Abstract
The application of systems biology methods in the emerging field of glycomics requires the collection and integration of glycosyltransferase data at the gene and enzyme level for the purpose of hypothesis generation. We systematically examined the relationship between gene expression, glycosyltransferase activity, glycan expression, and selectin-binding function in different systems, including human neutrophils, undifferentiated HL-60 (human promyelocytic cells), differentiated HL-60, and HL-60 synchronized in specific growth phases. Results demonstrate that 1) the sLeX (sialyl-Lewis-X) epitope is expressed in P-selectin glycoprotein ligand-1 (PSGL-1) from neutrophils at higher levels compared with HL-60. This variation may be due to differences in the relative activities of α1,3-fucosyltransferases and α2,3-sialyltransferases in these two cell types. 2) HL-60 cell differentiation along granulocyte lineage increased the activity of β1,4GalT and β1,3GlcNAcT by 1.6- to 3.2-fold. This may contribute to LacNAc chain extension as evidenced by the 1.7-fold increase in DSA-lectin (lectin recognizing LacNAc) binding to cells after differentiation. 3) The activity of enzymes contributing to sLeX formation in leukocytes likely varies as ST3[Galβ1,4GlcNAc] ≤ α1,3FT[sialyl-LacNAc] < β1,3GlcNAcT. 4) O-glycan specific glycosyltransferase activity does not undergo periodic variation with cell cycle phases. Overall, gene expression and enzyme activity data combined with knowledge of biochemistry can predict the resulting glycan structures and yield viable experimentally testable hypothesis.—Marathe, D. D., Chandrasekaran, E. V., Lau, J. T. Y., Matta, K. L., Neelamegham, S. Systems-level studies of glycosyltransferase gene expression and enzyme activity that are associated with the selectin binding function of human leukocytes.
Keywords: systems biology, glycan, PSGL-1, inflammation, hydroxyurea
Selectins are key adhesion molecules that contribute to inflammatory ailments and to certain types of cardiovascular disorders (1). P-selectin glycoprotein ligand-1 (PSGL-1), expressed on leukocytes, is a high affinity ligand for all three selectins (E-, P-, and L-selectin). This glycoprotein has 71 Ser/Thr sites for O-linked glycosylation and 3 sites for N-glycans. During disease, carbohydrate epitopes displayed on PSGL-1 bind selectins expressed on blood leukocytes, activated platelets, and stimulated endothelial cells. This molecular interaction enhances cell adhesion in the vasculature. The major carbohydrate epitope of PSGL-1 that recognizes selectins is O-linked, and it is located at the N-terminal anionic polypeptide segment of the protein in close proximity to the sulfated tyrosine residues (2). Sialyl-Lewis-X (sLeX)-type structures expressed on PSGL-1 represent the prototypic oligosaccharides that bind selectins (3,4,5).
Human promyelocytic leukemia HL-60 cells are often used as a model system to study leukocyte and selectin function. The core-2 tetrasaccharide Galβ1, 4GlcNAcβ1,6(Galβ1,3)GalNAcα (structure 1, Fig. 1) is a prominent structural constituent of O-glycans in granulocytes and promyelocytes, including PSGL-1 in HL-60 cells (6,7,8). Nine to 14% of PSGL-1 O-glycans are fucosylated, and of these only a minor fraction (2–4.5%) are thought to participate in selectin recognition. This minor structure is disialylated and monofucosylated with a terminal sLeX epitope. Glycosyltransferase enzymes involved in the construction of such glycans include α2,3-sialyltransferases (sialylTs); α1,3-fucosyltransferases (FTs); β1,3- and β1,4-galactosyltransferases (GalTs); and β1,3- and β1,6-GlcNActransferases (GlcNAcTs). The functional importance of these enzymes is supported by studies with various knockout mice that demonstrate that the lack of key enzymes like core-2 β1,6GlcNAcT-I, ST3Gal-IV, FT-VII, and GalT-I can lead to partial or complete lack of selectin-mediated leukocyte adhesion (9).
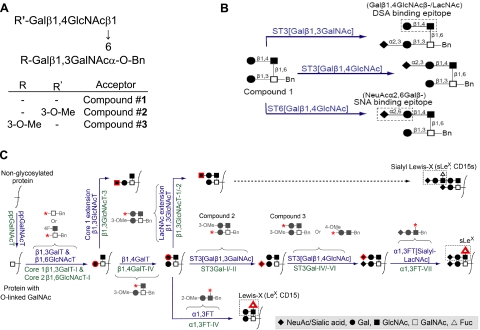
Figure 1.
Chemical structures of acceptors and schematic of potential O-linked glycosylation pathway in HL-60 cells. A) Compounds 2 and 3 are methylated acceptors based on the core-2 tetrasaccharide, structure 1. B) Compound 1 can be sialylated by at least 3 different enzyme activities, shown in blue above reaction arrows. Sialylated products formed can be assayed by monitoring lectin binding to cells, DSA-lectin for Galβ1,4GlcNAcβ/LacNAc and SNA-lectin for NeuAcα2,6Galβ. ST3[Galβ1,3GalNAc] catalyzes α2,3-sialylation of Galβ1,3GalNAcα. ST3[Galβ1,4GlcNAc] and ST6[Galβ1,4GlcNAc] catalyze α2,3/α2,6-sialylation of Galβ1,4GlcNAc. C) Schematic of biochemical pathway leading to sialyl Lewis-X (sLeX, NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAcβ−) formation in HL-60 cells. Enzyme activities are shown in blue above reaction arrows; candidate genes responsible for this activity are shown in green below the arrow. Red outline around selected monosaccharides indicates molecules added in corresponding steps. Acceptor substrates used to assay specific enzyme activities are shown in gray. Many of these acceptors are modified with fluoro (F) or methyl (Me) groups to distinguish between closely related enzyme activities. They are also modified with Benzyl (Bn) groups at the anomeric position to simplify postreaction separation of products. Red asterisk denotes fee reaction site available on the acceptor substrate. Monosaccharides in B and C are represented using symbolic nomenclature shown in symbol legend.
This study quantitatively examines the role of carbohydrate glycosylating enzymes in regulating the formation of selectin ligands in human leukocytes. It is significant for the emerging field of glycomics, since it is among the first to take a “systems approach” in studies of cellular glycosylation. Although traditional studies have focused individually on selected classes of glycosyltransferases in HL-60 cells and neutrophils (10,11,12,13,14,15,16,17,18), no study has comprehensively studied all enzymes contributing to O-linked glycosylation simultaneously in a systematic manner. We addressed this issue here by performing experiments at multiple levels including the gene, protein, glycan, and functional levels. Here, various hypotheses are generated by monitoring the gene expression and enzyme activity of a range of glycosyltransferases in either primary human neutrophils or HL-60 cells cultured under a variety of conditions. Enzymatic studies are performed with methylated and fluorinated synthetic, carbohydrate acceptors that were designed to discriminate a particular enzyme activity in a complex mixture. These hypotheses are then tested by examining selected glycan structures at the whole-cell and individual-protein level using flow cytometry. In the later case, PSGL-1 is immunoprecipitated onto polystyrene beads. Glycan structures on the immobilized protein are then quantitatively probed using carbohydrate-binding monoclonal antibodies and a lectin. The studies demonstrate that gene expression and enzyme activity data, when combined with knowledge of biochemistry, yield viable experimentally testable hypothesis.
MATERIALS AND METHODS
HL-60 cell and neutrophil experiments
HL-60 cells were cultured in Iscove’s modified Dulbecco’s media containing 20% fetal bovine serum as suggested by the American Type Culture Collection (ATCC; Manassas, VA). In some runs, these cells were differentiated along the granulocytic lineage by addition of 1.25% dimethyl sulfoxide (DMSO) to culture media (19). Differentiation was monitored by measuring cell-surface Mac-1 (CD11b) expression daily using anti-Mac-1 antibody (phycoerythrin conjugated clone D11, BD Pharmingen, San Diego, CA, USA) and flow cytometry (using FACSCalibur, BD Biosciences). Viability of differentiated cells was >90% as determined by trypan blue. Control runs did not contain DMSO.
In some studies, HL-60 cells were synchronized in the G0/G1 phase by addition of 1 mM hydroxyurea (HU) for 13 h. Other cell cycle inhibitors tested (3 μg/ml aphidicoline, 400 ng/ml nocodazole) were not as effective at synchronization. The Supplemental Materials describe cell cycle analysis methods using propidium iodide. After synchronization, cells were washed and resuspended in fresh media without HU. In controls, HL-60 cells were grown in the absence of HU.
Human polymorphonuclear leukocytes were isolated from freshly collected human blood obtained by venipuncture in 10 U/ml heparin (Elkins-Sinn, Cherry Hill, NJ, USA). After gradient separation (4), erythrocytes were removed by hypotonic lysis. Polymorphonuclear leukocytes were then stored in Ca2+-free HEPES buffer at 4°C. Cell viability was >99%, and >90% of the isolated leukocytes were neutrophils. Thus, we term these isolated cells as neutrophils. For glycosyltransferase activity assays, ∼50 × 106 neutrophils were further purified by sorting using a FACS-Vantage instrument. Here, gating based on granulocyte characteristic forward side-scatter profile along with differential staining using anti-CD16-FITC (BD Biosciences) was used to distinguish neutrophils from eosinophils and other residual leukocyte populations.
Live cell samples obtained at fixed time points were used for flow cytometry analysis (described next). Alternatively, cell samples were pelleted and stored frozen at −80°C for the remaining assays.
