mapped [22]. Unlike typical imprinted genes, Impact appears to be solitary; it is likely that the regulatory elements are confined to this locus. Hence, at least for this imprinted locus, the result clearly negates a hypothesis that tandem repeats play an important role in genomic imprinting [16]. To pursue other structural features of imprinted Impact, elucidating the genomic sequences of many other rodent and lagomorph species was of interest.
Figure 1.
Schematic representation of the first intron of eutherian Impact. The GenBank accession number and length are listed to the right of the common names. Horizontal lines show the relative lengths of the first intron. All sequences begin with GT and end with AG. Short vertical lines and gray boxes represent single CpG sites and CpG islands, respectively, which were detected by GrailEXP 3.31. Characteristic tandem repeats are exclusively found in the CpG islands of murids (mouse, rat, and wood mouse). Glires species are sorted by NCBI Taxonomy ID. The Impact gene is assumed to be imprinted in Glires species [GenBank:EF470590-EF470605] but not in other species [GenBank:AY574202-AY574212]. Asterisks indicate species whose monoallelic expression or methylation of the Impact gene have been experimentally confirmed [12,17].
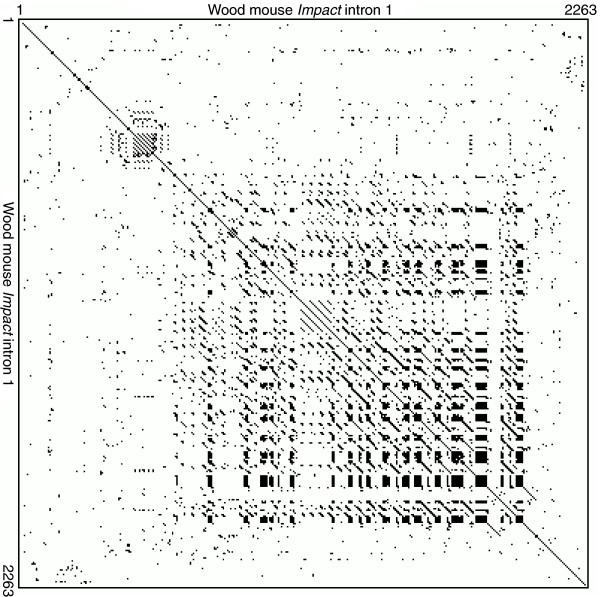
The genomic sequences determined in the current study are shown along with previous results (Figure 1). While lagomorphs (rabbit and cottontail) have similar intronic sequences to those of primates, artiodactyls, and carnivores, rodents have diversified structures. Although the porcupine, beaver, and sciurids (prairie dog and chipmunk) bear a CpG island at the 5' end like lagomorphs, murids (mouse, rat, and wood mouse) bear a longer one at the 3' side. Others unexpectedly bear no CpG islands. The lengths of these introns vary from 625 bp to more than 2 kb. The characteristic tandem repeat was found exclusively in murids (Figure 2). A homology search using the repetitive regions as queries did not hit any other sequences but themselves, suggesting that these sequences are unique to this locus in murids.
Figure 2.
Direct tandem repeat of wood mouse Impact. Self-Harr plot of the first intron of wood mouse Impact shows nested structure of direct tandem repeats around the CpG island. A dot was plotted when it satisfied the condition that there were more than 8 bases matching in a 10-bp window. While mouse and rat Impact also show quite similar plots, other eutherians apparently do not have this tandem repeat.
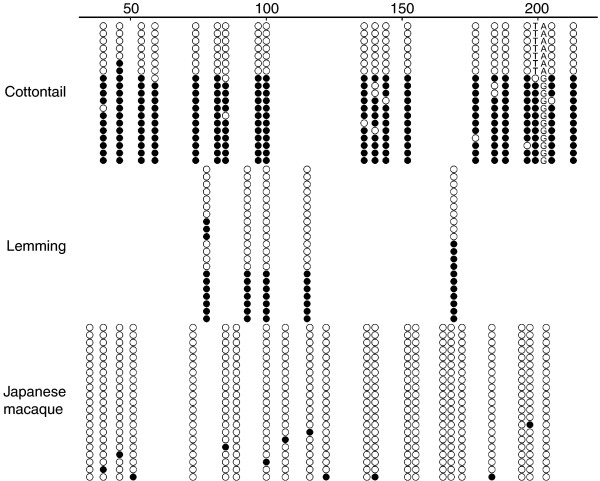
The scarcity of CpG dinucleotides in several rodents made us wonder whether they bear the DMR in this region and whether they are imprinted or not. We therefore chose lemming as one of those species, cottontail from lagomorphs, and Japanese macaque from the non-imprinted group for DNA methylation analysis by bisulfite cloning and sequencing [23]. For both mouse and rabbit Impact, the 5' portion of the first intron was shown to be subject to allele-specific methylation; the maternal and paternal alleles are hyper- and hypomethylated, respectively [12,17,19]. We decided to analyze the equivalent region for these three species (Figure 3).
