Abstract
The crystal structures of the cytoplasmic domain of the putative zinc transporter CzrB in the apo- and zinc-bound forms reported herein are consistent with the protein functioning in vivo as a homodimer. NMR, X-ray scattering and size exclusion chromatography provide support for dimer formation. Full-length variants of CzrB in the apo and zinc-loaded states were generated by homology modelling with the Zn2+ / H+ antiporter YiiP. The model suggests a way in which zinc binding to the cytoplasmic fragment creates a docking site to which a metallochaperone can bind for delivery and transport of its zinc cargo. Since the cytoplasmic domain may exist in the cell as an independent, soluble protein a proposal is advanced that it functions as a metallochaperone and that it regulates the zinc-transporting activity of the full-length protein. The latter requires that zinc binding becomes uncoupled from the creation of a metallochaperone-docking site on CzrB.
Keywords: function, high-resolution, membrane protein, metal chaperone, X-ray structure, YiiP
Introduction
Zinc plays an essential role in the life of the cell in at least two important ways. The first concerns catalysis; the second structure stabilization (Alberts et al., 1998). In catalysis, its involvement is ubiquitous in that each of the six major enzyme groups has a representative that requires zinc. The fact that zinc is a strong Lewis acid and an electrophile explains its catalytic function: stabilizing anionic reaction intermediates in the case of certain peptidases and hydrogenases, and generating reactive nucleophiles such as hydroxyls in the case of carbonic anhydrase. The ability of zinc to stabilize structure is clearly evident in the zinc finger motif of nucleic acid binding proteins, many of which play important regulatory roles. Stabilizing supramolecular structure is another case in point as obtains in the hexameric form of insulin located in secretory vesicles. Further, by contributing to the three-dimensional form of certain enzymes, zinc stabilizes structure in a way that facilitates catalysis without being involved directly in the reaction.
The most common ligands for zinc in performing its diverse catalytic and structural functions are water and the side chains of histidine, cysteine, aspartate and glutamate. Coordination numbers are typically 4 and 6 displaying tetrahedral and octahedral geometry, respectively (Alberts et al., 1998). Because of its essential nature, zinc homeostasis is an important issue for all cells. This is particularly true in light of the fact that zinc can also be toxic, leading to cell death and to β-amyloid deposition. The most extensively studied organism from the point of view of zinc homeostasis must surely be Escherichia coli. It has a zinc quota (number of zinc atoms per cell) of just under a million, a value that is very tightly regulated (Outten and O'Halloran, 2001). Remarkable however is the fact that essentially all of the cellular zinc exists in a bound state with less than a single zinc atom on average remaining free in the cytosol. It appears therefore that zinc ions are shuttled about in the cell in the bound form.
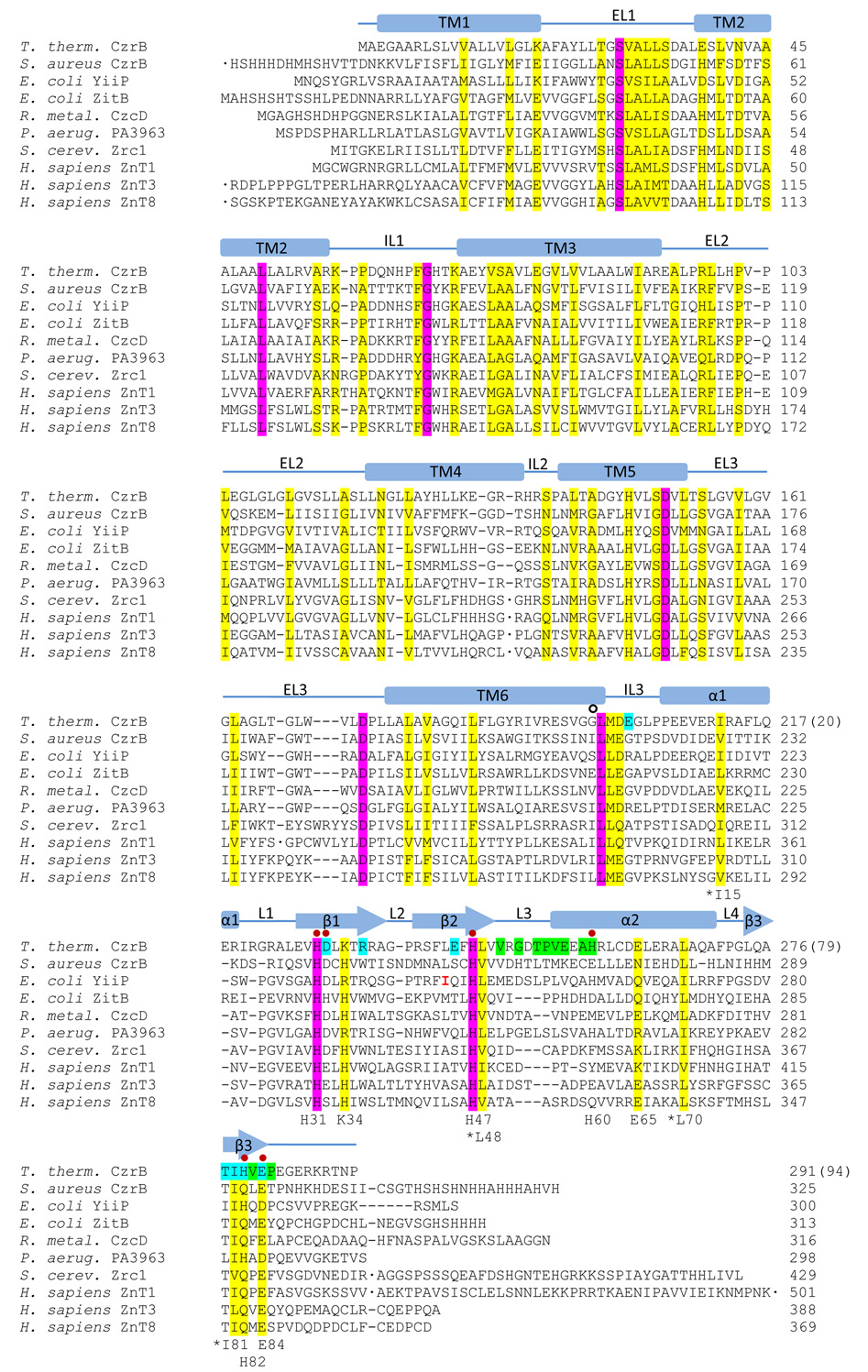
Part of the cellular homeostatic machinery involves transporter proteins that move zinc ions in and out of the cytosol. The cation diffusion facilitator (CDF) proteins constitute one such transporter family with members in all phyla (Haney et al., 2005; Montanini et al., 2007). In eukaryotes, for example, representatives include zinc tranporter-3 (ZnT-3) and ZnT-8 that function respectively to channel zinc into presynaptic and insulin secretory vesicles. Homologs in the bacterial world include CzrB in Thermus thermophilus, YiiP and ZitB in E. coli, CzcD in Ralstonia metallidurans, and PA3963 in Pseudomonas aeruginosa (Fig. 1). Of this group, YiiP has been the most extensively studied and shown to be a homodimer with Zn2+ / H+ antiporter activity (see Lu and Fu, 2007, and references therein). Our interest is in CzrB from Thermus and how it functions to transport zinc in this extreme thermophile (Spada et al., 2002). A long-term objective is to solve the high-resolution structure of the full-length protein in the apo and zinc-bound forms.
Figure 1.
Sequence alignment of CzrB from Thermus thermophilus with CDF proteins from other organisms that are involved in zinc transport. Identical and homologous residues are highlighted in magenta and yellow, respectively. Red dots mark residues involved in direct binding to zinc in CzrBsf. Secondary structure elements are indicated as α-helical (bars) and β-strand (arrows) using the zinc-CzrBsf structure. Residues involved in apo-CzrBsf dimerization are highlighted in green. Residues that make additional contacts subsequent to zinc binding are highlighted in blue. Numbers used to identify residues in the soluble fragment CzrBsf that are described in the text are shown at the bottom of each block. For reference, residue number 1 in CzrBsf corresponds to residue number 198 in the full-length protein and is marked with an open circle. The side chains of the conserved apolar residues (Ile15, Leu48, Leu70, Ile81) are all located in the hydrophobic core of CzrBsf. The conserved Glu65 has its side chain on the exposed surface of the protomer. Transmembrane helices are labeled TM1 – TM6 based on homology modelling using the known structure of YiiP (Lu and Fu, 2007).