Cell surface antigens, lectin, and P-selectin binding
Flow cytometry measurements were performed with HL-60 cells and neutrophils as described previously (4, 20), with minor modifications. Briefly, cells obtained at fixed time points were washed and resuspended in 30 mM HEPES buffer (pH=7.2) containing 1.5 mM CaCl2 and 0.1% human serum albumin at 0.5 × 106 cells/ml. In some runs, fluorescent monoclonal antibodies (mAbs) or lectins were incubated with these cells for 15 min at room temperature before reading in a flow cytometer. mAbs used include reagents against Lewis-X/LeX/CD15 (clone HI98-FITC), sialyl-Lewis-X/sLeX/CD15s (HECA-452-FITC or purified CSLEX-1 along with appropriate fluorescent secondary Ab), PSGL-1/CD162 (KPL-1-phycoerythrin/purified) and CD45 (C11-FITC, Ancell, Bayport, MN, USA). All mAbs including isotype control reagents were from BD Pharmingen unless mentioned otherwise. FITC conjugated lectins used include DSA (Datura stramonium) and SNA (Sambucus nigra) (EY Laboratories, San Mateo, CA, USA). In other runs, P-selectin fusion protein with a mouse IgG2a tail was incubated with 1:200 dilution of FITC-conjugated F(ab′)2 goat-anti-mouse secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA) and cells for 30 min at room temperature. Cells were washed before cytometry analysis in all studies performed with mAbs. Samples were diluted in 20-fold excess buffer before cytometry analysis in lectin binding assays. In control runs, P-selectin binding could be abrogated by 10 μg/ml blocking antibodies against P-selectin, anti-PSGL-1, 5 mM EDTA, or 2 mM soluble sLeX (4).
In some cases, HL-60 cells were treated with 100 U/ml chymotrypsin in HEPES buffer for 15 min at room temperature, washed twice, and resuspended at 0.4–1 × 106 cells/ml before cytometry analysis. Control runs were performed without chymotrypsin.
Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis
Total RNA isolation from cells, quantitative RT-PCR, and quantification of relative mRNA levels of glycosyltransferase genes were performed as described elsewhere (see ref. 21 and Supplemental Materials). Supplemental Table S1 lists individual primer pairs. PCR products for selected genes (indicated in the table) were purified from agarose gel and sequenced to confirm that the unique desired product was generated.
Cytometry beads and Western blotting
Glycan structures in PSGL-1 from HL-60 cells and neutrophils were assessed by immunoprecipitating this protein from cell lysate both for quantitative cytometry bead and qualitative Western blot analysis. In both cases, HL-60/neutrophils were lysed at 4°C in isotonic Tris-HCl lysis buffer containing 2% Nonidet P-40, 2 mM EDTA, 2 mM PMSF, and complete mini protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Protein concentration in lysate was determined by Bradford/Coomassie assay kit (Pierce, Rockford, IL, USA).
For cytometry bead assays, carbodiimide chemistry was used to covalently link anti-PSGL-1 mAb TB5 (mouse IgG1, BioVendor, Candler, NC, USA) to 6 μm carboxyl polystyrene beads (Polysciences, Warrington, PA, USA; ref. 22; see Supplemental Materials). Beads thus obtained are termed TB5 beads. The density of Ab on these beads was ∼19,000/μm2.
In each cytometry bead experiment, 10–400 μg of protein from cell lysate was incubated with 30,000–50,000 TB5 beads for 1 h at room temperature with gentle agitation. After washes with PBS containing 0.025% Tween-20, ∼7000–10,000 beads in separate vials were incubated with either 10–20 μg/ml of HECA-452-FITC (anti-sLeX), DSA-FITC lectin, or unconjugated anti-PSGL-1 polyclonal antibody (H-300, Rabbit IgG, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 20 min at room temperature. In the last case, 1:200 FITC-conjugated F(ab′)2 mouse-anti-rabbit IgG (Jackson ImmunoResearch) was subsequently added for 20 min after a washing step. Samples were washed before flow cytometry analysis. Flow cytometry geometric mean fluorescence intensity data are presented after subtraction of TB5-bead background signal obtained in the absence of cell lysate. Competitive binding assays and isotype-control beads described in Results were used along with Western blotting to validate these studies.
For the Western blots, 1–2 mg of protein from HL-60 and neutrophil lysate was precleared using protein A/G beads. PSGL-1 was then immunoprecipitated using 6 μg/ml of anti-PSGL-1 mAb (KPL-1, mouse IgG) and protein A/G beads. Immunoprecipitate equivalent to 300 μg lysate loaded in each lane was separated using an 8% SDS-PAGE gel, transferred onto nitrocellulose membrane, and probed using either KPL-1 or anti-sLeX Ab (HECA-452, rat IgM). Appropriate HRP-conjugated secondary Abs (Jackson ImmunoResearch) were subsequently added before developing signal with Lumiglo chemiluminescent substrate (Cell Signaling, Danvers, MA, USA).
Glycosyltransferase assays
HL-60/neutrophil cell pellets were homogenized in 0.1 M Tris-maleate buffer (pH 7.2) containing 2% Triton X-100 using a Dounce all-glass hand-operated grinder. The homogenate was centrifuged at 20,000 g for 1 h at 4°C. The protein concentration in the supernatant, measured using the BCA method (Pierce), was adjusted to 5–7.5 mg/ml by addition of extraction buffer. Glycosyltransferase activity in the cell lysate was determined by mixing the lysate (which contains the enzyme) with acceptor and radiolabeled donor sugar nucleotide under the reaction conditions detailed in Supplemental Materials for sialylTs (23), sulfotransferases (sulfoTs; ref. 24), GalTs (25), ppGalNAcTs, GlcNAcTs, and FTs (10, 26). Previous studies and other rationale outlined in the Supplemental Materials guided the choice of acceptors used in individual assays.
Many of the carbohydrate acceptors used are based on the core-2 tetrasaccharide structure (compound 1, Fig. 1A). Specific methyl modifications of this compound (compounds 2 and 3) were also applied to distinguish between closely related enzyme activities that may act on compound 1 (Fig. 1A, B). For example, although α2,3-sialylation is possible at the three position of Gal in both the Galβ1,3GalNAc and Galβ1,4GlcNAc arms of compound 1, only one of the two reactions is possible in each of the methyl analogs. Figure 1C presents potential biochemical pathways that lead to sLeX synthesis on PSGL-1 in leukocytes (23). Names of genes (below individual reaction arrows, green) that most likely catalyze specified enzymatic activity (above arrows, blue) are listed here. The figure also presents structures of selected acceptors (in gray) used to assay individual enzyme activities.
After reaction for 2- to 4-h (depending on assay design), the radiolabeled product for each run was separated from unreacted donor using either anionic (Dowex-1-Cl) or hydrophobic (Sep-Pak C18) chromatography or MagneHis Nickel particles (Promega, Madison, WI, USA). For GalT, GlcNAcT, FT, and sulfoT assays, the reaction mixture was diluted with 1 ml water and passed through a 1 ml Dowex-1-Cl column and the column was washed twice with 1 ml water. The breakthrough and the water wash contained the [14C]-galactosylated or [14C]-fucosylated products from neutral acceptors. Three milliliters of 0.2 M NaCl was used to elute [14C]-fucosylated products of sialylated acceptor and [35S]-sulfated compounds. For sialylT assays, the radioactive products from benzylglycosides and monosialylated benzylglycosides were separated on Sep-Pak C18 cartridges using elution of product by 3 ml methanol. In the ppGalNAcT assay, Ni-particles were used to isolate radioactive product, since PSGL-1 peptide with a 6X-His tag was used as an acceptor substrate. The radioactive content of isolated products was determined using Beckman LS9000 scintillation counter. Thin layer chromatography was performed with products from all reactions for which significant conclusions were drawn (Supplemental Fig. S1). These validate that unique products were formed in individual experiments as anticipated by the experimental design.
Controls for each assay contained the reaction mixture with everything except the exogenous acceptor. Radioactivity of control was subtracted from that of product for the results presented here. Radioactivity of controls was typically 50–100 counts per min (cpm)/assay, while efficient reactions result in counts >103 cpm/assay. All samples were tested in duplicate. Results from duplicate runs did not vary by more than 5%. Duplicate determination does not allow statistical analysis. Product formation rate varied approximately linearly with time during the course of our assays, with at most 12.7% of the sugar nucleotide being consumed over the time course (column 10 of Supplemental Table S2; Supplemental Fig. S2). Control experiments with varying concentrations of Triton-X (0.2–1%, Supplemental Fig. S3) show that product formation is not affected by the presence of detergent (0.5–0.66%) in our assays. The presence of glycosidases that can affect our measurements was not evident.
First-order rate constant from biochemical data
In the enzymatic assays for sialylT, FT, and GlcNAcT, the acceptor concentration exceeds monosaccharide concentration by >330-fold (column 4 vs. columns 7+9; Supplemental Table S2). Thus, in the Michaelis-Menten rate expression, v = Vm[S]/(Km+[S]), both the limiting substrate [S] and equilibrium constant Km are calculated with respect to the monosaccharide. Further, since typically Km > monosaccharide concentration ([S] for nucleotide-sugar donor) under our experimental conditions (column 11 vs. 7+9; Supplemental Table S2), the rate expression can be approximated to a first-order rate expression v∼k[S] (with k=Vm/Km, units of time−1). This apparent first-order rate constant, k, can then be calculated using reaction velocity (v) data obtained in the above experiments [k=(dpm of product/volume/time)/(specific activity of sugar nucleotide)/(sugar nucleotide concentration)]; k thus obtained is a measure of enzyme activity under our assay conditions and it includes enzyme concentration information.
A detailed derivation on how k measured in our experiments is related to the rate constant in the Golgi/in vivo (kvivo) is provided in Supplemental Materials.
Statistics
Student’s t test was performed for dual comparisons. P < 0.05 was considered significant.
RESULTS
Glycosyltransferase activity
Using a panel of unique, well-defined, chemically synthesized carbohydrate acceptors, we compared the glycosyltransferase activities of isolated human neutrophils with those of undifferentiated HL-60 cells, and HL-60 differentiated along the granulocytic lineage, (Table 1; Fig. 1). All data are normalized on the basis of cellular protein mass. Assuming that the protein concentration in cells (wt/vol) is constant between different cell types, the results effectively represent volume-averaged concentrations of active glycosyltransferases.
TABLE 1.