Figure 3.
DNA methylation analysis by bisulfite cloning and sequencing. The analysis was carried out for three species. Cottontail is one of the lagomorphs. Lemming (a rodent) contains fewer CpG sites in the first intron of Impact. Japanese macaque is a primate in which the IMPACT gene is not imprinted. Numbers indicate distances in base-pairs from the 3' end of the first exon. Each row represents an individual cloned allele. Circles represent CpG sites and their spacing reflects the CpG density of the region. Filled and open circles represent methylated and unmethylated sites, respectively. The single nucleotide polymorphism in cottontail fortunately provided allele-specific methylation data, although the parental origin is unknown. Note that this single nucleotide substitution has caused a coexistence of TpG and CpG; only the latter is subject to deamination.
We used one individual from each species. Fortunately, the cottontail has one A/G heterozygous site (position 201 of the sequence deposited under [GenBank:EF470591]) in this region, which allowed us to distinguish the two alleles. Although the parental origin could not be ascertained, one of the parental alleles is unmethylated and the other is heavily methylated. Possibly, the paternal allele of cottontail Impact may be exclusively expressed like rabbit Impact [12]. Unlike cottontail Impact, the lemming gene has only five CpG sites with no heterozygous sites in this region. However, the result suggests that the region is a DMR because there were unmethylated clones and fully methylated clones. It is likely that lemming Impact is also imprinted like other rodent orthologues despite the scarceness of CpG dinucleotides in the corresponding region. Macaque IMPACT has a CpG island in this region like the cottontail gene. In support of the fact that primate Impact exhibits biallelic expression [12], the 5' portion of the intron escapes DNA methylation in both alleles in Japanese macaque. Establishment of the DMR seems to be independent, not only of tandem repeats, but also of local CpG density. This raises another question: what then causes the difference in DNA methylation status between Glires and other mammals?
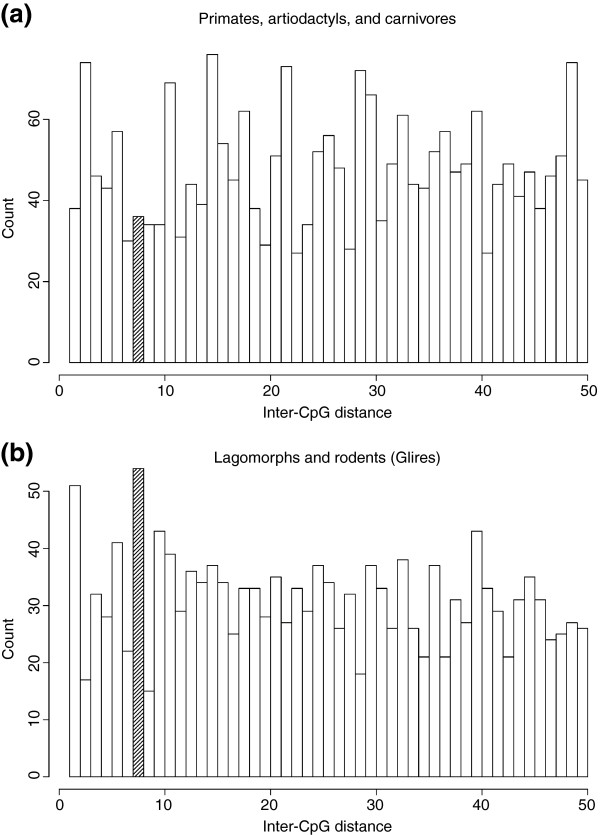
Recently, crystallography of a complex consisting of Dnmt3a and Dnmt3L revealed a correlation between its enzymatic activity and methylated CpG sites at distances of eight to ten base pairs [24]. Dnmt3a is a DNA methyltransferase and Dnmt3L is its regulatory factor; both of these proteins are needed for the de novo DNA methylation of imprinted genes [25-27]. Accordingly, periodicity of CpG dinucleotide locations is found in the DMRs of 12 imprinted genes that are subject to maternal methylation. Mouse Impact is one of these genes, bearing a large number of CpG dinucleotides spaced with 10-bp periodicity [24]. However, this periodicity originates in the direct repeats found only in murids. In order to search for other CpG periodicity that may be related to the de novo DNA methylation of the Impact DMR, we examined only the 500-bp region at the 5' end of the intron in the eutherians. Frequencies of CpG pairs at a given distance with respect to all pairs are separately shown for Glires species (putative imprinted group) and other eutherians in Figure 4. While a conspicuous 8-bp CpG interval, but neither a 7- nor 9-bp interval, is observed in species of the Glires clade, the frequency of 8-10-bp intervals in other eutherian species is low (p = 2.46 × 10-3; see Materials and methods). Additionally, the periodic occurrence of CpG sites 9.5 bp apart on average was not observed in this region [24] (Additional data file 1). These results suggest that the CpG periodicity of 8 bp plays an important role in imprinting and that the accumulation of this periodicity might relate to acquisition of imprinting in the common ancestor of extant Glires species.