By analogy to YiiP, CzrB is predicted to consist of an N-terminal transmembranal domain with six helices and a cytosolic C-terminus. There are indications that the C-terminal domain of CzrB may be expressed independently in vivo although the role played by this soluble fragment is not known (Spada et al., 2002). In the absence of crystals of the full-length protein it was considered worthwhile to establish the structure of the cytosolic domain in its apo and zinc-loaded form. Accordingly, the C-terminal domain, consisting of about 100 residues, was over-expressed in E. coli and the soluble fragment (CzrBsf) was crystallized with and without zinc (Höfer et al., 2007). The crystal structures of the apo and zinc-bound forms, solved to 1.7 and 1.8 Å, respectively, suggest that the cytosolic domain functions as a homodimer. The tendency of the water-soluble fragment to exist as dimers in solution has been verified in the current study by a variety of solution-based methods including small-angle X-ray scattering, 1H-nuclear magnetic resonance (NMR) spectroscopy, and size-exclusion chromatography. Whilst the current work was in progress the crystal structure of the zinc-bound form of YiiP at a resolution of 3.8 Å was reported (Lu and Fu, 2007). We have used this, in combination with the structures of apo and zinc-CzrBsf, to produce structural homology models for the full-length CzrB with and without bound zinc. The results suggest that the protein dimer functions as a fully integrated machine complete with sensor, actuator and transporter parts. Thus, the cytosolic zinc concentration is sensed by the exposed C-terminal domain and ion binding to it triggers a conformational change that facilitates uploading of zinc, presumably from a metallochaperone, for transport across the membrane. The model relies on the protein working as a dimer with the transmembrane domains oriented in the membrane in a zinc-independent fashion.
Results
Crystal Structures
The crystal structure of the apo and zinc forms of CzrBsf has been solved to 1.7 Å and 1.8 Å resolution, respectively. Refinement statistics are presented in Table 1.
Table 1.
Data collection and processing statistics
| Zinc-CzrBsf | Apo-CzrBsf | |||
|---|---|---|---|---|
| Data collection | ||||
| Data set | Crystal 2, set x2a | Crystal 1, set xtal 1c | Crystal 1, set x2c | |
| Wavelength (Ǻ) | 1.2827 | 1.2818 | 1.2567 | 0.9124 |
| Resolution (Ǻ) | 30 – 1.80 (1.90 – 1.80)a | 30 – 1.90 (2.00 – 1.90)a | 30 – 1.80 (1.90 – 1.80)a | 50 – 1.70 (1.76 – 1.70)a |
| Reflections (total/unique) | 56,909/7,849 | 46,123/6,667 | 50,341/7,811 | 270,944/21,179 |
| R(merge) | 0.072 (0.389)a | 0.126 (0.529)a | 0.099 (0.644)a | 0.042 (0.393)a |
| I/σ(I) | 20.1 (4.6)a | 16.0 (3.7)a | 14.8 (2.6)a | 54.8 (4.4)a |
| Completeness (%) | 98.2 (96.4)a | 97.5 (93.2)a | 98.1 (96.3)a | 98.9 (92.4)a |
| Redundancy | 7.3 (7.4)a | 6.9 (6.6)a | 6.4 (6.6)a | 12.8 (6.2)a |
| Space group, unit cell | C2221; 50.8 × 74.5 × 43.9 Ǻ3 90° 90° 90° | P3212; 60.33 × 60.33 × 92.14 Ǻ3 90° 90° 120° | ||
| Structure solution | ||||
| Resolution / reflections used for MAD | 30 - 2.2 Ǻ / 4,414 | |||
| Zn sites | 3 (relative occupancies 1.0, 0.9, 0.4) | |||
| Z-score, FOM | 13.4, 0.56 | |||
| Residues built by RESOLVE | 86; 72 placed in sequence | |||
| R / Rfreeb after RESOLVE | 0.331 / 0.336 | |||
| Refinement | ||||
| Resolution | 30 - 1.80 (1.85 - 1.80)a | 50 - 1.70 (1.74 - 1.70)a | ||
| Final model | Residues 3–91, 4 Zn, 99 HOH | Residues 6–87 (A,B chains), 3 SO4, 301 HOH | ||
| Missing structure | Res. 1–2, 92–94; N of Met 3; sidechains of Met 3, Glu 86 | Res. 1–5, 88–94; side chains of Glu 10, Glu 21 | ||
| Multiple conformations | Glu 10, Arg 16, Arg 61, Arg 68 | Gln A20, Gln B20, Arg B22, Asp B32, Glu B65, Arg B68 | ||
| Model atoms | 836 | 1650 | ||
| Reflections (total / test set) | 7397 / 378 | 20053 / 1086 | ||
| R / Rfreeb | 0.207 / 0.250 (0.254 / 0.365)a | 0.187 / 0.224 (0.225 / 0.319)a | ||
| RMSD (bond lengths / angles) | 0.008 Ǻ / 1.109° | 0.012 Å / 1.202° | ||
| TLS groups | 5 | 2 | ||
Numbers in parentheses refer to the highest resolution shell
Rfree was calculated using 5 % of data randomly excluded from refinement
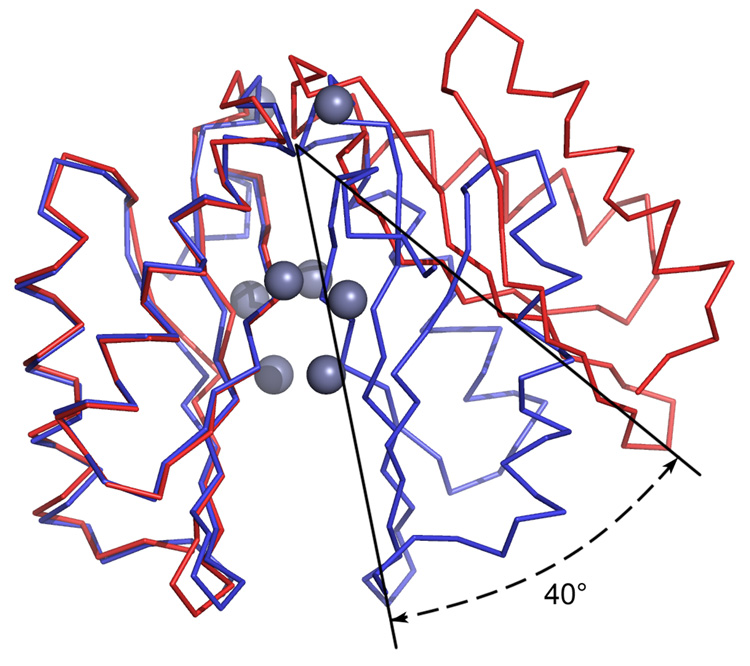
The apo form of the protein crystallized as a dimer in the asymmetric unit (Figure 2). With zinc bound the asymmetric unit contains a single protein molecule. However, application of crystallographic symmetry produces a dimer (Figure 2). Our view is that both forms of the protein exist as dimers under physiological conditions. Support for this conclusion is presented below. Henceforth in this paper they will be treated as dimers.
Figure 2.
Ca traces of apo- (red) and zinc-CzrBsf (blue). Zinc ions are shown as bluish gray spheres. The protomers on the left were overlain to highlight the change in relative orientation of the second protomer upon zinc binding. The figure shows that metal binding triggers a dramatic reorientation of the two monomers with respect to one another as illustrated by the 40° angular shift.
The apo form of CzrBsf includes residues 6 – 87 in each protomer; the zinc form includes 3 additional N-and 4 additional C-terminal residues. The apo and zinc models include 3 sulfate and 4 zinc ions, respectively. One each of the sulphate and zinc ions (Zn4) sits on a special position with occupancy of 0.5. About 100 and 300 structured waters, respectively, have been placed in the zinc and apo forms of the protein.