Glycosyltransferase activity in neutrophils, undifferentiated HL-60 cells, and differentiated HL-60 cells
| Enzyme activity | Probable genes | Acceptor | Glycosyltransferase activity (cpm × 10−3/mg protein)a
|
Glycosyltransferase activity ratio (differentiated HL-60/undiff. HL-60)b
|
|
|---|---|---|---|---|---|
| Neutrophils | Undiff. HL-60 | ||||
| GalNAc transferase ppGalNAcT | Truncated PSGL-1 N-terminal sequence peptide | 4.14 | 5.96 | 3.31 | |
| Sialyltransferase | |||||
| ST3[Galβ1,3GalNAc] | ST3Gal-I/-II | Galβ1,3(3-O-MeGalβ1, 4GlcNAcβ1,6)GalNAcα-O-Bn (#2) | 72.8 | 50.6 | 1.54 |
| ST3[Galβ1,3GalNAc] | ST3Gal-I/-II | Galβ1,3GalNAcα-O-Bn | ND | 68.5 | 0.69 |
| ST3[Galβ1,4GlcNAc] & ST6[Galβ1,4GlcNAc] | ST3Gal-IV/-VI & ST6Gal-I | 3-O-MeGalβ1,3(Galβ1, 4GlcNAcβ1,6)GalNAcα-O-Bn (#3) | 9.7 | 44.6 | 0.26 |
| ST3[Galβ1,4GlcNAc] & ST6[Galβ1,4GlcNAc] | ST3Gal-IV/-VI & ST6Gal-I | Galβ1,4GlcNAcβ-O-Bn | ND | 36.5 | 0.19 |
| ST3[Galβ1,4GlcNAc] | ST3Gal-IV/-VI | 4-O-MeGalβ1,4GlcNAcβ-O-Bn | 1.7 | 7.9 | 1.01 |
| ST3[Galβ1,3GlcNAc] | ST3Gal-III | Galβ1,3GlcNAcβ-O-Bn | 1.9 | 17.0 | 0.27 |
| ST3[Galβ1,3GlcNAc] | ST3Gal-III | 2-O-MeGalβ1,3GlcNAcβ-O-Bn | 0.9 | 9.3 | 0.65 |
| Galactosyltransferase | |||||
| β1,4GalT & β1,3GalT | β1,4GalT-I/-IV & core 1 β1,3GalT-I | GlcNAcβ1,6GalNAcα-O-Bn | 173.9 | 351.7 | 1.74 |
| β1,3GalT | core 1 β1,3GalT-I | GalNAcα-O-Bn | 75.8 | 41.3 | 0.92 |
| β1,4GalT | β1,4GalT-I/-IV | Galβ1,3(GlcNAcβ1,6)GalNAcα-O-Al | ND | 271.7 | 1.68 |
| β1,4GalT | β1,4GalT-I/-IV | 3-O-MeGalβ1,3(GlcNAcβ1,6) GalNAcα-O-Bn | 103.1 | 291.4 | 1.59 |
| GlcNAc transferase | |||||
| LacNAc extension β1,3GlcNAcT | β1,3GlcNAcT-1/-2 | 3-O-MeGalβ1,3(Galβ1, 4GlcNAcβ1,6)GalNAcα-O-Bn (#3) | 35.7 | 196.9 | 3.03 |
| LacNAc extension β1,3GlcNAcT | β1,3GlcNAcT-1/-2 | Galβ1,4GlcNAcβ-O-Bn | 83.6 | 116.4 | 3.16 |
| LacNAc extension β1,3GlcNAcT | β1,3GlcNAcT-1/-2 | Galβ1,4GlcNAcβ1,3Galβ1, 4GlcNAcβ-O-Bn | ND | 53.4 | ND |
| Core 1 extension β1,3GlcNAcT | β1,3GlcNAcT-3 | Galβ1,3(3-O-MeGalβ1, 4GlcNAcβ1,6)GalNAcα-O-Bn (#2) | 2.2 | 2.5 | ND |
| Globo H extension GlcNAcT | Galβ1,3GalNAcβ1,3Galα-O-Me | 1.9 | ND | ND | |
| β1,2GlcNAcT | β1,2GlcNAcT-I | Fetuin triantennary asialo agalacto glycopeptide devoid of terminal GlcNAc | 15.1 | 258.4 | ND |
| Fucosyltransferase | |||||
| α1,3FT | α1,3FT-IV/-IX | 2-O-MeGalβ1,4GlcNAcβ-O-Bn | 19.9 | 628.4 | 1.34 |
| α1,3FT | α1,3FT-IV/-IX | GalNAcβ1,4GlcNAcβ-O-Bn | ND | 745.2 | 0.55 |
| α1,3FT | α1,3FT-IV/-IX | Galβ1,4GlcNAcβ1,3Galβ1, 4GlcNAcβ-O-Bn | ND | 820.6 | 1.12 |
| α1,3FT[sialyl-LacNAc] | α1,3FT-VII | NeuAcα2,3Galβ1,4GlcNAcβ-O-Bn | 38.1 | 110.3 | 1.12 |
| α1,3FT | α1,3FT-VI | GlcNAcβ1,4GlcNAcβ-O-Bn | 0.3 | ND | ND |
| α1,4FT | α1,3/4FT-III | 2-O-MeGalβ1,3GlcNAcβ-O-Bn | 0.4 | 0.20c | ND |
| α1,2FT | Galβ-O-Bn | 0.7 | 0.15c | ND | |
| α1,6FT | Fetuin triantennary asialo agalacto glycopeptide | 33.5 | 271.8 | 1.73 | |
Enzyme activity measured using specific acceptors is listed along with likely candidate gene-products/enzymes that contribute to this activity in HL-60 and neutrophils. Other enzymes not listed here may also contribute to this activity. ND, not done.
The cpm can be converted to dpm by dividing these values by scintillation counter efficiency of 0.64 for 3H- and 0.97 for 14C-labeled sugars. Data are representative of 3 experiments. In the case of neutrophils, cells from 3 different, normal donors were used.
Glycosyltransferase activity ratio represents activity of DMSO differentiated HL-60 cells divided by activity of undifferentiated control HL-60 cells.
Data from Chandrasekaran et. al., 1996 (26).
ppGalNAcT activity, which initiates O-linked glycan formation, was comparable in neutrophils and HL-60 cells. Differentiation increased ppGalNAcT activity in HL-60 cells by ∼3.3-fold.
In the case of sialyltransferases, on comparing the activity of neutrophil enzymes on compounds 2 and 3, we observed that the sialylation of Galβ1,3GalNAc− in the core-2 tetrasaccharide is more prominent over Galβ1, 4GlcNAc−. Compound 3 is an acceptor for both ST3[Galβ1,4GlcNAc] and ST6[Galβ1,4GlcNAc] (23). The specific methylated acceptor 4-O-MeGalβ1, 4GlcNAcβ-O-Bn was used to distinguish between these two activities, since it is acted upon by ST3[Galβ1,4GlcNAc] but not cloned mammalian ST6Gal-I (23). SialylTs in neutrophil lysates were 17.5% active on this acceptor (1700 cpm/mg) compared with compound 3 (9700 cpm/mg). Together, these studies suggest that the activity of neutrophil sialyltransferases likely follow the sequence: ST3[Galβ1,3GalNAc] > ST6[Galβ1, 4GlcNAc] > ST3[Galβ1,4GlcNAc]. The activity of these three sialylTs in HL-60 cells decreased in the same sequence, only ST3[Galβ1,4GlcNAc] and ST6[Galβ1, 4GlcNAc] activities were 3- to 4.5-fold higher in this cell line compared with the primary cells. ST3[Galβ1, 3GalNAc] activity was at similar levels in HL-60 cells relative to neutrophils. HL-60 cell differentiation markedly reduced the activity of ST3[Galβ1,3GlcNAc] and cumulative activity of ST6[Galβ1,4GlcNAc] and ST3[Galβ1,4GlcNAc], but it did not affect α2,3-sialylation of either type II (Galβ1,4GlcNAc-) or type III (Galβ1,3GalNAc-) chains.
We observed that β1,4GalTs were more active in HL-60 cells compared with neutrophils. In contrast, core-1 β1,3GalT activity was higher in neutrophils. Differentiation of HL-60 cells moderately increased β1,4GalT activity by 59–74%.
β1,3GlcNAcT activity assays reveal that neutrophils and HL-60 cells have high activity for LacNAc extension (i.e., extension of the Galβ1,4GlcNAc-chain). Core-1 extension by GlcNAcT on the Galβ1,3GalNAc chain was low. β1,3GlcNAc addition to LacNAc occurred at 1.4- to 5.5-fold higher rates in HL-60 cells compared with neutrophils. HL-60 cell differentiation further increased β1,3GlcNAcT activity in the LacNAc chain by ∼3-fold.
Among the fucosyltransferases, α1,3FT and α1,6FT activities were observed in HL-60 cells and neutrophils, while α1,2FT activity was absent. Among the α1,3FTs (FT-IV, -IX, and -VII), the α1,3FT-VII prefers to fucosylate sialyl-LacNAc (NeuAcα2,3Galβ1,4GlcNAc) -based substrates, whereas α1,3FT-IV and -IX preferentially fucosylate nonsialylated LacNAc substrates. Using the appropriate substrates, thus, we observed that fucosylation of sialyl-LacNAc (α1,3FT[sialyl-LacNAc]) mostly brought about by α1,3FT-VII, was the dominant α1,3FT activity in neutrophils. Conversely, α1,3FT activity in nonsialylated LacNAc (mostly mediated by α1,3FT-IV) was greater in HL-60 cells (Note: transcripts for FT-IX are absent in HL-60 cells; ref. 12). Both α1,3FT activities were 2.8- to 30-fold greater in HL-60 cells compared with neutrophils. Differentiation of HL-60 cells did not result in alteration of α1,3FT[sialyl-LacNAc] activity. Fucosylation activity on the nonsialylated LacNAc acceptor was still dominant after differentiation. Finally, there was a 73% increase in α1,6FT activity upon HL-60 cell differentiation, consistent with observations by others (27).
In sulfotransferase assays, we did not detect carbohydrate sulfotransferase activity in neutrophils using either compound 2, compound 3, or 3-O-MeGalβ1, 3(GlcNAcβ1,6)GalNAcα-O-Bn as acceptors. This indicates the absence of Gal:3-O- and GlcNAc:6-O-sulfotransferase activities in human neutrophils.