Figure 4.
Periodicity of CpG sites in the 500-bp region at the 5' portion of the intron. Counts of each distance from 2-50 bp are shown for (a) non-Glires eutherians and (b) Glires species. The 8-bp periodicity is evident only in Glires (p = 2.46 × 10-3; see Materials and methods), which bears the DMR in this region.
Discussion
Whereas the possible importance of tandem repeats in genomic imprinting is still disputed [28-30], several lines of evidence negate the hypothesis [31-34]. The present study also argues against the proposed role of repetitive elements in the imprinting of Impact. Since it is suggested that imprinting has evolved randomly at various times in different lineages [7], molecular mechanisms that achieve monoallelic gene expression may vary from locus to locus. Tandem repeats can be observed almost everywhere in mammalian genomes [18]. Hence, it seems unreasonable to assume that tandem repeats per se have a role in genomic imprinting in general. What we should address is a specific role of each tandem repeat, such as offering a high concentration of insulator binding sites [35], rather than presence or absence of any repeats.
The tandem repeat in murid Impact has a complex structure with nested repetitive elements, but the shortest sequence element is 5'-TCGGC-3'. This 5-bp directed element is concatenated to constitute the long stretch in mouse, rat, and wood mouse genomes. It is possible that 10-bp periodicity, which is caused by juxtaposition of the element, is so stable for nucleosome positioning that it allows the region to expand the repeat. It is reported that 10-bp periodic GpC, which corresponds to one DNA helical repeat, is often found in regions that form nucleosome structure well [36]. The shortest element definitely contains GpC dinucleotide (note that this is not CpG dinucleotide). It is also likely that tandem repeats near imprinted genes are just a consequence, rather than a cause, of the epigenetic regulation [37]. The 3' portion of the CpG island appears to be just such a product of expansion of an element containing a single CpG, resulting in high frequency of CpG. This region in the field mouse, another murid, failed to amplify by PCR. Possibly, a large repeat expansion in the intron impedes the PCR amplification of the field mouse genome; however, we have not tested this. Similarly, neither agouti nor paca, closely related caviomorph rodents, could be amplified by PCR at this locus. Possibly, they have a unique shared substitution or insertion that prevented amplification. The chicken intron also could not be amplified by this method.
It was suggested that CpG content per se could be recognized by methylation machinery to give rise to primary DMRs [19]. Contrary to this hypothesis, the CpG content in the Impact DMR turned out to vary considerably among species of the Glires clade, also suggesting necessity to search for DMRs other than CpG islands. Rather than discern the CpG dinucleotide density, the de novo methylation complex seemingly prefers to interact with CpG sites arranged at an interval of 8 bp. The 8-bp CpG periodicity was preferentially observed in Glires, in which the Impact gene is imprinted (Figure 4). In a broad sense, the periodicity 5'-CGNNNNNNCG-3' can be considered as a DNA motif or protein-biding site that is targeted by the Dnmt3a-Dnmt3L complex. It is possible that accumulation of the motif in the common ancestor of Glires was related to the acquisition of the Impact imprinting. In fact, the short genomic sequence of lemming shown here does not contain the 8-bp periodicity. We do not insist that the periodicity is the necessary and sufficient factor for the genomic imprinting; however, it seems to have a role (Figure 4 and Additional data file 1).