Apo CzrBsf
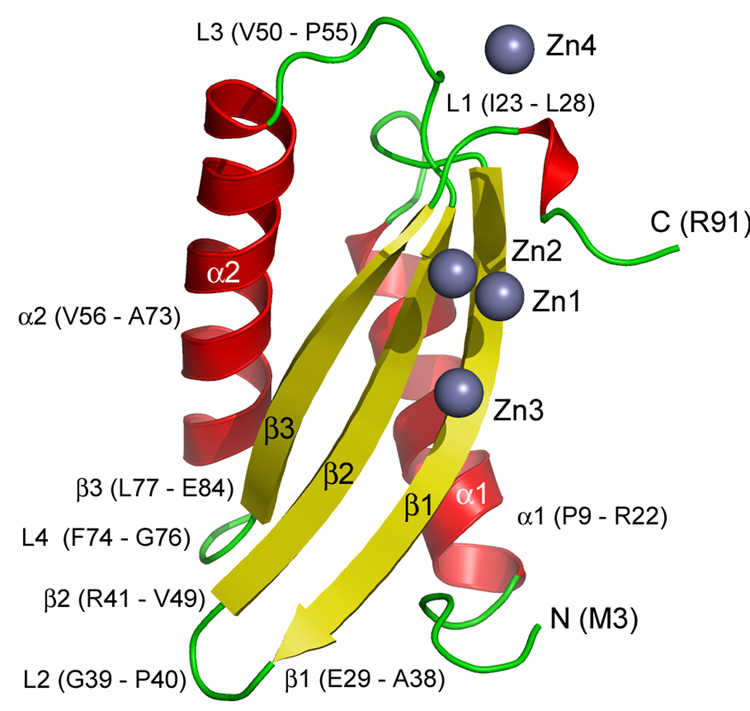
The overall shape of the protomer within the apo homodimer is that of an ellipsoid with a flattened face paralleling its long axis (Figure 2 and Figure 3). The protomer is highly structured with a distinctly polar surface and a hydrophobic core (Figure 4). It consists of three β-strands and two α-helices. The former exist as a mixed β-sheet with the central strand running parallel and anti-parallel to its neighbor on either side. It creates the planar surface of the flattened ellipsoid. The rounded part of the domain is made up of the two helices that run anti-parallel to one another and are slightly displaced from each other along their long axes. They sit on top of the β-sheet. The connectivity of the different elements in the protomer from the N-to the C-terminus is α1-β1-β2-α2-β3 (Figure 3). The N-terminus of CzrBsf connects to the transmembrane domain presumably by an extension of its C-terminal helix.
Figure 3.
Cartoon representation of secondary structure elements (α-helix, red; β-strand, yellow; loops and disordered regions, green) in the zinc-CzrBsf model. Zinc ions are shown as bluish gray spheres. Parts of the protein referred to in the text are labeled. The one-letter code is used to identify amino acids.
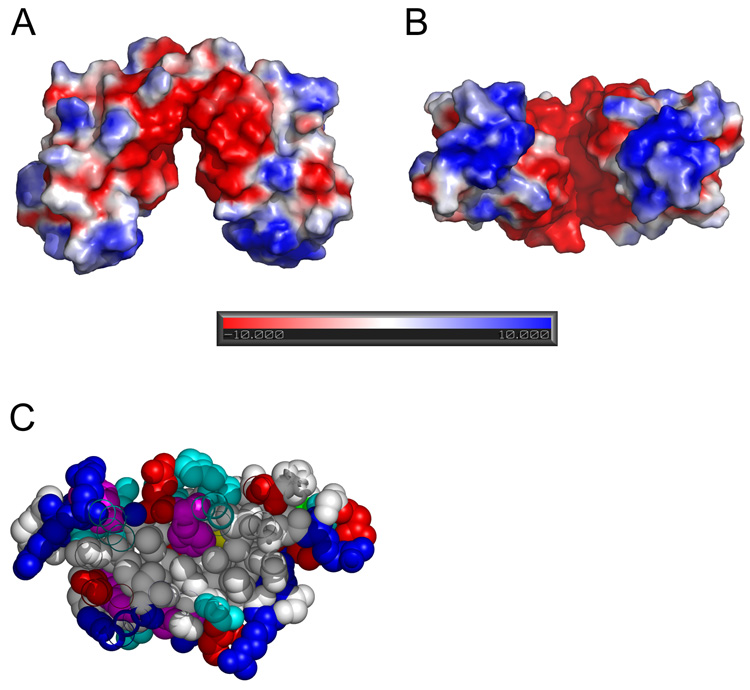
Figure 4.
Surface potential (A, B) and hydrophobic core of apo-CzrBsf (C). The surface potential (negative, red; positive, blue; neutral, white) was calculated using the Adaptive Poisson-Boltzmann Solver (APBS) program. The pronounced negative charge in the cleft between the two protomers likely contributes to keeping the apo-form of the dimer in the ‘open’ splayed apart conformation. The negatively charged cleft is viewed from the side (parallel to the membrane surface) in A and from below (normal to the membrane surface) in B. A cross-section through one of the apo-CzrBsf monomers in C clearly shows the micelle-like nature of this cytosolic fragment with its apolar interior and polar surface. Residues are colored according to side chain type: hydrophobic – white, polar – cyan, aromatic – magenta, positive – blue, negative – red.
The apo form of the protein exists as a dimer in the asymmetric unit (Figure 2). Contact between the monomers along one end of the flattened ellipsoid involves particularly the loop connecting β2 and α2 and residues toward the C-terminus (hydrogen bonds (two of each per dimer): Glu57(N) – Gly52(O) 3.0 Å, Val56(N) – Thr54(O) 2.8 Å; hydrophobic contacts involve Val50, Pro55, Val56, Ala59, His60, Val83, and Pro85. This gives the dimer a distinct inverted V-shaped appearance with the membrane-anchored ends of each protomer splayed apart (Figure 2). Facing one another from either arm of the V are the flattened β-sheets of the two protomers.
Zinc CzrBsf
The zinc form of the protein is also considered to exist as a homodimer (Figure 2) with very minor changes in protomer secondary and tertiary structure associated with metal binding. A superposition of the apo-CzrBsf and Zn-CzrBsf models (Figure 2) reveals an RMS difference of 0.7–0.8 Å over 82 common Cα atoms; the RMS difference between protomers in the apo-dimer is 0.45 Å. The largest zinc-induced changes occur in the backbone of the β1-β2 and β2-α2 loops.(RMSD, 1–2 Å). In the vicinity of the zinc binding sites the backbone is mostly unaltered compared to the apo form (RMSD < 1 Å). However, the side chains of residues that interact with zinc reorient to provide better coordination (Figure 5). Further, with zinc bound the density for several additional residues at both the C- and N-termini are revealed, some of which participate in crystal contacts. The extended C-terminus contains a 310 helical turn. The most dramatic effect of zinc binding is that the two protomers that are splayed apart in the apo form snap together along the flat surfaces of the opposing ellipsoids (Figure 2). The effect comes about as a result of an action that brings the two arms of the inverted V together with a hinge-like motion about the contact surface at the β2-α2 loop region. The net effect is to bring the N-terminus ends of the two protomers into very close proximity. The distance between N-termini in the apo-dimer is 33.5 Å. The distance between corresponding residues (Gly6) in the zinc form is 15.2 Å. Since these are connected to the transmembrane domain it is likely that this large zinc-triggered motion will impact on the relative disposition of the cytosolic and transmembrane domains, as discussed below.
Figure 5.
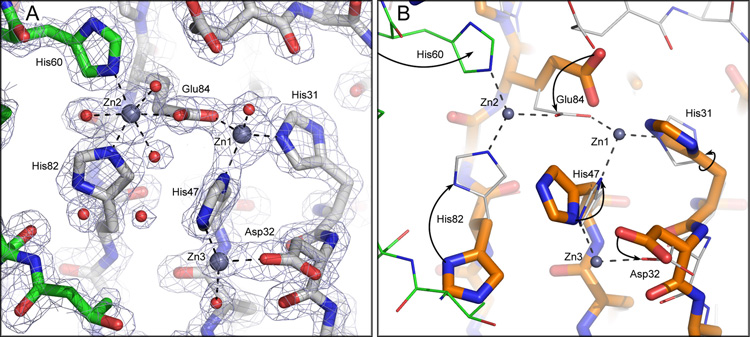
Zinc coordination at the dimer interface in CzrBsf. (A) Zinc-CzrBsf protomers in different colors. (B) An overlay of zinc and apo protomers. The two protomers in zinc-CzrBsf are distinguished in A by having carbon atoms colored gray and green, respectively. Amino acid oxygens and nitrogens are shown as red and blue sticks, respectively. Zinc ions are represented as bluish gray spheres. In A water oxygens are shown as red spheres and electron density (2Fo-Fc) is contoured at 1.5 sigma. The overlay in B shows apo-CzrBsf with thick sticks and orange carbon atoms and zinc- CzrBsf with thin sticks and gray carbon atoms. Arrows highlight the changes side chains undergo upon binding zinc.