Overall, we observed that mature neutrophils display similar enzyme activities as HL-60 cells, although the absolute level of glycosyltransferase activity is markedly different. HL-60 cell differentiation resulted in an increase in β1,3GlcNAcT and ppGalNAcT activity, and it decreased ST6[Galβ1,4GlcNAc]. The effects of cell differentiation on β1,4GalT and α1,3FT activity were only modest. On combination of the radioactivity measurements of Table 1 with additional data from Supplemental Table S2, the apparent first order reaction rate constants for selected enzymes can be computed (Table 2). These calculations suggest that the in vitro glycosyltransferase activity for human leukocytes (k) follow the sequence: ST3[Galβ1,4GlcNAc] < α1,3FT[sialyl-LacNAc] ≤ β1,3GlcNAcT.
TABLE 2.
Apparent first-order rate constant (k)
| Enzyme activity | Probable genes | Acceptor | Apparent first-order rate constant × 103 (h−1)a
|
||
|---|---|---|---|---|---|
| Neutrophils | Undifferentiated HL-60 | Differentiated HL-60 | |||
| Sialyltransferase
|
|||||
| ST3[Galβ1,3GalNAc] | ST3Gal-I/-II | Galβ1,3(3-O-MeGalβ1,4GlcNAcβ1,6) GalNAcα-O-Bn (#2) | 4.69 | 3.26 | 5.19 |
| ST3[Galβ1,4GlcNAc] | ST3Gal-IV/-VI | 4-O-MeGalβ1,4 GlcNAcβ-O-Bn | 0.11 | 0.51 | 0.53 |
| ST3[Galβ1,4GlcNAc] & ST6[Galβ1,4GlcNAc] | ST3Gal-IV/-VI & ST6Gal-I | 3-O-MeGalβ1,3(Galβ1,4GlcNAcβ1,6) GalNAcα-O-Bn (#3) | 0.62 | 2.87 | 0.77 |
| GlcNAc transferase | |||||
| LacNAc extension β1,3GlcNAcT | β1,3GlcNAcT-1/-2 | 3-O-MeGalβ1,3(Galβ1,4GlcNAcβ1,6) GalNAcα-O-Bn (#3) | 4.71 | 25.95 | 85.74 |
| Fucosyltransferase | |||||
| α1,3FT | α1,3FT-IV/-IX | 2-O-MeGalβ1,4 GlcNAcβ-O-Bn | 2.14 | 67.72 | 96.25 |
| α1,3FT[sialyl-LacNAc] | α1,3FT-VII | NeuAcα2,3Galβ1,4 GlcNAcβ-O-Bn | 5.99 | 17.33 | 20.01 |
Reaction velocity (v) values used for these calculations are from Table 1.
Glycosyltransferase gene expression
Although the chemical substrates used in our assays are designed to discriminate between closely related enzyme activities, they do not unambiguously reveal the precise genes responsible for a given measured activity. To determine the relation between gene expression and enzyme activity, quantitative RT-PCR studies were thus performed with RNA from neutrophils and undifferentiated/differentiated HL-60 cells (Fig. 2). Figure 3 summarizes and compares glycosyltransferase gene expression data with enzyme activity measurements.
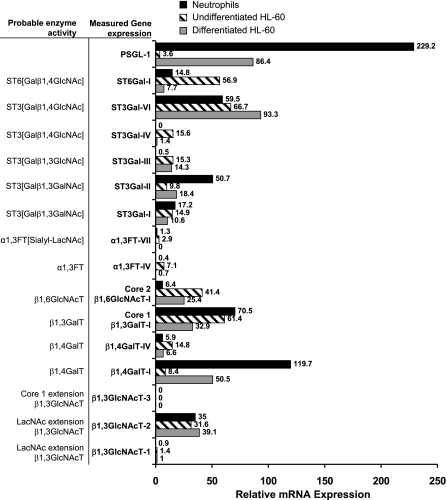
Figure 2.
RT-PCR analysis of glycosyltransferases: Relative mRNA levels of glycosyltransferase gene expression in isolated human neutrophils, undifferentiated HL-60 cells, and DMSO differentiated HL-60 using quantitative RT-PCR. Gene names appear in bold fonts; their corresponding enzyme activity is in plain font in an adjacent column.
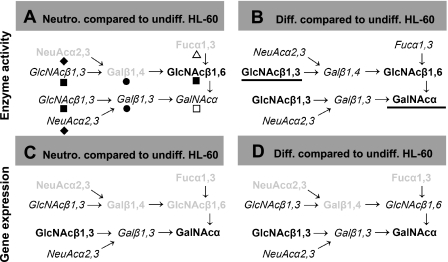
Figure 3.
Glycosyltransferase activity and gene expression. The expression of the sialyl lewis-X epitope on the core-2 trisaccharide structure Galβ1,3(GlcNAcβ1,6GalNAcα) located at the N terminus of PSGL-1 is thought to be critical for selectin recognition. Enzyme activity (A, B) and gene expression (C, D) studies were performed to quantify levels of various glycosyltransferases contributing to formation of this structure. In A and C, enzyme (activity/gene expression) levels in human neutrophils are quantified relative to undifferentiated (undiff.) HL-60 cells (i.e., Neutro/undiff HL-60). In B and D, enzymes that were altered on differentiation (diff.) of HL-60 cells along granulocyte lineage are highlighted (i.e., diff HL-60/undiff HL-60). >2-fold increase and <50% decrease in enzyme activity or gene expression are highlighted using underline and faded/gray notation, respectively. Gene/enzyme levels that changed to a lesser extent are shown in italics. Gene/enzyme levels that were not measured appear in plain font without any highlighting. In A, monosaccharides added by different enzyme activities are shown using the symbolic notations of Fig. 1C.
In agreement with results from the enzymology studies that demonstrate that enzymes contributing to the addition of outer nonreducing residues in O-glycans (ST3[Galβ1,4GlcNAc], α1,3FTs, and β1,4GalT) are higher in HL-60 cells compared with neutrophils, we also observed in RT-PCR runs that ST3Gal-IV, FT-VII, FT-IV, and β1,4GalT-IV were higher in the former cells (Fig. 2). Transcript levels of ST3Gal-I and -II, enzymes that sialylate the Galβ1,3GalNAc− sequence, were either equal to or lower in HL-60 cells compared with neutrophils. This finding is consistent with the enzymatic studies. Transcript levels for β1,3GlcNAcT-1 and -2, enzymes that contribute to the formation of polyLacNAc structures in O-glycans, were similar in neutrophils and HL-60 cells. In comparison, in enzymatic studies, a 1.4- to 5.5-fold higher β1,3GlcNAcT enzyme activity was noted for HL-60 cells. mRNA level of core-2 forming β1,6GlcNAcT was 6-fold higher, while PSGL-1 gene expression was 65-fold lower in undifferentiated HL-60 cells compared with neutrophils. The enzymatic activity of core-2 β1,6GlcNAcT was not measured in this study. Overall, there is agreement between enzyme activity and gene expression for many enzymes. Differences were observed in the activity of selected enzymes in undifferentiated HL-60 cells vs. neutrophils, and these may translate to differences in cellular glycan expression.
On differentiation, the gene expression pattern of HL-60 cells was altered to more closely resemble the transcript levels in neutrophils. There was qualitative agreement in gene expression of most enzymes, though some quantitative differences were still evident. For example, transcripts of several enzymes contributing to the formation of sLeX (ST3Gal-IV, FT-VII, FT-IV, and β1,4GalT-IV) were down-regulated on differentiation and their gene expression was quantitatively similar to that of neutrophils. Gene expression of PSGL-1 and β1,4GalT-I were also up-regulated 6- to 24-fold to levels similar to that of neutrophils. Although the gene expression data of differentiated HL-60 cells resembled that of terminally differentiated neutrophils for most enzymes (Fig. 2), the enzyme/glycosyltransferase activities of differentiated HL-60 cells more closely resembled the undifferentiated cells (Table 1). This last observation is consistent with the proposition that while gene expression changes rapidly to more closely resemble the cell differentiation state, enzyme levels may take longer times due to the slower turnover rates of proteins.
Hypothesis testing at the cellular and molecular level
Gene expression and enzymatic studies together suggest two hypotheses: 1) that glycan structures on HL-60 cells may differ from those on neutrophils. In this regard, the enzymology studies show that activities contributing to the formation of sLeX structure on O-glycans are expressed at higher levels in HL-60 cells. Thus, higher sLeX expression is expected in these cells. 2) Differentiation of HL-60 cells along granulocytic lineage does not make them more closely resemble neutrophils in terms of their glycosyltransferase activities. However, cell differentiation augmented β1,4GalT and β1,3GlcNAcT activity, suggesting that extended LacNAc chains may be more prevalent after HL-60 cell differentiation. The suppression of ST6[Galβ1,4GlcNAc] and an increase in ppGalNAcT activity after differentiation were also noted.
We examined these hypotheses at the cell level by monitoring selected antigens expressed on leukocyte cell surface using standard flow cytometry (Fig. 4). Quantitative molecular level measurements were also performed by assaying O-linked glycans on PSGL-1 using a novel cytometry bead assay (Fig. 5). The later studies use mAb bearing polystyrene beads to capture PSGL-1 from cell lysates. Immunoprecipitated protein on beads was then probed with antibodies and lectin to determine the following: PSGL-1 number, sLeX epitope, and LacNAc expression on immobilized proteins. Selected results were validated using Western blot analysis.
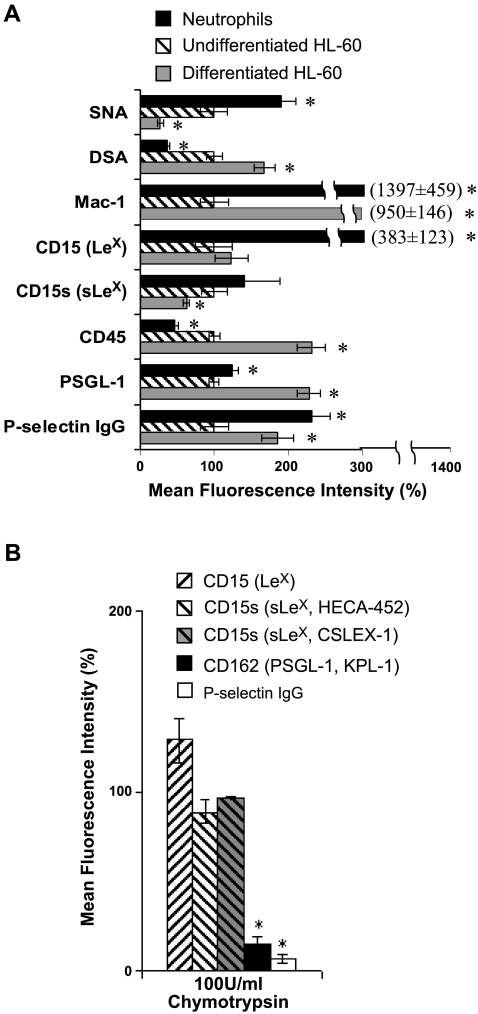
Figure 4.