One possible hypothesis is that, in the common ancestor, tandem duplication of a short fragment containing 8-bp CpG periodicity occurred repeatedly, resulting in recruitment of methylation machinery during oogenesis. In this model, critical sites for the interaction with the enzymatic complex are CpG dinucleotides at an interval of 8 bp. The other nucleotides could have been neutrally mutated or diverged because the change does not affect the DNA-protein interaction. In any case, the present study also suggests a limit to the usefulness of conventional homology search algorithms for detecting imprinted genes. It may be important to investigate unexplored features of genomic sequences like the latent periodicity suggested by our studies. Each de novo DNA methyltransferase seems to have a specific genomic context associated with methylation, although functional redundancy is also observed [38]. In our additional analysis of the mouse genome, obvious, moderate, and much lower 8-bp periodicities were observed in SineB1, IAP, and Line1 repeats, respectively (data not shown; see Materials and methods). These results seem consistent with the experiment using Dnmt3-mutant mice [38]. The most parsimonious explanation is that the 12 maternally methylated DMRs are methylated by the same protein complex. By this expanded comparative analysis, we could successfully exclude the potential role of the 10-bp periodicity in the Impact imprinting described above [24]. For the other 11 DMRs, further analysis of the kind presented here may facilitate the understanding of genomic imprinting. Considering the molecular mechanisms that are needed, characteristic features of genomic sequences in imprinted genes should be identified in order to elucidate the true nature of genomic imprinting.
Conclusions
As a step towards a better understanding of the establishment of DMRs, we took the unique approach of using comparative genomics. Only one species-specific imprinted gene was chosen, but various mammalian genomic DNAs were collected. The results are summarized by the following three points. First, direct tandem repeats, which are found only in murids, are dispensable for the imprinting. Second, establishment of the DMRs does not rely on of G+C content and CpG density. Finally, a CpG periodicity of 8 bp, but neither 9 nor 10 bp, may play an important role in the establishment of this imprinting. Serial duplication of this region could have resulted in the accumulation of this periodicity, which might be related to establishment of imprinting at this locus in the common ancestor of rodents and lagomorphs. These three are apparently true at least for the Impact gene. Nevertheless, the method and implication documented in the present study should be applied to many other loci in order to help understand the general molecular mechanisms of genomic imprinting.
Materials and methods
Animal resources
Rodent and lagomorph tissues (livers or spleens) were generous gifts from the Royal Ontario Museum (ROM) in Toronto, Ontario, Canada. Rabbits and rats were derived from closed colonies maintained by Kitayama Labes (Ina, Nagano, Japan) and Clea Japan (Tokyo, Japan), respectively. The Japanese macaque (Macaca fuscata) brain was a gift from Dr Hiroyuki Okuno at University of Tokyo.
Sequencing the first intron of Impact
Genomic DNA was extracted from livers or spleens of rodents and cottontail, and from brains of a rabbit, rat, and macaque. The first round of PCR was performed using primers 5'- ATG GCT GAR GDG GAM KYA GGG A -3' (forward) and 5'- CAA AGT GTC CAT TTG GGG TCA TC -3' (reverse). The second round of PCR was performed using a pair of nested primers: 5'- AGG GAR CRR CCA GAG GCA G -3' (forward) and 5'- ACA CAC CAC TCC TCG CCA TA -3' (reverse). Both PCR reactions were performed in the presence of 3.5% dimethyl sulfoxide (DMSO). PCR products were treated with exonuclease I and shrimp alkaline phosphatase (Amersham, London, UK) for subsequent direct sequencing. Sequence data from this article has been deposited as [GenBank:EF470590-EF470605].
DNA methylation analysis
We used the EpiTect Bisulfite Kit (Qiagen, Germantown, MD, USA) for the bisulfite treatment of genomic DNA. Primers used for lemming, cottontail, and macaque were 5'- GTG AGG TTT TTY GGG TAG GGA AYG G -3' (forward), 5'- CAA TAA ACT CCA AAC CAA CCA CAA C TAT AC -3' (reverse), 5'- GTG AGG TTT YGG YGG GGY GTT GTT -3' (forward), 5'- CTA CCT ACA ACC CAC TAC TAC TCA ATC -3' (reverse), 5'- GTG AGG TTT YGG YGG GGT GTT GAT -3' (forward), and 5'- CAC CRT CCR AAA CAA ACC CAA CCC -3' (reverse), respectively. For each species, the amplified positions are 1-224, 1-240, and 1-227, respectively. Position 1 corresponds to the first nucleotide (the 5' end) of the intron and also to the position in the GenBank data.