The snapping shut of the V upon metal ion binding has the effect that the contact surface area between the two protomers goes from ~400 Å2 in the absence of zinc to ~1,050 Å2 in its presence. The additional contact area is stabilized by hydrogen bonds (two of each per dimer) Glu45(OE1) – Thr80(OG1), 2.9 Å; His82(ND1) – Ile81(O), 2.8 Å), salt bridges (two of each per dimer) (Arg36(NH1) – Glu5(OE2), 3.0 Å; Arg36(NH2) – Glu5(OE1), 2.6 Å), and by the coordination of a zinc ion (Zn2, two per dimer).
Zinc Coordination Sites
Each protomer is associated with four zinc ions (Zn1-Zn4). The first three exist at or close to the interface between protomers in the dimer and may be physiologically relevant. Zn4 is likely only present in the crystal. It sits at a special position shared by two symmetry-related protomers from different dimers. It is coordinated by two Glu57(OE1) from different dimers and by 4 waters, and is 2.4 Å distant from Glu57(OE1).
Zn1 is tetra-coordinated by His47(ND1) (bond length, 2.0 Å), His31(NE2) (2.1 Å), and Glu84(OE2) (1.9 Å), all from the same protomer, and by HOH517 (2.0 Å) (Figure 5). Liganding atoms in the primary coordination sphere around Zn1 are arranged tetrahedrally.
Zn2 is hexa-coordinated by His82(NE2) (2.1 Å) and Glu84(OE1) (2.3 Å) in protomer A, by His60(NE2) (2.2 Å) in protomer B, and by 3 water molecules: HOH585(O) (2.3 Å), HOH527(O) (2.3 Å) and HOH589(O) (2.5 Å) (Figure 5). The geometric arrangement of the six ligand atoms with respect to zinc is octahedral. All residues that are involved in coordinating Zn2 from protomer B are part of helix α2 whereas those from protomer A originate in strand β3. Given that Zn2 is clearly coordinated by residues in both monomers it presumably is part of the glue that binds the two protomers together in the dimer. Its symmetry partner does the same thing on the other side of the dimer effectively pinning the monomers together.
Zn3 is likely to be tetra-coordinated. It is clearly coordinated by His47(NE2) (2.4 Å), Asp32(OD2) (2.0 Å), and HOH516(O) (2.5 Å) (Figure 5). A fourth ligand (possibly water of weak density) however, is not apparent. The lack of density for a fourth ligand can be explained by weaker interactions and higher disorder at this site. In fact, the B-factor of Zn3 at 80 Å2 is substantially higher than that of the other two Zn sites (17 and 18 Å2 for Zn1 and Zn2, respectively). In vivo this site is likely unoccupied at lower concentrations of zinc.
Zn1 and Zn2 are separated by a distance of 4.9 Å and are bridged by the carboxyl oxygens of Glu84. Zn1 and Zn3 are 5.9 Å apart and are bridged by the imidazole of His47.
The distances and angles of liganding atoms in the primary coordination spheres of Zn1, Zn2 and Zn3 are close to ideal and are in agreement with zinc serving a structural role in the zinc-CzrBsf dimer (Alberts et al., 1998). The geometry of zinc coordinating residues is close to ideal and does not appear to be distorted by interactions with zinc (Table S1 in Supplemental Material).
Characteristics of the Protein in Solution
The crystal structure data suggest that both the apo and zinc forms of the CzrBsf protein exist as homodimers. It is possible that the constraints imposed by the crystal lattice artifactually give rise to dimer formation. Since the CzrBsf part of the full-length transporter is expected to be extramembranal and to be exposed to the cytosol, it was considered worthwhile to evaluate the tendency of the soluble fragment to oligomerize and to characterize its structure in aqueous solution. Accordingly, its solution properties have been quantified by size exclusion chromatography, small-angle X-ray scattering, 1H-NMR spectroscopy and circular dichroism, as follows.
Size Exclusion Chromatography
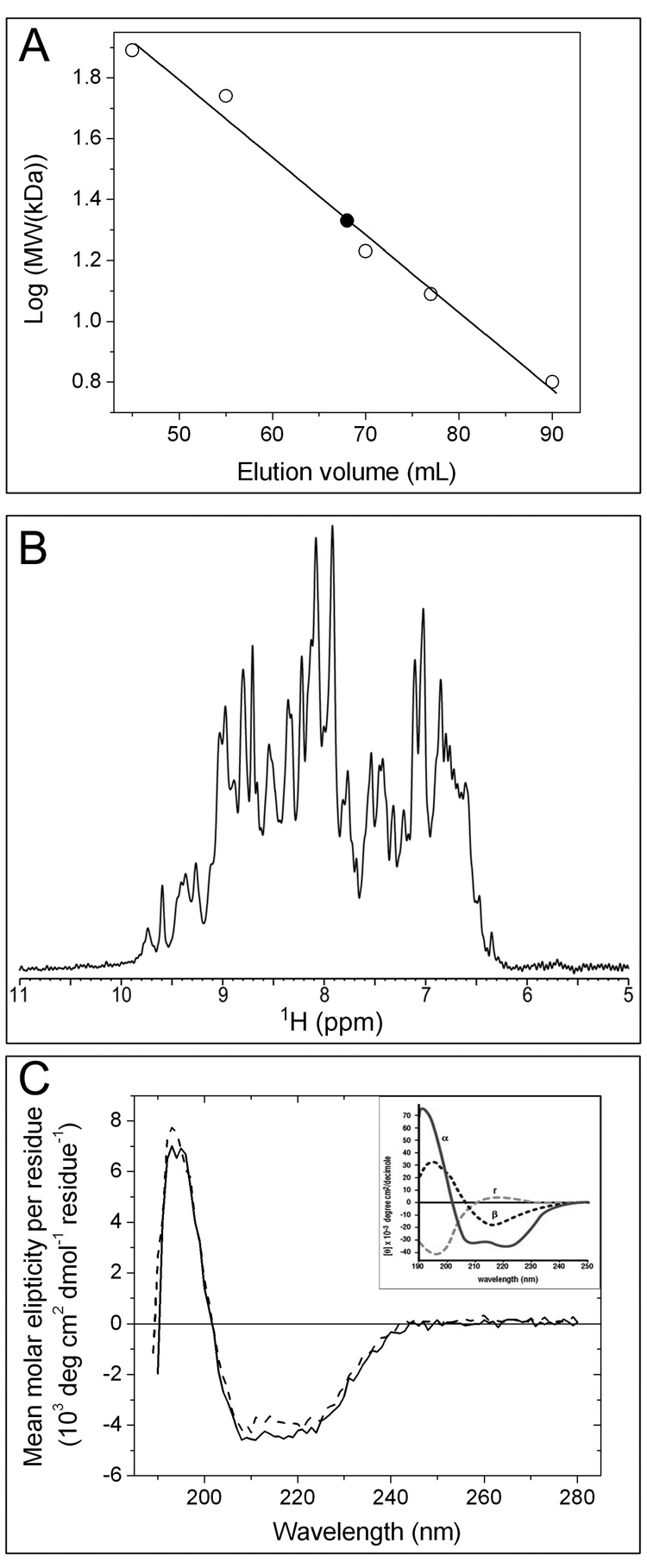
The hydrodynamic volume and state of oligomerization of apo-CzrBsf in solution were investigated using size exclusion chromatography. Five water-soluble proteins, whose molecular weights bracketed that of the test protein, were employed as standards. The data (Figure 6A) show that apo-CzrBsf elutes with an apparent molecular weight of 21.6 kDa. The calculated molecular weight of the protein, based on its amino acid composition, is 10.8 kDa. These data are consistent with the apo form of the protein existing in solution as a dimer.
Figure 6.