Cell surface expression and selectin binding assay for HL-60 cells and human neutrophils. A) SNA (binds NeuAcα2,6Galβ-) and DSA (binds LacNAc) lectin binding to cells, cell surface expression of Mac-1 (CD11b), CD15 (LeX), CD15s (sLeX), CD45, and PSGL-1 (CD162), and binding of P-selectin fusion protein to the cells was measured with isolated human neutrophils, undifferentiated HL-60 cells and DMSO differentiated HL-60 using flow cytometry. PSGL-1 expression was 24% higher in neutrophils compared with HL-60 cells, while P-selectin binding was 2.3-fold higher. On differentiation of HL-60 cells, PSGL-1 expression increased by 2.3-fold and this was accompanied by a 1.9-fold increase in selectin binding. B) Undifferentiated HL-60 cells were treated with 100U/ml chymotrypsin for 15 min at room temperature before above measurements. Chymotrypsin treatment drastically reduced cell-surface PSGL-1 expression and P-selectin binding, but this did not affect sLeX (CD15s) expression by >5%. In A and B, mean fluorescence intensity was normalized with respect to undifferentiated HL-60 cells without chymotrypsin. Data are means ± se for 3–5 experiments. *P < 0.05 vs. undifferentiated HL-60 cells.
Figure 5.
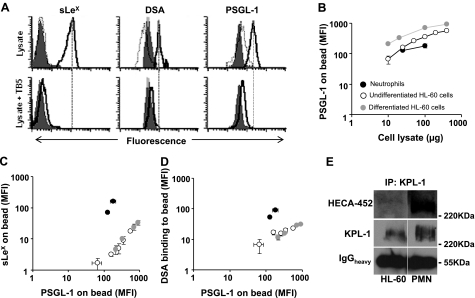
Glycan structures on PSGL-1 assayed using quantitative cytometry bead and qualitative Western blot analysis: antigen-sandwich experiments were performed using flow cytometry (A–D). Here, PSGL-1 from undifferentiated/differentiated HL-60 cell or neutrophil lysate was immunoprecipitated onto beads bearing anti-PSGL-1 mAb (TB5 beads). These beads were then probed with anti-sLeX mAb (HECA-452), DSA-lectin (which binds LacNAc), or an anti-PSGL-1 rabbit Ab (H-300). A) Representative cytometry histograms for neutrophil lysates. Top row: anti-sLeX, DSA-lectin, and anti-PSGL-1 binding were observed when 25 μg cell lysate (100 μl vol) was incubated with TB5 beads (empty histogram with bold outline) but not when isotype control beads (IB4 beads, empty histogram with plain outline) were used. Absence of cell lysate resulted in low Ab/lectin binding to TB5 beads (filled gray histograms). Bottom row: binding of anti-sLeX, DSA-lectin and anti-PSGL-1 could be abolished by preincubating lysates with excess TB5 Ab (27 μg/ml) for 15 min before addition of TB5 beads under conditions identical to the studies described in the top row. Similar controls were performed with undifferentiated and differentiated HL-60 cells. B) Flow cytometry geometric mean fluorescence intensity (MFI) corresponding to the amount of PSGL-1 captured by TB5 bead when cell lysate amount was varied from 10–400 μg. C, D) sLeX (C; anti-CD15s/HECA-452) and LacNAc (D; DSA-lectin) expression on PSGL-1 quantified on TB5 beads in the presence of varying amounts of cell lysate for the three cell types. Data are plotted for sLeX and LacNAc structures vs. corresponding PSGL-1 bound to beads. Background fluorescence intensity obtained in the absence of cell lysate was subtracted for all data in B–D. E) PSGL-1 from neutrophil and HL-60 cells was immunoprecipitated using anti-PSGL-1 mAb (KPL-1). Western blots probed with either KPL-1 or HECA-452 support cytometry bead experiments that suggest higher sLeX expression on neutrophil PSGL-1 compared with that from HL-60 cells. The IgG heavy chain (IgGheavy) band shows that the same amount of immunoprecipitating Ab (KPL-1) was loaded in each lane. Error bars in all plots are means ± se for ≥3 experiments.
Figure 5A demonstrates that the cytometry beads specifically report on glycan structures on the captured PSGL-1. In support of this, the anti-PSGL-1 beads (TB5 beads), but not the isotype control anti-CD18 mAb beads (IB4 beads), captured PSGL-1 from lysate. The former beads also bound Abs and lectins that recognize glycan epitopes, while the control beads did not exhibit this feature. In additional controls, the background signal for anti-PSGL-1, anti-sLeX, and DSA-lectin binding to TB5 beads was low when the cell lysate was absent (Fig. 5A, top panels). Finally, the specific binding interactions could be competed away to background levels by incubating the cell lysate with excess soluble TB5 mAb, before TB5-bead addition (Fig. 5A, bottom panels). Although data for neutrophils are presented in Fig. 5A, similar specificity was also noted with HL-60 cell lysate. Anti-LeX/CD15 mAb did not bind beads bearing immobilized PSGL-1. Finally, increasing cell lysate concentration increases the amount of PSGL-1 captured on TB5 beads (Fig. 5B).
Cell-level studies compared glycans on human neutrophils with undifferentiated HL-60 cells (Fig. 4A). Here, flow cytometry reports on carbohydrate structures on both glycoproteins and glycolipids without discrimination (Fig. 4B). As seen, only a relatively small fraction of HL-60 cell CD15 and CD15s is expressed at the P-selectin binding site located at the N terminus of PSGL-1, since treatment of these cells with protease chymotrypsin led to an 80% excision of this site without dramatically reducing overall cell surface CD15 or CD15s expression. In cytometry studies (Fig. 4A), we observed that neutrophils express 14-fold higher Mac-1 (CD11b) and 24% greater PSGL-1 levels compared with undifferentiated HL-60 cells (Fig. 4A). The latter finding is reasonable since PSGL-1 mRNA is 60-fold higher in neutrophils, although a linear relationship between RNA and protein expression is absent. DSA (which binds Galβ1,4GlcNAc/LacNAc structure) binding to neutrophils was lower by 64% compared with undifferentiated HL-60 cells, and this is in agreement with the enzyme activity and gene expression measurements that show that neutrophils have a lower activity of enzymes contributing to LacNAc formation. CD15 (LeX) was 3.8-fold higher in neutrophils, and this is an unexpected finding since enzyme activity data show that α1,3-fucosylation activity on the nonsialylated substrate is 31-fold lower in neutrophils compared with HL-60 cells (Table 2). As elaborated in the Discussion, one possibility is that the neutrophil CD15 detected in our assay were formed during the early-myeloid stage before cell maturation. Even though the overall cellular expression of CD15s/sLeX was comparable in neutrophils vs. HL-60 cells, the ratio of CD15s to CD15 (i.e., sLeX/LeX) was higher in the case of HL-60 cells. This is in agreement with the higher levels of α2,3-sialylT activity (ST3[Galβ1,4GlcNAc]) observed in HL-60 cells, which may also sialylate terminal LeX structures on N-linked glycans.
Cytometry bead assays were performed to discriminate between glycan structures on PSGL-1 in HL-60 cells vs. neutrophils. These studies reveal that neutrophil PSGL-1 exhibits 10-fold higher sLeX expression and 3- to 5-fold higher DSA-lectin binding compared with both undifferentiated and differentiated HL-60 cells (Fig. 5C, D). Immunoprecipitation followed by Western blot analysis (Fig. 5E) supports the observation that sLeX antigen associated with PSGL-1 is indeed higher in neutrophils compared with HL-60 cells (Fig. 5E). This suggests the possibility that higher levels of P-selectin may bind neutrophils compared with HL-60 cells. This proposition is in agreement with selectin binding data (Fig. 4A), which show that neutrophils bind P-selectin at 2.3-fold higher levels compared with HL-60 cells even though their PSGL-1 expression is comparable.
Differentiation of HL-60 cells resulted in a 9.5-fold increase in the differentiation marker Mac-1 and a 2.3-fold increase in CD45 expression (Fig. 4A). A 68% increase in DSA and 73% decrease in SNA lectin binding of differentiated HL-60 cells over undifferentiated cells was also observed. This is consistent with the observed up-regulation of LacNAc extension enzyme activity (β1,4GalT and β1,3GlcNAcT) and down-regulation in ST6[Galβ1,4GlcNAc] activity upon HL-60 cell differentiation. Cell surface CD15 was not altered on differentiation, while there was a slight decrease in cellular CD15s expression. Cell surface PSGL-1 protein levels increased by 2.3-fold, and this is consistent with increased mRNA expression. A proportional 1.9-fold increase in P-selectin binding to cells was observed, and this suggests only relatively minor alterations in the PSGL-1 specific glycosylation on differentiation. This last observation is consistent with molecular-level investigations (Fig. 5C, D) where the expression of sLeX and DSA-lectin binding to PSGL-1 in differentiated HL-60 cells is similar to that in undifferentiated cells.
Overall, glycan structures and function measured at the cell and molecule level are largely consistent with hypothesis generated using enzymatic studies of HL-60 cells before and after differentiation.
Cell cycle regulation of glycosyltransferase activity, glycan expression, and selectin binding function
Previous studies (28) suggest that the activity of N-glycan specific glycosyltransferases, GlcNAcT-III and -V, vary with cell cycle phases. We examined if a similar relationship exists between the proliferating state of leukocytes and O-glycan-specific enzymes. In particular, we determined if the pattern of HL-60 cell glycosyltransferases in any particular growth phase more closely resembles neutrophils. To this end, HL-60 cell growth was synchronized using HU. Treatment with 1 mM HU was found to synchronize 84–89% of the cells in the G0/G1 phase (Table 3). After their release from G0/G1 arrest by removal of HU from the media at 13 h, a majority of HL-60 cells were observed in the S-phase, the G2/M phase, and back in the G1 phase at 20, 24, and 31 h, respectively.