Computational analysis of DNA sequences
CpG islands were detected with GrailEXP 3.31 [39]. Mouse repetitive elements, that is, SineB1, IAP, and Line1, were identified by RepeatMasker Open-3.1.9 using a modified library [40]. Other analyses, such as showing each CpG site and determining the frequencies of intervals between two CpG dinucleotide sites, were performed using Perl scripts, which are available upon request from KO.
Statistical tests for CpG periodicity
We evaluated the statistical significance of the periodicities between imprinted and nonimprinted groups at distances from 2-50 bp using the one-tailed Fisher's exact test. We also employed the Bonferroni method for multiple testing correction of the p-values estimated from the tests [41]. Among distances from 2-50 bp, 8 bp is the only periodicity that has a significantly higher count in the imprinted group than in the nonimprinted group at a significance level of 0.01 (Additional data file 1).
Abbreviations
DMR, differentially methylated region.
Authors' contributions
KO conceived of the study, performed experiments, analyzed data, and drafted the manuscript. RFW and SWS participated in the coordination of the study, interpretation of data, and helped draft the manuscript. All authors had the opportunity to discuss the results and comment on the final manuscript.
Additional data files
The following additional data are available. Additional data file 1 is a table showing the numeric data used to draw Figure 4 and the p-values of Fisher's exact test and Bonferroni correction for the periodicity of CpG sites.
Supplementary Material
Numeric data used to draw Figure 4 and the p-values of Fisher's exact test and Bonferroni correction for the periodicity of CpG sites.
Acknowledgments
Acknowledgements
We thank Dr Burton K Lim (Royal Ontario Museum, Toronto, Ontario, Canada) and Dr Hiroyuki Okuno (University of Tokyo) for the generous gift of tissue samples, and Dr Layla Parker-Katiraee (University of Toronto), Dr Kenta Nakai (University of Tokyo), and Dr Takashi Ito (University of Tokyo) for discussions. We acknowledge The Centre for Applied Genomics for DNA sequencing and Mr Pingzhao Hu for statistical analyses. Supported by KAKENHI (20870008), Genome Canada/Ontario Genomics Institute, the Canadian Institutes for Health Research (CIHR), the Canadian Institutes for Advanced Research, the McLaughlin Centre for Molecular Medicine, the Canadian Foundation for Innovation, the Ontario Ministry of Research and Innovation, and The Hospital for Sick Children Foundation. SWS holds the GlaxoSmithKline-CIHR Pathfinder Chair in Genetics and Genomics at the University of Toronto and The Hospital for Sick Children.
Contributor Information
Kohji Okamura, Email: o-cuniculus@umin.ac.jp.
Richard F Wintle, Email: rwintle@sickkids.ca.
Stephen W Scherer, Email: steve@genet.sickkids.on.ca.
References
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci USA. 2002;99:327–332. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fan H, Yang HH, Hu Y, Buetow KH, Lee MP. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics. 2004;83:395–401. doi: 10.1016/j.ygeno.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib H, Zaitoun I, Kim ES. Comparative analysis of sequence characteristics of imprinted genes in human, mouse, and cattle. Mamm Genome. 2007;18:538–547. doi: 10.1007/s00335-007-9039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hore TA, Rapkins RW, Graves JA. Construction and evolution of imprinted loci in mammals. Trends Genet. 2007;23:440–448. doi: 10.1016/j.tig.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Rens W, Clarke O, Mungall AJ, Hore T, Graves JA, Dunham I, Ferguson-Smith AC, Ferguson-Smith MA. The evolution of imprinting: chromosomal mapping of orthologues of mammalian imprinted domains in monotreme and marsupial mammals. BMC Evol Biol. 2007;7:157. doi: 10.1186/1471-2148-7-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara Y, Hirai M, Nishiyama K, Kanazawa I, Ueda T, Sakaki Y, Ito T. Screening for imprinted genes by allelic message display: identification of a paternally expressed gene impact on mouse chromosome 18. Proc Natl Acad Sci USA. 1997;94:9249–9254. doi: 10.1073/pnas.94.17.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Sakaki Y, Ito T. GI domain-mediated association of the eukaryotic initiation factor 2alpha kinase GCN2 with its activator GCN1 is required for general amino acid control in budding yeast. J Biol Chem. 2000;275:20243–20246. doi: 10.1074/jbc.C000262200. [DOI] [PubMed] [Google Scholar]