Characterization of the solution properties of CzrBsf. (A) Size exclusion chromatography. Apo-CzrBsf eluted at an included volume of 68 mL (filled square) corresponding to a molecular weight of 21.6 kD. The identity and molecular weight of the standard proteins used to calibrate the column (open squares) follow: conalbumin (77 kDa), hyaluronidase (55 kDa), myoglobin (16.9 kDa), cytochrome c (12.4 kDa), and GB1 (6.2 kDa). The line of best fit to the data is shown as a solid line., (B) 1H Nuclear magnetic resonance. The amide and aromatic region of the 1H NMR spectrum of ~1 mM apo-CzrBsf at 25 °C and pH 6.5 is shown. (C) Circular dichroism spectra for secondary structure analysis of the His-tagged protein are shown before (solid line) and after lyophilization (dashed line). The inset shows for comparison the circular dichroic behavior of proteins exclusively in the α-helical (α), antiparallel β-sheet (β) and random coil (r) conformations (from ‘Protein Structure and Function’, ed. Petsko GA and Ringe D, 2004).
Small-angle X-ray Scattering
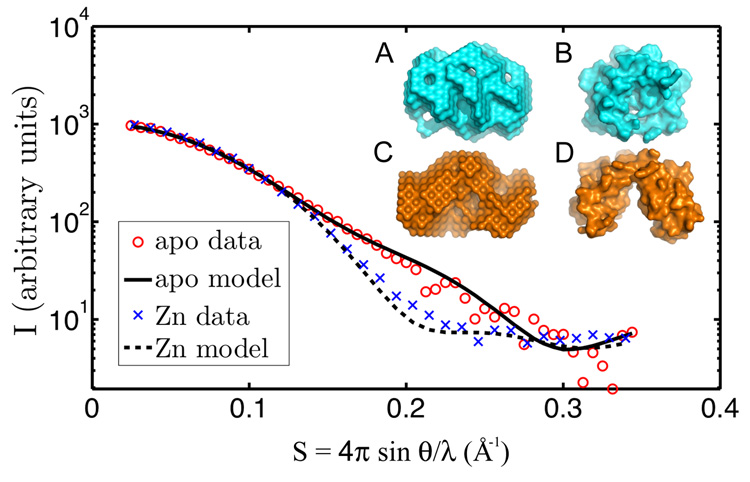
Small-angle X-ray scattering (SAXS) of protein solutions can yield information about the oligomeric state of the solute and can also provide low-resolution envelopes and other structural information (Svergun, 1999). Accordingly, the SAXS behavior of the apo and zinc forms of CzrBsf in solution was investigated and the corresponding scattering profiles are presented in Figure 7. For comparison, the expected profiles calculated using the X-ray crystallographic models (section Crystal Structures) in their dimeric configurations are included in the figure. The experimental solution scattering data are in excellent agreement with the predicted curves and support the view that both apo and zinc forms of the protein exist as dimers in solution. At low resolution (S < 0.1 Å−1), the apo- and zinc-CzrBsf scattering profiles overlap. However, they diverge significantly in the vicinity of S = 0.2 Å−1. This is consistent with a change in dimer shape upon zinc binding. The radius of gyration for apo-CzrBsf calculated from the scattering data is 18.2 Å. The corresponding value for the zinc form of the protein is 15.7 Å. Similarly, the maximum diameters, Dmax, which optimize the match of the observed and calculated profiles are 60 Å and 55 Å, respectively, for the apo and zinc forms of CzrBsf. These data are consistent with the formation of a more compact dimeric structure upon binding zinc. Low-resolution envelopes constructed from the scattering curves using DAMMIN (Svergun, 1999) are shown in Figure 7 (inset) along with the corresponding envelopes from the crystal structures. These results support the contention that the crystal structures indeed reflect both the configuration and oligomeric state of the apo and zinc forms of the protein in solution.
Figure 7.
Small-angle X-ray scattering from CzrBsf in solution with and without added zinc. The scattering data are shown as individual data points (with zinc, blue x; without added zinc, red circle). Predicted scattering profiles are shown as solid or dashed lines along with the corresponding apo- (inset A) and zinc-CzrBsf (inset C) models. For comparison, the crystal structures of zinc- and apo-CzrBsf are shown in insets B and D, respectively.
NMR Spectroscopy
A small section from a one-dimensional 1-1 echo 1H NMR spectrum of apo-CzrBsf corresponding to the amide and aromatic proton signals is shown in Figure 6B. The dispersion of the amide proton resonances in the ~8–10 ppm region indicates that the protein is folded under the experimental conditions used. Moreover, a series of 1-1 echo 1H spectra with increasing echo delays were recorded and used to determine the average transverse relaxation time, T2, of the amide protons (HN). As discussed by (Anglister et al., 1993), the average HN T2 can be used to estimate the effective protein rotational correlation time (τc) according to τc ≈ 1/(5T2) ns, where T2 is the average HN T2 value in seconds. For the ~1 mM apo-CzrBsf solution at 25 °C the average amide proton T2 was 13.3 ms, which gives an effective τc of ~15 ns. The rotational correlation time of a protein is determined by both molecular size and shape, and the estimated τc value of ~15 ns is characteristic of proteins having molecular weights in the ~20 – 30 kDa range (Cavanagh et al., 2007). This clearly indicates that apo-CzrBsf oligomerizes in solution, most likely into dimeric form. Additional NMR spectra (data not shown), recorded at 25 °C and pH 6.5 as a function of protein concentration (between ~0.25 and 1 mM protein) and ionic strength (between 0 and 100 mM NaCl) revealed effectively identical HN resonance frequencies and average T2 values, indicating the high stability of CzrBsf dimers in solution.
Circular Dichroism
Circular dichroism in the UV region provides useful information on the secondary structure make-up of a protein in solution. Apo-CzrBsf with the His-tag in place was subjected to this form of analysis and the data are shown in Figure 6C. The spectrum reveals the characteristic negative peaks in ellipticity in the vicinity of 208 nm and 222 nm associated with α-helices and at 217 nm associated with β-strands. Deconvolution of the spectrum using three different algorithms in the CDPro program suite, as described under Experimental Procedures, provided an average probable secondary structure composition of 18 % α-helix, 30 % β-strand, 25 % turn and 27 % disordered region (Table S2). The corresponding values obtained based on the X-ray structure are 35, 30, 12 and 23 %, respectively (DSSP; Kabsch and Sander, 1983). The disparities should be viewed in light of the fact that the CD values are based on a polypeptide containing 111 residues, the N-terminal segment of which includes a His-tag and is presumably disordered. In contrast, the crystal structure is based on an analysis of 82 ‘ordered’ residues in the apo form of CzrBsf. Lyophilization of the protein had little effect on its secondary structure as revealed by CD analysis (Figure 6C).
Discussion
The crystal structure of the cytoplasmic domain of the putative zinc transporter CzrB in Thermus determined in this study is consistent with the protein functioning in vivo as a dimer. This was corroborated by solution studies with CzrBsf that involved size exclusion chromatography, 1H NMR and small-angle X-ray scattering. In what follows, the zinc binding characteristics of the cytoplasmic domain are examined and a proposal is advanced for how CzrBsf may function as a metallochaperone and as an independent regulator of CzrB zinc transport activity.
Zinc Binding
We anticipate that three of the four zinc ions observed in the zinc-CzrBsf structure are physiologically relevant (Figure 2). The fourth, Zn4, likely plays a role in crystallogenesis and crystal packing.
The Zn1 and Zn2 sites are homologous with the two zinc sites in the cytosolic domain of the E. coli zinc transporter, YiiP (Figure S1; (Lu and Fu, 2007)). As in YiiP, they straddle the three centrally located β-strands of the domain with Zn2 acting to bridge protomers in the dimer. The two zinc ions are 4.2 Å apart in YiiP. The corresponding separation in CzrBsf is 4.9 Å. The zinc coordination is similar between the two proteins but there are notable differences. For example, the carboxyl of the highly conserved Glu84 in CzrBsf is coordinated to Zn1 and Zn2. The corresponding residue in YiiP is Asp285 where just one of the carboxyl oxygen atoms appears to engage in bidentate coordination to bridge the two zinc ions. One other notable difference arises at His47 where the imidazole side chain of this conserved residue bridges Zn2 and Zn3 in CzrBsf. In YiiP the corresponding residue is Ile245. We speculate that the disparity between CzrBsf and YiiP at this residue and in this region of the structure arises from a 3-residue frame shift in the region between loops L2 and L3 that includes strand β2 in YiiP (Figure S2). The misalignment is perhaps understandable given that the YiiP structure is reported at 3.8 Å.