TABLE 3.
Cell cycle analysis
| Cell phase | Time (h)
|
|||||
|---|---|---|---|---|---|---|
| 0 | 13 | 20 | 24 | 31 | 38 | |
| G0/G1 (%) | 51.4 ± 1.1 | 84.3 ± 2.4 | 15.6 ± 2.1 | 30.1 ± 4.7 | 73.9 ± 1.8 | 29.7 ± 5.9 |
| S (%) | 28.2 ± 1.1 | 9.4 ± 2.0 | 80.7 ± 2.7 | 9.8 ± 1.2 | 15.5 ± 1.3 | 56.4 ± 6.3 |
| G2/M (%) | 20.4 ± 0.9 | 6.2 ± 0.6 | 3.7 ± 1.5 | 60.2 ± 5.1 | 10.6 ± 1.3 | 13.9 ± 3.5 |
HU (1 mM) was added at 0 h and removed at 13 h. Italic numbers highlight the cell phase in which a majority of cells are found at a given time. Data represent percentage of nonapoptoic cells. Values are means ± se; n ≥ 4.
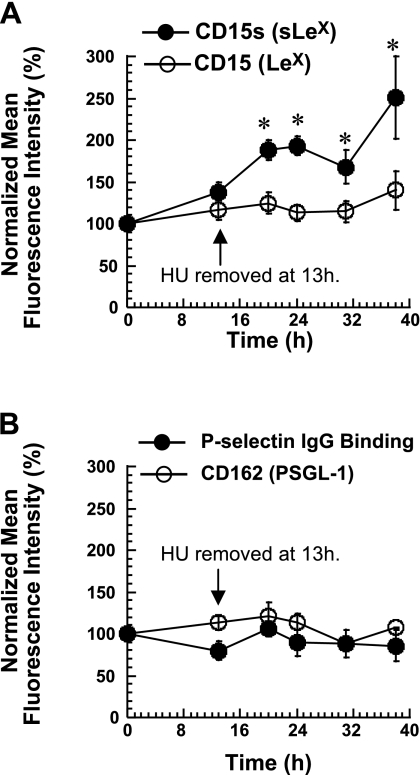
Periodic cycling of glycosyltransferase activities with cell cycle phases was not evident in these experiments (Supplemental Table S3). However, within 13 h of HU treatment, there was a 25–45% decrease in several enzyme activities that acted on the Galβ1,4GlcNAc arm of the core-2 tetrasaccharide, including ST3[Galβ1,4GlcNAc], β1,4GalT, and β1,3GlcNAcT. All the enzymes gradually approached their baseline activity after removal of HU. This increased activity of enzymes in the Galβ1,4GlcNAc arm after HU removal is consistent with our observation that HL-60 cell-surface sLeX expression increased by 2.5-fold within 38 h of HU removal (Fig. 6). During this time, LeX expression, PSGL-1 expression, and P-selectin binding were not altered.
Figure 6.
HU treatment of HL-60 cells. HL-60 cells were incubated with 1 mM HU for 13 h to synchronize cells in the G0/G1 phase. HU was then removed. CD15s (sLeX), CD15 (LeX) (A), and PSGL-1 expression levels (B) were monitored for cells synchronized in different phases. P-selectin IgG binding to cells was also measured (B). Data are presented as the percentage of mean fluorescence intensity of control HL-60 cells (without HU), which were cultured in parallel. CD15s increased after HU removal. Data are means ± se for ≥3 experiments. *P < 0.05 vs. control without HU.
DISCUSSION
There is growing interest in applying systems-level knowledge to predict cellular glycan epitopes, since such analysis complements more conventional structural investigations. Although structural studies are typically complex and they do lead to the identification of the more abundant glycans, functionally important, less abundant epitopes may be missed. To this end, in the area of systems biology, attempts have been made to predict glycan structures based on glycosyltransferase microarray data (29) and using mathematical models based on a reaction network framework (30). Complementing such approaches, in the current study, we examined the degree to which knowledge of glycosyltransferase gene expression and enzyme activity allows both qualitative and quantitative prediction of cell-level and protein-level glycan structures. The focus was on enzymes modifying the outer nonreducing residues of core-2 linked O-glycans since these modifications play a key role in regulating selectin-mediated cell adhesion.
Distinct pathways and enzymes with overlapping specificity may contribute to the formation of glycans in neutrophils vs. HL-60 cells
In studies that compared human neutrophils with HL-60 cells, although similar mRNA and glycosyltransferase activities were identified in both cells, differences were noted regarding the relative expression and activity of these enzymes. In both cell types, β1,4Gal attachment to GlcNAcβ proceeded at 1.4- to 7-fold greater rates than β1,3Gal linkage to GalNAcα. Further, the rates of α2,3 sialylation of the Galβ1,3GalNAc moiety in the core-2 tetrasaccharide (Fig. 1B) occurred 6- to 43-fold more rapidly compared with the rates of Galβ1,4GlcNAc sialylation. α1,3FT enzymes that fucosylate the sialylated LacNAc structure (NeuAcα2, 3Galβ1,4GlcNAc) were the dominant α1,3FT in neutrophils, while fucosyltransferases acting on Galβ1, 4GlcNAc were greater in HL-60 cells. In agreement with this, Northern blot analysis of RNA levels from these two cell types (10) also suggests that FT-IV is dominant in HL-60 cells, while FT-VII is more prominent in neutrophils. Finally, the activity of enzymes contributing to LacNAc extension β1,3GlcNAcT; α1,3FT; and ST3[Galβ1,4GlcNAc] activity was considerably lower in neutrophils compared with HL-60 cells. Of the three members of the α2,3sialylT family that contribute to ST3[Galβ1,4GlcNAc] activity (ST3Gal-III, -IV, and -VI; ref. 13), mRNA levels of ST3Gal-III and -IV were lower in neutrophils compared with HL-60 cells. Thus, it is possible that although ST3Gal-VI is important for sialylation in neutrophils, ST3Gal-III and -IV play dominant roles in HL-60 cells.
Overall, our observations predict that both distinct and overlapping pathways may result in sLeX formation in HL-60 cells vs. neutrophils. Although FT-IV/-VII and ST3Gal-III/IV contribute to the formation of sLeX epitopes in HL-60 cells, FT-VII and ST3Gal-VI may be more important in neutrophils. Functional/structural data that measure P-selectin binding to PSGL-1 and sLeX expression associated with PSGL-1 corroborate enzymatic studies that suggest differences in the O-linked glycan structure and/or their distribution in neutrophils vs. HL-60 cells. In this regard, both the expression of sLeX and P-selectin binding function are higher in neutrophils compared with HL-60. That changes in glycosyltransferase activity can result in different glycan structures in PSGL-1 is also in agreement with animal studies conducted with ST3Gal-IV knockouts, where it is shown that knocking out this enzyme results in PSGL-1 that binds P-selectin but not L-selectin (31). All predicted differences in glycan structures based on glycosyltransferase measurements can be explained using glycan/function measurements except for the high levels of LeX in neutrophils compared with HL-60 cells. We speculate that this may be due to the increased α1,3-fucosyltransferase activity during neutrophil maturation. In support of this, Le Marer et al. (32) show that α1,3-fucosyltransferase is 3- to 10-fold higher during the promyelocytic/early-myeloid developmental stages compared with mature neutrophils. These authors also show that FT-IV is the dominant α1,3-FT during the early-myeloid stage, while FT-VII is dominant during the late-myeloid and neutrophil stages. Thus, the abundant LeX structures found in neutrophils may have been formed early in the developmental process.
sLeX expression in leukocytes may be limited by low α2,3-sialylT activity
A comparison of the first-order glycosyltransferase reaction rate constants (Table 2) suggests that enzyme activity in our in vitro assays follows the sequence ST3[Galβ1, 4GlcNAc] < α1,3FT[sialyl-LacNAc] ≤ β1,3GlcNAcT. In addition, a proportionality relation is suggested between the rate constant measured in our assays, k, and that expected in situ/in vivo in cells, kvivo. According to this, kvivo varies in proportion to k · (Km,mono/Km,acc) (full derivation in Supplemental Materials). Taking into account this relation, the results in Table 2, and Km data (Supplemental Table S2), we conclude that enzyme activities in situ in human leukocytes likely vary as ST3[Galβ1,4GlcNAc] ≤ α1,3FT[sialyl-LacNAc] < β1, 3GlcNAcT. Thus, α2,3-sialylation, and to a lesser extent α1,3-fucosylation, may represent the rate-limiting step that controls sLeX expression.
These above findings are in agreement with the results of Wilkins et al. (8), who show that 52% of O-linked glycans on PSGL-1 of HL-60 cells are the nonfucosylated, nonsialylated core-2 tetrasaccharide. Since α1,3 fucosylation is thought to follow α2,3 sialylation during core-2 glycan biosynthesis (23), our findings suggest that low levels of ST3[Galβ1,4GlcNAc] in these cells may limit the formation of terminal structures on the core-2 tetrasaccharide. Further, higher relative activity of α1,3FT[sialyl-LacNAc] over ST3[Galβ1, 4GlcNAc] in neutrophils (ratio of rate constants=54.5) as compared with that in undifferentiated HL-60 cells (ratio=34) may account for the greater prevalence of sLeX structures expressed on PSGL-1 in the former cells.
Differentiation of HL-60 cells results in altered glycosyltransferase activity and associated cell-surface glycan structures
In studies that examined the effect of cell differentiation on glycosyltransferases, we observed that although the gene expression profile of differentiated HL-60 cells resembled that of human neutrophils, the glycosyltransferase activities of differentiated HL-60 cells more closely resembled those of undifferentiated HL-60 rather than mature neutrophils. In this regard, both α2,3sialylT and α1,3FT activities remained high in differentiated HL-60 cells, similar to undifferentiated cells. This suggests slower turnover rates of enzymes responsible for glycosylation compared with corresponding transcripts.