- Pereira CM, Sattlegger E, Jiang HY, Longo BM, Jaqueta CB, Hinnebusch AG, Wek RC, Mello LE, Castilho BA. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J Biol Chem. 2005;280:28316–28323. doi: 10.1074/jbc.M408571200. [DOI] [PubMed] [Google Scholar]
- Okamura K, Sakaki Y, Ito T. Comparative genomics approach toward critical determinants for the imprinting of an evolutionarily conserved gene Impact. Biochem Biophys Res Commun. 2005;329:824–830. doi: 10.1016/j.bbrc.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weidman JR, Murphy SK, Nolan CM, Dietrich FS, Jirtle RL. Phylogenetic footprint analysis of IGF2 in extant mammals. Genome Res. 2004;14:1726–1732. doi: 10.1101/gr.2774804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- Neumann B, Kubicka P, Barlow DP. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagiwara-Takeuchi Y, Li T, Vu TH, Hirai M, Hattori M, Sakaki Y, Hoffman AR, Ito T. Comparative genome analysis of the mouse imprinted gene Impact and its nonimprinted human homolog IMPACT: toward the structural basis for species-specific imprinting. Genome Res. 2000;10:1878–1889. doi: 10.1101/gr.139200. [DOI] [PubMed] [Google Scholar]
- Gelfand Y, Rodriguez A, Benson G. TRDB—the Tandem Repeats Database. Nucleic Acids Res. 2007;35(Database issue):D80–D87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- Laird CD, McConaughy BL, McCarthy BJ. Rate of fixation of nucleotide substitutions in evolution. Nature. 1969;224:149–154. doi: 10.1038/224149a0. [DOI] [PubMed] [Google Scholar]
- Okamura K, Ito T. Lessons from comparative analysis of species-specific imprinted genes. Cytogenet Genome Res. 2006;113:159–164. doi: 10.1159/000090828. [DOI] [PubMed] [Google Scholar]
- Okamura K, Yamada Y, Sakaki Y, Ito T. An evolutionary scenario for genomic imprinting of Impact lying between nonimprinted neighbors. DNA Res. 2004;11:381–390. doi: 10.1093/dnares/11.6.381. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Holmes R, Chang Y, Soloway PD. Timing and sequence requirements defined for embryonic maintenance of imprinted DNA methylation at Rasgrf1. Mol Cell Biol. 2006;26:9564–9570. doi: 10.1128/MCB.00058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter B, Helms V, Paulsen M. Tandem repeats in the CpG islands of imprinted genes. Genomics. 2006;88:323–332. doi: 10.1016/j.ygeno.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Paoloni-Giacobino A, D'Aiuto L, Cirio MC, Reinhart B, Chaillet JR. Conserved features of imprinted differentially methylated domains. Gene. 2007;399:33–45. doi: 10.1016/j.gene.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunahara S, Nakamura K, Nakao K, Gondo Y, Nagata Y, Katsuki M. The oocyte-specific methylated region of the U2afbp-rs/U2af1-rs1 gene is dispensable for its imprinted methylation. Biochem Biophys Res Commun. 2000;268:590–595. doi: 10.1006/bbrc.2000.2189. [DOI] [PubMed] [Google Scholar]
- Reed MR, Riggs AD, Mann JR. Deletion of a direct repeat element has no effect on Igf2 and H19 imprinting. Mamm Genome. 2001;12:873–876. doi: 10.1007/s00335-001-2027-9. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Mann MR, Nwoko O, Duran KL, Bartolomei MS. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol Cell Biol. 2002;22:2450–2462. doi: 10.1128/MCB.22.8.2450-2462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Constancia M, Reik W. Tandem repeat hypothesis in imprinting: deletion of a conserved direct repeat element upstream of H19 has no effect on imprinting in the Igf2-H19 region. Mol Cell Biol. 2004;24:5650–5656. doi: 10.1128/MCB.24.13.5650-5656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto R, Kinoshita Y, Kawabe A, Kinoshita T, Takashima K, Nordborg M, Nasrallah ME, Shimizu KK, Kudoh H, Kakutani T. Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 2008;4:e1000048. doi: 10.1371/journal.pgen.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Xu Y, Uberbacher EC. Automated gene identification in large-scale genomic sequences. J Comput Biol. 1997;4:325–338. doi: 10.1089/cmb.1997.4.325. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Abdi H. Bonferroni and Sidak corrections for multiple comparisons. In: Salkind N, editor. Encyclopedia of Measurement and Statistics. Thousand Oaks, CA: Sage; 2007. pp. 103–107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Numeric data used to draw Figure 4 and the p-values of Fisher's exact test and Bonferroni correction for the periodicity of CpG sites.