In the absence of zinc the surface potential of CzrBsf in the cleft between the two protomers is highly negative (Figure 4A and B). By charge repulsion this will contribute to holding the two protomers apart and to stabilizing the inverted V-shape of the apo form of the protein. The negative potential arises primarily from the side chains of Glu45, Glu67, Glu84, Asp32 and Asp64. Upon zinc binding the side chain carboxyls of these residues form hydrogen bonds (Glu45 – Thr80), coordinate directly with zinc (Asp32, Glu84) or do so via a water molecule (Glu67, Asp64).
The large negative surface potential between protomers in the apo-CzrBsf dimer will serve to direct zinc into the cleft region as intracellular concentrations of the metal ion rise. We speculate that the Zn1 site will be the first to fill because three of its coordinating ligands (His31, His47, Glu84) already exist on the surface of apo-CzrBsf with side chains oriented in such a way that only small changes are required to properly coordinate Zn1, as illustrated in Figure 5. The rotation of the imidazole ring of His47 to ligand Zn1 may in turn facilitate the repositioning of Asp32 to coordinate with the second zinc, Zn3, via its side chain carboxyl. It may also facilitate the relatively large movement of the His82 imidazole which contributes to creating a binding pocket for Zn2. The latter is stabilized also by the carboxyl of the Glu84 side chain, already suitably positioned following the locking in of Zn1. With this ‘partial pocket’ in place, the stage is set for the neighboring protomer to rotate into position and to bring its surface exposed His60 in on top of the anchored Zn2. Along with three solvent molecules this then completes the hexacoordination of the protomer-bridging Zn2. The net effect of this sequential zinc binding is to snap the open end of the dimer shut and to change the relative disposition of the cytosolic and transmembranal domains with possible consequences for interactions with metallochaperones and zinc transport, as outlined below.
Homology Modelling and Mode of Action
YiiP is a Zn2+ / H+ antiporter in E. coli. The zinc-bound form of the full-length transporter was solved recently to a resolution of 3.8 Å (Lu and Fu, 2007). CzrB is a homolog with an overall sequence identity of 32.6 %. The corresponding values for the cytoplasmic and transmembranal domains are 31.5 % and 33.2 %, respectively. Despite the relatively low sequence homology in the cytoplasmic domains, the structural homology between them is impressive. Thus, for example, zinc-CzrBsf overlays the cytoplasmic domain of YiiP with a Cα-RMSD of 1.8 Å over 79 residues (Figure S2).
The transmembranal domain of YiiP is made up of a bundle of six tightly packed helices. Five of these extend across the membrane and encircle a sixth short helix in a way that creates two cavities at either end of the bundle. One cavity is exposed to the periplasm (and to the outer leaflet of the membrane) and contains a zinc ion at its base. The second is exposed to the cytosol and access to it is presumably by way of a space that exists between the cytoplasmic and transmembranal domains. It has been proposed that a metallochaperone delivers zinc to this intracellular cavity. The ion then makes its way across the membrane into the extracellular cavity where effectively it is out of the cell (Lu and Fu, 2007). The model shows the YiiP dimer in the form of a Y. The stem of the Y is the cytoplasmic dimer and the two arms represent the transmembrane domains that diverge from one another as they angle across the bilayer. In the current study, we have solved the structure of the apo and zinc-bound forms of CzrBsf. The YiiP structure, which is of the full-length transporter, is only available in the zinc-bound state. By homology modelling to the latter we set about exploring the changes in the full-length form of CzrB that might accompany zinc binding to its cytoplasmic domain, CzrBsf, and that may provide clues to the mode of action of the protein as a cation facilitator transporter. Accordingly, a structural homology model of the transmembrane domain of CzrB (CzrBtm) was created using the corresponding domain of YiiP (YiiPtm). This was spliced to the high-resolution structure of CzrBsf in both its apo and zinc-bound states with a view to producing a picture of the full-length protein. In this modelling exercise, the orientation of the transmembrane domain in the bilayer ‘observed’ with YiiP (Lu and Fu, 2007) was preserved in the CzrBsf-CzrBtm/YiiPtm ‘chimera’.
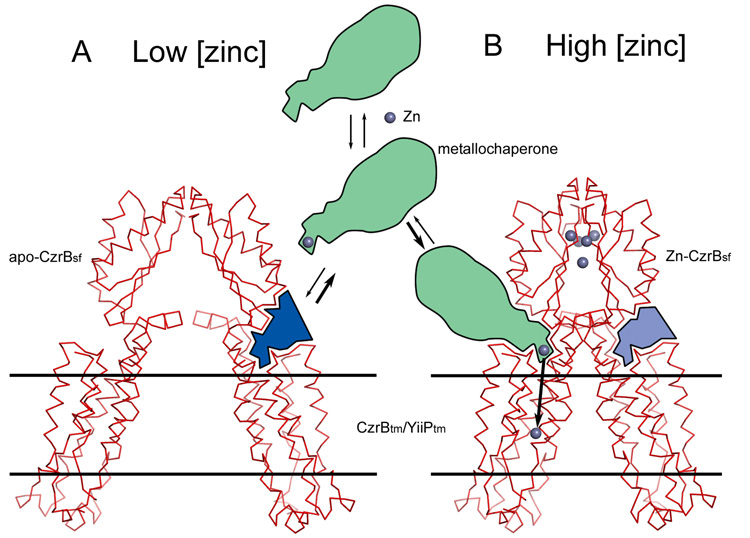
As noted, in the apo form of CzrBsf the protomers of the dimer are splayed apart. They snap together into close proximity upon zinc binding. When this ion-binding event happens in the chimera the angle and the space between the cytoplasmic and transmembrane domains changes dramatically (shaded blue in Figure 8). This is the region where the holo-metallochaperone (green in Figure 8) is proposed to interact with the transporter and to transfer its cargo of metal ions to the intracellular cavity for translocation across the membrane. In this model, zinc binding to the CzrBsf triggers the conformational change that enables chaperone docking and zinc delivery for transport through the transmembrane part of the protein.
Figure 8.
A model for CzrB action as a zinc transporter. The proposed mechanism is based on the changes induced in the dimer conformation of CzrBsf upon zinc binding (Figure 2) combined with homology modelling from the YiiP (Lu and Fu, 2007) to the CzrB transmembrane domains. The structures shown are chimeras of the apo-CzrBsf and CzrBtm/YiiPtm (A) and zinc-CzrBsf and CzrBtm/YiiPtm (B) as described in the text. At low zinc levels the relative orientation of the CzrBsf and CzrBtm domains is such that the zinc-loaded metallochaperone (green) fails to dock productively (see site represented by light blue in A) with CzrB to deliver its cargo of metal. Binding of zinc to CzrBsf causes the dimer to snap shut bringing the two transmembrane domains into close proximity and creating sites (represented by the dark blue area in B) conducive to zinc-metallochaperone docking and to zinc ion delivery.
This proposal requires that the orientation of the transmembrane domain of the full-length transporter in the membrane remains fixed and independent of zinc status. It also requires that zinc binding trigger a clamping together of the cytosolic domains in the dimer. The net effect is to alter the angle between the cytosolic and transmembrane domains in a way that enables interaction with a zinc-bearing chaperone. High-resolution X-ray structures of the full-length CzrB with and without zinc will go a long way toward evaluating this proposal.
It is interesting to consider the possibility that CzrBsf, which may be expressed in the cell as an independent protein, itself acts as a metallochaperone. Further, given that the full-length protein would appear to function as a homodimer of form CzrB-CzrB, the monomeric form of CzrBsf could destabilize the active full-length species by competing with the partnered cytoplasmic domains. This would lead to an inactive heterodimer of form CzrB-CzrBsf (Figure S3). In this hybrid, it is possible for the two CzrBsf’s to snap shut upon zinc binding but without any attendant change in the relative orientations of the cytoplasmic and transmembrane domains in the full-length monomer. As a result no docking site is created for the metallochaperone to bind and to deliver its cargo. Regardless of zinc status then the CzrB-CzrBsf complex would be incompetent as a transporter. In this way, CzrBsf could regulate the transport activity of the protein.