On differentiation, ST6[Galβ1,4GlcNAc] activity decreased and this correlated well with the observed decrease in ST6Gal-I mRNA levels and SNA lectin binding to α2,6 sialylated epitopes on HL-60 cells on differentiation. An increase in β1,4GalT and β1, 3GlcNAcT activity was also observed (Table 1), and this suggests the presence of greater amounts of LacNAc structures in differentiated cells. In agreement with this, DSA-lectin binding was increased by 68% in differentiated HL-60 cells. A 2.3-fold increase in PSGL-1 protein expression was noted along with an increase in the corresponding mRNA levels. Accompanying this change was a proportional 1.9-fold increase in P-selectin binding to differentiated HL-60 cells. This increase in selectin binding is consistent with a previous study that DMSO-differentiated HL-60 cells have augmented binding to human umbilical vein endothelial cells bearing E- and P-selectin (33). The direct correlation between PSGL-1 expression and P-selectin binding suggests minimal alterations in PSGL-1 glycan structures on differentiation. This prediction from cell-based assays is consistent with cytometry bead experimental results (Fig. 5).
Transient HU treatment increases leukocyte sLeX expression
HU is a common drug used for the treatment of sickle cell anemia, chronic myeloid leukemia, and HIV. In these diseases, HU is thought to have beneficial effects due to its effect in inhibiting cell growth, reducing the number of blood neutrophils in circulation, regulating cell-surface L-selectin expression, inhibiting increased H2O2 production, increasing fetal hemoglobin levels, and reducing cell adhesion to subendothelial matrix proteins (34, 35). Selectin-ligand recognition is known to contribute to at least some of these pathologies, e.g., sickle cell anemia (34). In the current studies, the addition of 1 mM HU to synchronize cells into a single growth phase resulted in the reduced activity of enzymes that acted on the Galβ1,4GlcNAc arm of the core-2 tetrasaccharide to form the sLeX epitope. The treatment time for this study was short (13 h), and this was insufficient to change the protein-specific glycan structure, as evidenced by selectin binding measurements. However, on removal of HU, the glycosyltransferase levels were observed to return to their baseline level and this was accompanied by increased ST3[Galβ1,4GlcNAc] activity and expression of sLeX on HL-60 cells. In other experiments performed with prolonged HU addition (data not shown), we observed that HU reduced P-selectin binding by 50%. Together, our studies with short- and long-term addition of HU suggest a potential role of HU in altering selectin binding function by altering cellular glycosyltransferase activity/expression.
Gene expression and glycosyltransferase activity assays provide complementary information
Glycosyltransferase gene expression data complement enzymatic assays. Although RT-PCR studies allow efficient and quantitative comparison of a given gene between different cell types (e.g., differences in FT-VII mRNA levels between neutrophils and HL-60 cells), they do not allow relative quantification of different enzymes (e.g., FT-IV vs. FT-VII) within a single cell. Glycosyltransferase assays allow the latter comparison, although it is possible that multiple enzymes may act on a given acceptor. Thus, specific acceptors for a given enzyme have to be designed.
A comparison of results from these two assays reveals that enzyme activities correlate well with gene expression, although their relation is not always linear. In an example that demonstrates the superiority of the glycosyltransferase assays over gene expression, our enzymatic studies show that GlcNAc:6-O-sulfotransferase activity is absent in neutrophils even though genes encoding for this transferase have been reported in neutrophils based on RNA measurements (17, 36). In another example, although our RT-PCR studies predict the presence of FT-VI in HL-60 cells (data not shown), enzyme activity corresponding to this gene was negligible in our enzymatic study. As discussed elsewhere (37), besides gene expression, other factors like correct folding of enzymes, their appropriate post-translational modification, and homo/hetero-dimerization are important features contributing to enzyme activity. Defects in any of these processes can lead to the absence of enzyme activity despite the presence of RNA.
In an example highlighting the use of the RT-PCR studies, we noted increased β1,4GalT activity after HL-60 cell differentiation using the core-2 trisaccharide acceptor GlcNAcβ1,6(Galβ1,3)GalNAcα. This suggests that the formation of polyLacNAc structures is likely to be enhanced after leukocyte differentiation. The key question then is to determine if such extension is expected in O-glycans (by β1,4GalT-IV) or N-glycans (by β1,4GalT-I). In this regard, the gene expression data reveal a 500% increase in β1,4GalT-I and a 55% decrease in β1,4GalT-IV expression. This suggests that differentiation likely promotes polyLacNAc formation in N-glycans rather than O-glycans. In agreement with this proposition, data in the literature (38) show that granulocytic differentiation of HL-60 cells increases polyLacNAc structures in the N-linked glycans of the lysosomal membrane glycoproteins lamp-1 and -2. Our cytometry bead assay also demonstrated minimal changes in LacNAc structures associated with the O-glycans of PSGL-1, even though DSA-lectin binding (LacNAc structure) was augmented at the whole-cell level.
Predicting glycan structures based on enzyme activity and transcript levels
In addition to enzyme activity and gene expression, other features also contribute to glycan structures. Foremost, the sequential spatial organization of enzymes in the Golgi regulates glycan formation. The existence of multienzyme glycosyltransferase complexes (39) and the competition between multiple enzymes (like ST3Gal-I and C2GnT for the same substrate; ref. 40) are additional parameters regulating in vivo glycosyltransferase activity. Finally, the primary structure of the peptide substrate regulates the function and activity of polypeptide GalNActransferases (ppGalNAcTs), which initiate O-linked glycosylation (41). This can subsequently affect glycan extension. Due to this site-specific nature of glycosylation, a different distribution of proteins in two cell types may result in different densities of measured overall cell surface glycan levels. Although comprehensive prediction of cell-surface glycan structures must account for the detailed features listed above, the current study suggests that semiqualitative predictions are possible using measurements of glycosyltransferase activity combined with the existing knowledge of biochemical pathways. Viable experimentally testable hypotheses that can be validated at the whole-cell and single-protein levels can also be generated using this approach. Such systems-level measurements of glycosyltransferase enzyme and gene activity may be helpful not only in the areas of inflammation research as shown here but also in the context of other diseases that are associated with aberrant glycosylation.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL63014 and HL76211 to S.N., CA35329 to K.L.M. and AI56082 to J.T.Y.L.). Carbohydrate acceptors used in this work were synthesized by various members of Matta Laboratory.
References
- Neelamegham S. Transport features, reaction kinetics and receptor biomechanics controlling selectin and integrin mediated cell adhesion. Cell Commun Adhes. 2004;11:35–50. doi: 10.1080/15419060490471793. [DOI] [PubMed] [Google Scholar]
- Spertini O, Cordey A S, Monai N, Giuffre L, Schapira M. P-selectin glycoprotein ligand 1 is a ligand for L-selectin on neutrophils, monocytes, and CD34+ hematopoietic progenitor cells. J Cell Biol. 1996;135:523–531. doi: 10.1083/jcb.135.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W S, Tang J, Shaw G D, Camphausen R T. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- Beauharnois M E, Lindquist K C, Marathe D, Vanderslice P, Xia J, Matta K L, Neelamegham S. Affinity and kinetics of sialyl Lewis-X and core-2 based oligosaccharides binding to L- and P-selectin. Biochemistry. 2005;44:9507–9519. doi: 10.1021/bi0507130. [DOI] [PubMed] [Google Scholar]
- Leppanen A, Penttila L, Renkonen O, McEver R P, Cummings R D. Glycosulfopeptides with O-glycans containing sialylated and polyfucosylated polylactosamine bind with low affinity to P-selectin. J Biol Chem. 2002;277:39749–39759. doi: 10.1074/jbc.M206281200. [DOI] [PubMed] [Google Scholar]
- Aeed P A, Geng J G, Asa D, Raycroft L, Ma L, Elhammer A P. Characterization of the O-linked oligosaccharide structures on P-selectin glycoprotein ligand-1 (PSGL-1) Glycoconj J. 1998;15:975–985. doi: 10.1023/a:1006985825141. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Carlsson S R, Klock J C, Dell A. Structures of O-linked oligosaccharides isolated from normal granulocytes, chronic myelogenous leukemia cells, and acute myelogenous leukemia cells. J Biol Chem. 1986;261:12796–12806. [PubMed] [Google Scholar]
- Wilkins P P, McEver R P, Cummings R D. Structures of the O-glycans on P-selectin glycoprotein ligand-1 from HL-60 cells. J Biol Chem. 1996;271:18732–18742. doi: 10.1074/jbc.271.31.18732. [DOI] [PubMed] [Google Scholar]
- Lowe J B. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol. 2003;15:531–538. doi: 10.1016/j.ceb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Clarke J L, Watkins W. Alpha1,3-L-fucosyltransferase expression in developing human myeloid cells. Antigenic, enzymatic, and mRNA analyses. J Biol Chem. 1996;271:10317–10328. doi: 10.1074/jbc.271.17.10317. [DOI] [PubMed] [Google Scholar]
- Ellies L G, Sperandio M, Underhill G H, Yousif J, Smith M, Priatel J J, Kansas G S, Ley K, Marth J D. Sialyltransferase specificity in selectin ligand formation. Blood. 2002;100:3618–3625. doi: 10.1182/blood-2002-04-1007. [DOI] [PubMed] [Google Scholar]
- Nakayama F, Nishihara S, Iwasaki H, Kudo T, Okubo R, Kaneko M, Nakamura M, Karube M, Sasaki K, Narimatsu H. CD15 expression in mature granulocytes is determined by alpha 1,3-fucosyltransferase IX, but in promyelocytes and monocytes by alpha 1,3-fucosyltransferase IV. J Biol Chem. 2001;276:16100–16106. doi: 10.1074/jbc.M007272200. [DOI] [PubMed] [Google Scholar]
- Okajima T, Fukumoto S, Miyazaki H, Ishida H, Kiso M, Furukawa K, Urano T. Molecular cloning of a novel alpha2,3-sialyltransferase (ST3Gal VI) that sialylates type II lactosamine structures on glycoproteins and glycolipids. J Biol Chem. 1999;274:11479–11486. doi: 10.1074/jbc.274.17.11479. [DOI] [PubMed] [Google Scholar]
- Ogawa J I, Inoue H, Koide S. alpha-2,3-Sialyltransferase type 3N and alpha-1,3-fucosyltransferase type VII are related to sialyl Lewis(x) synthesis and patient survival from lung carcinoma. Cancer. 1997;79:1678–1685. doi: 10.1002/(sici)1097-0142(19970501)79:9<1678::aid-cncr7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Watanabe E, Kawashima K, Sekine S, Dohi T, Oshima M, Hanai N, Nishi T, Hasegawa M. Expression cloning of a novel Gal beta (1-3/1-4) GlcNAc alpha 2,3-sialyltransferase using lectin resistance selection. J Biol Chem. 1993;268:22782–22787. [PubMed] [Google Scholar]
- Sasaki K, Kurata K, Funayama K, Nagata M, Watanabe E, Ohta S, Hanai N, Nishi T. Expression cloning of a novel alpha 1,3-fucosyltransferase that is involved in biosynthesis of the sialyl Lewis x carbohydrate determinants in leukocytes. J Biol Chem. 1994;269:14730–14737. [PubMed] [Google Scholar]
- Uchimura K, Muramatsu H, Kaname T, Ogawa H, Yamakawa T, Fan Q W, Mitsuoka C, Kannagi R, Habuchi O, Yokoyama I, Yamamura K, Ozaki T, Nakagawara A, Kadomatsu K, Muramatsu T. Human N-acetylglucosamine-6-O-sulfotransferase involved in the biosynthesis of 6-sulfo sialyl Lewis X: molecular cloning, chromosomal mapping, and expression in various organs and tumor cells. J Biochem (Tokyo) 1998;124:670–678. doi: 10.1093/oxfordjournals.jbchem.a022164. [DOI] [PubMed] [Google Scholar]
- Brockhausen I, Kuhns W, Schachter H, Matta K L, Sutherland D R, Baker M A. Biosynthesis of O-glycans in leukocytes from normal donors and from patients with leukemia: increase in O-glycan core 2 UDP-GlcNAc:Gal beta 3 GalNAc alpha-R (GlcNAc to GalNAc) beta(1–6)-N-acetylglucosaminyltransferase in leukemic cells. Cancer Res. 1991;51:1257–1263. [PubMed] [Google Scholar]
- Watson R W, Rotstein O D, Parodo J, Bitar R, Hackam D, Marshall J C. Granulocytic differentiation of HL-60 cells results in spontaneous apoptosis mediated by increased caspase expression. FEBS Lett. 1997;412:603–609. doi: 10.1016/s0014-5793(97)00779-5. [DOI] [PubMed] [Google Scholar]
- Beauharnois M E, Neelamegham S, Matta K L. Quantitative measurement of selectin-ligand interactions: assays to identify a sweet pill in a library of carbohydrates. Methods Mol Biol. 2006;347:343–358. doi: 10.1385/1-59745-167-3:343. [DOI] [PubMed] [Google Scholar]
- Nasirikenari M, Segal B H, Ostberg J R, Urbasic A, Lau J T. Altered granulopoietic profile and exaggerated acute neutrophilic inflammation in mice with targeted deficiency in the sialyltransferase ST6Gal I. Blood. 2006;108:3397–3405. doi: 10.1182/blood-2006-04-014779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Goldsmith H L, McIntosh F A, Shankaran H, Neelamegham S. Biomechanics of P-selectin PSGL-1 bonds: shear threshold and integrin-independent cell adhesion. Biophys J. 2006;90:2221–2234. doi: 10.1529/biophysj.105.065789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran E V, Xue J, Xia J, Chawda R, Piskorz C, Locke R D, Neelamegham S, Matta K L. Analysis of the specificity of sialyltransferases toward mucin core 2, globo, and related structures. identification of the sialylation sequence and the effects of sulfate, fucose, methyl, and fluoro substituents of the carbohydrate chain in the biosynthesis of selectin and siglec ligands, and novel sialylation by cloned alpha2,3(O)sialyltransferase. Biochemistry. 2005;44:15619–15635. doi: 10.1021/bi050246m. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran E V, Lakhaman S S, Chawda R, Piskorz C F, Neelamegham S, Matta K L. Identification of physiologically relevant substrates for cloned Gal: 3-O-sulfotransferases (Gal3STs): distinct high affinity of Gal3ST-2 and LS180 sulfotransferase for the globo H backbone, Gal3ST-3 for N-glycan multiterminal Galbeta1, 4GlcNAcbeta units and 6-sulfoGalbeta1, 4GlcNAcbeta, and Gal3ST-4 for the mucin core-2 trisaccharide. J Biol Chem. 2004;279:10032–10041. doi: 10.1074/jbc.M311989200. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran E V, Jain R K, Matta K L. Mucin biosynthesis revisited. The enzymatic transfer of Gal in beta 1,3 linkage to the GalNAc moiety of the core structure R1-GlcNAc beta 1,6GalNAc alpha-O-R2. J Biol Chem. 1992;267:19929–19937. [PubMed] [Google Scholar]
- Chandrasekaran E V, Jain R K, Larsen R D, Wlasichuk K, DiCioccio R A, Matta K L. Specificity analysis of three clonal and five non-clonal alpha 1,3-L-fucosyltransferases with sulfated, sialylated, or fucosylated synthetic carbohydrates as acceptors in relation to the assembly of 3′-sialyl-6′-sulfo Lewis x (the L-selectin ligand) and related complex structures. Biochemistry. 1996;35:8925–8933. doi: 10.1021/bi952194e. [DOI] [PubMed] [Google Scholar]
- Liu A H, Liu F, Li Z, Gu J X, Chen H L. Alterations in glycosyltransferases during myeloid and monocytoid differentiation of HL-60 cells. Cell Biol Int. 1998;22:545–550. doi: 10.1006/cbir.1998.0293. [DOI] [PubMed] [Google Scholar]
- Guo H B, Jiang A L, Ju T Z, Chen H L. Opposing changes in N-acetylglucosaminyltransferase-V and -III during the cell cycle and all-trans retinoic acid treatment of hepatocarcinoma cell line. Biochim Biophys Acta. 2000;1495:297–307. doi: 10.1016/s0167-4889(99)00157-3. [DOI] [PubMed] [Google Scholar]
- Kawano S, Hashimoto K, Miyama T, Goto S, Kanehisa M. Prediction of glycan structures from gene expression data based on glycosyltransferase reactions. Bioinformatics. 2005;21:3976–3982. doi: 10.1093/bioinformatics/bti666. [DOI] [PubMed] [Google Scholar]
- Krambeck F J, Betenbaugh M J. A mathematical model of N-linked glycosylation. Biotechnol Bioeng. 2005;92:711–728. doi: 10.1002/bit.20645. [DOI] [PubMed] [Google Scholar]
- Sperandio M, Frommhold D, Babushkina I, Ellies L G, Olson T S, Smith M L, Fritzsching B, Pauly E, Smith D F, Nobiling R, Linderkamp O, Marth J D, Ley K. Alpha 2,3-sialyltransferase-IV is essential for L-selectin ligand function in inflammation. Eur J Immunol. 2006;36:3207–3215. doi: 10.1002/eji.200636157. [DOI] [PubMed] [Google Scholar]
- Le Marer N, Palcic M M, Clarke J L, Davies D, Skacel P O. Developmental regulation of alpha 1,3-fucosyltransferase expression in CD34 positive progenitors and maturing myeloid cells isolated from normal human bone marrow. Glycobiology. 1997;7:357–365. doi: 10.1093/glycob/7.3.357. [DOI] [PubMed] [Google Scholar]
- Sjogren F, Stendahl O, Ljunghusen O. The influence of retinoic acid and retinoic acid derivatives on beta2 integrins and L-selectin expression in HL-60 cells in vitro. Inflammation. 2000;24:21–32. doi: 10.1023/a:1006983824890. [DOI] [PubMed] [Google Scholar]
- Benkerrou M, Delarche C, Brahimi L, Fay M, Vilmer E, Elion J, Gougerot-Pocidalo M A, Elbim C. Hydroxyurea corrects the dysregulated L-selectin expression and increased H(2)O(2) production of polymorphonuclear neutrophils from patients with sickle cell anemia. Blood. 2002;99:2297–2303. doi: 10.1182/blood.v99.7.2297. [DOI] [PubMed] [Google Scholar]
- Hillery C A, Du M C, Wang W C, Scott J P. Hydroxyurea therapy decreases the in vitro adhesion of sickle erythrocytes to thrombospondin and laminin. Br J Haematol. 2000;109:322–327. doi: 10.1046/j.1365-2141.2000.02040.x. [DOI] [PubMed] [Google Scholar]
- Li X, Tu L, Murphy P G, Kadono T, Steeber D A, Tedder T F. CHST1 and CHST2 sulfotransferase expression by vascular endothelial cells regulates shear-resistant leukocyte rolling via L-selectin. J Leukoc Biol. 2001;69:565–574. [PubMed] [Google Scholar]
- Young W W., Jr Organization of Golgi glycosyltransferases in membranes: complexity via complexes. J Membr Biol. 2004;198:1–13. doi: 10.1007/s00232-004-0656-5. [DOI] [PubMed] [Google Scholar]
- Lee N, Wang W C, Fukuda M. Granulocytic differentiation of HL-60 cells is associated with increase of poly-N-acetyllactosamine in Asn-linked oligosaccharides attached to human lysosomal membrane glycoproteins. J Biol Chem. 1990;265:20476–20487. [PubMed] [Google Scholar]
- Giraudo C G, Maccioni H J. Ganglioside glycosyltransferases organize in distinct multienzyme complexes in CHO-K1 cells. J Biol Chem. 2003;278:40262–40271. doi: 10.1074/jbc.M305455200. [DOI] [PubMed] [Google Scholar]
- Priatel J J, Chui D, Hiraoka N, Simmons C J, Richardson K B, Page D M, Fukuda M, Varki N M, Marth J D. The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity. 2000;12:273–283. doi: 10.1016/s1074-7613(00)80180-6. [DOI] [PubMed] [Google Scholar]
- Pratt M R, Hang H C, Ten Hagen K G, Rarick J, Gerken T A, Tabak L A, Bertozzi C R. Deconvoluting the functions of polypeptide N-alpha-acetylgalactosaminyltransferase family members by glycopeptide substrate profiling. Chem Biol. 2004;11:1009–1016. doi: 10.1016/j.chembiol.2004.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.