Conclusions
The crystal structure of the cytoplasmic domain of the putative zinc transporter CzrB from Thermus thermophilus has been solved. Structures, determined to resolutions of 1.7 and 1.8 Å in the apo- and zinc-bound forms, respectively, are consistent with the protein functioning in vivo as a dimer. Support for dimer formation was provided by solution studies involving size exclusion chromatography, 1H NMR and small-angle X-ray scattering. The overall structure of the CzrBsf monomer, which consists of two antiparallel α-helices sitting atop a 3-stranded mixed β-sheet, is stabilized by an apolar core and is quite insensitive to zinc binding. However, the conformation of the dimer changes dramatically upon zinc binding. In the apo-form the two monomers associate at one end of the molecule and are splayed apart at the other. With zinc loading the monomers snap together along their full length. The zinc bound form of the protein includes four zinc ions, one of which plays a role in crystallogenesis. The other three are located in an anionic cleft at the monomer-monomer interface and are considered to be physiologically relevant. Zinc coordination involves highly conserved histidine, glutamate and aspartate residues, and water molecules. Full-length variants of CzrB in the apo and zinc-loaded states were generated by homology modelling with the known Zn2+ / H+ antiporter YiiP. The model suggests a way in which zinc binding to the cytoplasmic fragment creates a docking site to which a metallochaperone can bind for delivery and transport of its zinc cargo. Since the cytoplasmic domain may exist in the cell as an independent, soluble protein a proposal is advanced that it functions as a metallochaperone and that it regulates the zinc-transporting activity of the full-length protein. The latter requires that zinc binding becomes uncoupled from the creation of a metallochaperone-docking site on CzrB. A more complete evaluation of these assorted proposals awaits a high-resolution structure of the full-length protein in its apo and zinc-bound forms.
Experimental Procedures
Protein Expression and Purification
Full details of the expression and purification of the His-tagged (6His-CzrBsf) and His-tag-free cytosolic domain of CzrB (CzrBsf) have been reported (Höfer et al., 2007). Samples of the His-tag-free domain for NMR analysis were subjected to gel filtration using a column (volume: 120 mL, HiLoad™ 16/60, Superdex™ 75, preparative grade, Amersham Biosciences, GE, Piscataway, NJ) pre-equilibrated with 25 mM sodium phosphate buffer pH 6.5. The protein eluted as a single, Gaussian-shaped peak and had an estimated purity of 95 % based on SDS-PAGE analysis. Protein concentration was determined by the Bradford method (Höfer et al., 2007).
Crystallization, X-ray Diffraction, Structure Determination and Refinement
Crystallization of the apo and zinc-bound forms of CzrBsf was performed as described (Höfer et al., 2007). Apo-CzrBsf crystals were obtained by hanging drop vapor diffusion at 20 °C by combining 1 µL of a 15 mg protein/mL solution in 20 mM Tris-HCl pH 8.0 and 1 µL of precipitant (0.2 M ammonium sulfate, 15 %(w/v) PEG 4000, 0.1 M sodium acetate pH 4.6) and using 300 µL of the same precipitant in the reservoir. Crystals of the Zn-CzrBsf were grown in batch mode at 4 °C at final concentrations of 28 mg protein/mL, 25 mM zinc chloride, 0.1 M Tris-HCl pH 7.5.
X-ray data were collected on the F1 (apo-CzrBsf) and F2 beamlines (Zn-CzrBsf) at the Cornell High Energy Synchrotron Source (CHESS, Ithaca, NY). On F1, a 100 µm-diameter 0.9124 Å beam was used together with an ADSC Quantum-4 detector positioned 180 mm from the crystal. On F2, data were collected using a 20 µm-diameter beam produced by single-bounce capillary optics and an ADSC Quantum-210 detector placed 134 mm from the sample. Two crystals were used to obtain full datasets at the zinc edge (1.2827 Å), the zinc peak (1.2818 Å) and the zinc high energy remote (1.2567 Å).
Data on the Zn-CzrBsf crystals were integrated with MOSFLM (Leslie, 1992) and scaled with SCALA (CCP4, 1994). Multiple wavelength anomalous diffraction (MAD) phasing was carried out at 2.2 Å with SOLVE (Terwilliger and Berendzen, 1999) using the signal from the bound zinc, followed by density modification and automatic chain tracing by RESOLVE (Terwilliger, 2003). Three zinc ions were located and an interpretable map was produced; the iterative build procedure of RESOLVE placed almost the entire polypeptide chain, with unambiguous assignment of residues to the CzrBsf sequence. Refinement was performed using REFMAC (Murshudov et al., 1997), Coot (Emsley and Cowtan, 2004), and O (Jones et al., 1991); for the later stages all available data, to 1.8 Å resolution, were used. Five TLS groups were used for the last few refinement steps. The Zn3 zinc ion is apparently of low occupancy; it was located during refinement and its identity verified by an anomalous difference Fourier map. Waters were placed automatically by Coot, with a few manual adjustments. The final R and Rfree values were 0.207 and 0.250, respectively. Details of the structure determination are summarized in Table 1.
The apo-CzrBsf crystal diffraction data were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997). MOLREP (CCP4, 1994) was used to place the Zn-CzrBsf model without zinc ions in the apo unit cell. ARP/wARP (Perrakis et al., 2001) was then invoked and about 90 % of the model was built automatically. A few residues in the loops were manually built using O and were input into ARP/wARP to complete the model. During the last stages of model building, water molecules were added using ARP/wARP. A final restrained refinement with REFMAC using TLS parameters brought the R and Rfree values to 0.187 and 0.224, respectively.
The quality of both structures was examined with MolProbity (Davis et al., 2007). The total MolProbity score placed the Zn-CzrBsf structure in the 95th percentile (score 1.45) and the apo-CzrBsf model in the 97th percentile (score 1.31). The Ramachandran plot revealed that the Zn-CzrBsf structure has 98.9 % and 1.1 % of its residues occupying the most favoured and additionally allowed regions, respectively. The corresponding values for the apo-form are 98.8 % and 1.2 %.
Circular Dichroism
Secondary structure analysis was carried out using 6His-CzrBsf eluted from a Ni-NTA column in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 200 mM imidazole. The buffer was exchanged into 50 mM sodium phosphate, 200 mM NaCl, pH 7.4 by ultrafiltration (2 mL Centricon, 3 kDa cut-off) at 5,000 g and 4 °C. A 20 µL sample with a concentration of 3.3 mg protein/mL was transferred into a quartz cuvette with a path length of 0.1 mm. CD data were recorded at 20 °C using an AVIV-202 Circular Dichroism Spectrometer (AVIV Biomedical Inc., Lakewood, NJ) from 280 to 180 nm with 1 nm steps and 2 s integration time per point. A CD spectrum of the corresponding buffer was recorded under the same conditions and subtracted from that of the protein solution. CD data were analyzed by 3 popular methods from the CDPro program suite (Sreerama and Woody, 2000) using a reference set of 43 soluble proteins.
SAXS
Data Collection
Solution scattering data were collected on beamline G1 at CHESS (Ithaca, NY). A custom 1,024 × 1,024 (69.78 µm) pixel CCD detector fabricated by the Gruner group (Cornell University, Ithaca, NY) was used to measure sample scattering profiles. Two-dimensional images were integrated by Data Squeeze 2.07 (Datasqueeze Software, Wayne, PA) to give one-dimensional intensity profiles as a function of the momentum transfer S = 4πsinθ/λ (where 2θ is the scattering angle). Measurements were taken at 20 °C with a sample-to-detector distance of 880 mm. With a calibrated wavelength of 1.236 Å (10.03 keV), scattering profiles covered an S-range from 0.025 to 0.327 Å−1. The incident X-ray beam was collimated to a spot size measuring 0.5 × 0.5 mm2 which was significantly smaller than the opening of the sample cells. Commercial sample cells holding approximately 12 µl (ALine Inc., Redondo Beach, CA) were fitted with 25 µm thick scratch-free mica windows (Attwater, Lancashire, UK). Silver behenate powder (The Gem Dugout, State College, PA) was used to locate the beam center and to calibrate the sample-to-detector distance.
CzrBsf in 50 mM Tris-HCl pH 7.5 was concentrated to 28 mg/mL. All samples were centrifuged at 10,000 × g for 5 min prior to loading the sample cell. Matching buffer samples were used for solvent subtraction. The zinc-bound protein was prepared by gently rocking 100 µL of 28 mg protein/ml solution with 2.5 µL 1.0 M ZnCl2 solution at 4 °C for 5 min. To assess radiation-induced aggregation and possible denaturation, samples were exposed twice for 5 s, 20 s, and 120 s, comparing Guinier plots of successive curves for nonlinearity at low angle. Protein concentrations of 28 mg/mL and 2.8 mg/mL were also compared to assess concentration dependence. Best data for the apo-form of the protein were obtained by combining the low-angle portion of the first 20 s exposure (S < 0.1 Å−1) with the wide-angle portion (S > 0.1 Å−1) of the first 120 s exposure, both at 2.8 mg protein/mL. The zinc-bound protein solution became cloudy after the first 120 s exposure and showed signs of increased sensitivity to radiation damage. Accordingly, the scattering profile was obtained by combining a 5 s exposure at low angles (S < 0.1 Å−1) with a 120 s exposure at wider angles (S > 0.1 Å−1) using a sample solution that was 3.5 mg protein/mL.
Data Analysis
Pair-distance distribution functions were calculated from scattering profiles using the GNOM program (Svergun, 1992). The maximum diameter of the particle (Dmax) was adjusted in GNOM to obtain the best goodness-of-fit parameter (0.827 "good" for the apo state, and 0.93 "excellent" for the Zn-bound state). The distribution function fell naturally to near zero beyond Dmax but was constrained to zero for the final solution. Radius of gyration (RG) is also computed by GNOM. Expected scattering profiles were calculated from the crystal structures using the CRYSOL program (Svergun et al., 1995). Experimental profiles were fed into CRYSOL at the final stage of computation for automatic superposition. Low-resolution envelopes were obtained from the pair-distance distribution functions using the DAMMIN (Svergun, 1999) program in fast mode. No symmetry was imposed on the solution, so the symmetric appearance of the envelopes in both cases reflects the underlying symmetry of the complex. Envelopes were visualized with PyMOL (DeLano, 2002) using atomic radii set to the dummy atom packing radius determined by DAMMIN.
Size Exclusion Chromatography
Size exclusion chromatography was used to assess oligomer formation. For this purpose, the mobility of 10 – 30 mg CzrBsf on a column (volume: 120 mL, HiLoad™ 16/60, Superdex™ 75, preparative grade, Amersham Biosciences, GE, Piscataway, NJ) in a mobile phase consisting of 50 mM Tris-HCl pH 8.0 at a flow rate of 1 mL/min and at 24 °C was monitored. The column was calibrated using the following molecular weight marker proteins: myoglobin (16.9 kDa; Sigma), cytochrome c (13.4 kDa; Sigma), hyaluronidase (55 kDa; Sigma), conalbumin (77 kDa; Sigma), and the B1 immunoglobulin binding domain of protein G (GB1, 6.2 kDa; for details on protein expression and purification see (Nadaud et al., 2007)).
NMR Spectroscopy
Protein for use in the NMR studies was column purified as described above, and an Amicon Ultra-15 5,000 Molecular Weight Cut-Off device (Millipore, Billerica, MA) was used to exchange the buffer and concentrate the protein. The final NMR sample consisted of CzrBsf at a concentration of ~1 mM in an aqueous solution containing 50 mM sodium phosphate pH 6.5, 5 mM dithiothreitol (DTT), 7 %(v/v) deuterium oxide (99.96 % D20, Aldrich, Milwaukee, WI), and 0.02 %(w/v) sodium azide in a total volume of 0.5 mL. For the NMR measurements the sample was transferred to a 5 mm sample tube (Wilmad-Labglass, Buena, NJ). NMR experiments were carried out at 25 °C using a Bruker spectrometer (Bruker, Billerica, MA) operating at 600 MHz 1H frequency and equipped with a triple resonance three-axis pulsed field gradient probe, optimized for proton detection. The 1-1 pulse echo sequence (Sklenar and Bax, 1987) was used to qualitatively assess the structural integrity of CzrBsf in solution, and to determine the average transverse relaxation time of the amide protons.
Multiple Alignment and Homology Modelling
Multiple sequence alignment of full-length CzrB with its homologues from different organisms was performed using the M-coffee web-server (Moretti et al., 2007). Despite low strict homology, the alignment shows good consistency and an overall alignment score of 90. The alignment along with the recently solved YiiP structure (PDB ID 2QFI; Lu and Fu, 2007) were submitted to the Swiss-Model web-server (Schwede et al., 2003) to build a full-length CzrB homology model. The model corresponds to the zinc-bound form of CzrB. The cytoplasmic domain of the CzrB model was structurally aligned and replaced with the solved high-resolution zinc-CzrBsf structure. The zinc-CzrB dimer was generated by applying crystallographic symmetry in the zinc-CzrBsf structure to the full-length zinc-CzrB model. To produce a full-length model of the apo-CzrB dimer, we assumed that the orientation of the transmembrane part of CzrB (CzrBtm) in the membrane remains the same regardless of the zinc loading state. Accordingly, the dimeric form of zinc-CzrBsf in the zinc-CzrB model was replaced by the apo-CzrBsf dimer domain preserving the membrane orientation of the CzrBtm domains. This manipulation resulted in a moving apart of the two CzrBtm domains by ~18 Å and a changing in the angle at the presumed flexible linker between CzrBsf and CzrBtm by ~20°.
Supplementary Material
Supplemental Data including Materials, 2 tables and 3 figures can be found online at http://
ACKNOWLEDGEMENTS
The research was supported by grants provided by Science Foundation Ireland (02-IN1-B266), the National Institutes of Health (GM61070 and GM75915) and the National Science Foundation (IIS-0308078) to MC, Enterprise Ireland (CFTD/04/106) to JGW, and The Ohio State University (start up funds to CPJ). This work is based on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR 0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Coordinates and structure factors have been deposited in Protein Data Bank with accession numbers 3BYP for apo-CzrBsf and 3BYR for zinc-CzrBsf.
REFERENCES
- Alberts IL, Nadassy K, Wodak SJ. Analysis of zinc binding sites in protein crystal structures. Protein Sci. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglister J, Grzesiek S, Ren H, Klee CB, Bax A. Isotope-edited multidimensional NMR of calcineurin B in the presence of the non-deuterated detergent CHAPS. J. Biomol. NMR. 1993;3:121–126. doi: 10.1007/BF00242480. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy. San Diego: Academic Press; 2007. [Google Scholar]
- Collaborative Computational Project, N. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMol molecular graphic system. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Haney CJ, Grass G, Franke S, Rensing C. New developments in the understanding of the cation diffusion facilitator family. J. Industrial Microbiol. & Biotech. 2005;32:215–226. doi: 10.1007/s10295-005-0224-3. [DOI] [PubMed] [Google Scholar]
- Höfer N, Kolaj O, Li H, Cherezov V, Gillilan R, Wall JG, Caffrey M. Crystallization and preliminary X-ray diffraction analysis of a soluble domain of the putative zinc transporter CzrB from Thermus thermophilus. Acta Crystallogr. F. 2007;63:673–677. doi: 10.1107/S1744309107032277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992 [Google Scholar]
- Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317:1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics. 2007;8:107. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S, Armougom F, Wallace IM, Higgins DG, Jongeneel CV, Notredame C. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007;35:W645–W648. doi: 10.1093/nar/gkm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nadaud PS, Helmus JJ, Höfer N, Jaroniec CP. Long-range structural restraints in spin-labeled proteins probed by solid-state nuclear magnetic resonance spectroscopy. J. Am.Chem.Soc. 2007;129:7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Petsko GA, Ringe D. Protein Structure and Function. London: New Science Press; 2004. [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modelling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenar V, Bax A. Spin-echo water suppression for the generation of pure phase two-dimensional NMR spectra. J. Magn. Res. 1987;74:469–479. [Google Scholar]
- Spada S, Pembroke JT, Wall JG. Isolation of a novel Thermus thermophilus metal efflux protein that improves Escherichia coli growth under stress conditions. Extremophiles. 2002;6:301–308. doi: 10.1007/s00792-001-0257-0. [DOI] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analyt. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data including Materials, 2 tables and 3 figures can be found online at http://