SUMMARY
The ATPase p97/Cdc48 promotes degradation of ubiquitin-conjugated proteins and its ability to bind substrates is mediated by cofactors containing ubiquitin-binding domains, such as NPL4/UFD1 or the yeast UBX domain protein Ubx2. Here, we employed 'network proteomics' to show that p97 assembles with all of the thirteen mammalian UBX proteins. Remarkably, those UBX proteins that bind ubiquitin conjugates also interact with a broad spectrum of E3 ubiquitin ligases, only one of which has been previously linked to p97 function. In particular, UBXD7 links p97 to the ubiquitin ligase CUL2/VHL and its substrate hypoxia-inducible factor 1α (HIF1α). Depletion of p97 leads to accumulation of endogenous HIF1α and increased expression of the HIF1α target carbonic anhydrase IX, revealing an unexpected role for p97 in functional regulation of HIF1α. We propose that p97 plays a far broader role than previously anticipated in the regulation of protein turnover.
INTRODUCTION
p97, also known as Cdc48 in yeast, is a homohexameric AAA (ATPase associated with a variety of activities) ATPase, highly conserved from archaebacteria to mammals. As one of the most abundant proteins in the cell (Peters et al., 1990), p97/Cdc48 performs a variety of functions ranging from cell cycle regulation to membrane fusion and protein degradation (Ye, 2006). The role of p97/Cdc48 in the ubiquitin-proteasome system (UPS) was first revealed in a genetic screen for mutants defective in the turnover of ubiquitin fusion proteins (the UFD pathway) (Ghislain et al., 1996). However, the bulk of subsequent p97 studies have focused on its role in endoplasmic reticulum-associated protein degradation (ERAD). Typically, ERAD substrates are misfolded or misassembled proteins, but ER-resident enzymes like HMGR can also be regulated by this system (Doolman et al., 2004; Hampton et al., 1996). Dislocation from the ER back into the cytosol is a hallmark of ERAD and p97 provides the driving force for this process (Rouiller et al., 2002; Ye et al., 2001). Also at the ER, yeast Cdc48 participates in the activation of the transcription factors Spt23 and Mga2 by dissociating the active subunits from their membrane-bound partners (Rape et al., 2001; Shcherbik and Haines, 2007).
The most extensively studied p97 binding partners are p47 and the NPL4/UFD1 heterodimer, which form alternative complexes with p97 and direct its activity to different cellular processes. The NPL4/UFD1 adapter is needed for the function of p97 in UPS-dependent protein degradation, including the ERAD pathway, while p47 enables p97 to participate in homotypic membrane fusion. The yeast p47 ortholog, Shp1, is also involved in proteolytic events (Ye, 2006).
Many p97 functions, regardless of whether they are associated with proteolysis or not, involve recognition of ubiquitinated protein substrates. The p97/p47 complex can bind ubiquitinated proteins via the UBA domain in p47, whereas the NPL4/UFD1 heterodimer performs the same function through the NZF (NPL4 zinc finger) domain in NPL4 and the UT3 region in UFD1 (Meyer et al., 2002; Park et al., 2005; Ye et al., 2003).
While p47 and NPL4/UFD1 are substrate-recruiting cofactors, p97 also interacts with a variety of substrate-processing cofactors like the E4 enzyme Ufd2 (Richly et al., 2005) or the deubiquitinating enzymes VCIP135 (Uchiyama et al., 2002; Wang et al., 2004) and Otu1 (Rumpf and Jentsch, 2006). With the exception of Ufd2, all p97 cofactors enumerated above interact with the N-terminal domain of p97. Ufd2 and another yeast Cdc48 cofactor, Ufd3/Doa1, dock onto p97’s C-terminus (Yeung et al., 2008).
Irrespective of its bound cofactors, the underlying function of p97 is believed to be the conversion of the energy derived from ATP hydrolysis into mechanical force used to disassemble protein complexes or segregate polypeptides from intracellular structures such as the ER membrane. p97 comprises three domains, the N-terminal domain and two AAA ATPase domains (D1, D2). Structural studies revealed that the ATPase domains form two hexameric rings stacked on top of each other with a pore in the center. Multiple conformational changes have been observed in the p97 hexamer during its ATPase cycle and several models have been proposed to explain how p97 ATPase activity can be translated via the adaptors into tension applied on protein substrates (Pye et al., 2006).
We describe a comparative proteomic study of mammalian UBX domain-containing p97 cofactors. Our experiments unexpectedly revealed a large number of E3 ligases that interact with a subset of UBX-domain proteins. Among these interactions, we found that the UBX-domain protein UBXD7 bound subunits of the CUL2/VHL ubiquitin ligase complex and its substrate, HIF1α. We further show that HIF1α is a novel endogenous substrate of p97 and UBXD7 mediates its interaction with p97. Our results suggest that the role of p97 in ubiquitin-mediated proteolysis is far more pervasive than previously envisioned.
RESULTS
Mammalian p97 Interacts with Multiple UBX Domain-Containing Cofactors
To further understand the molecular basis for p97’s diverse functions, we analyzed p97-Myc immunoprecipitates from human 293 cells by MudPIT (Multidimensional Protein Identification Technology) (Link et al., 1999), searching for new p97 cofactors. This analysis revealed eight p97 binding partners (Table 1), all containing a UBX (structurally similar to ubiquitin) domain in their C-terminal region (Fig. 1A). At the time of our analysis, p47 was the only human UBX-domain protein known to interact with p97 (Kondo et al., 1997; Meyer et al., 2000) and the UBX domain of p47 was known to be required for binding the N-terminal domain of p97 (Uchiyama et al., 2002; Yuan et al., 2001).
Table 1.
UBX-Domain Proteins Identified by Mass Spectrometry in p97-Myc Immunoprecipitates (in order of sequence coverage)
| PROTEIN NAME | MW (Da) | Sequence count | Spectrum count | Sequence coverage |
|---|---|---|---|---|
| p47 | 40573 | 19 | 153 | 52.2% |
| UBXD8 | 52624 | 11 | 12 | 33.7% |
| p37 | 37077 | 6 | 7 | 32.3% |
| UBXD7 | 54862 | 8 | 12 | 29.0% |
| SAKS1 | 33325 | 2 | 2 | 19.5% |
| UBXD4 | 29278 | 4 | 5 | 19.3% |
| ASPL | 60183 | 7 | 13 | 17.2% |
| FAF1 | 73954 | 7 | 11 | 8.2% |
The number of unique peptides identified (sequence count) is indicated, as is the total number of peptides (spectrum count), which takes into account that some peptides were identified multiple times.
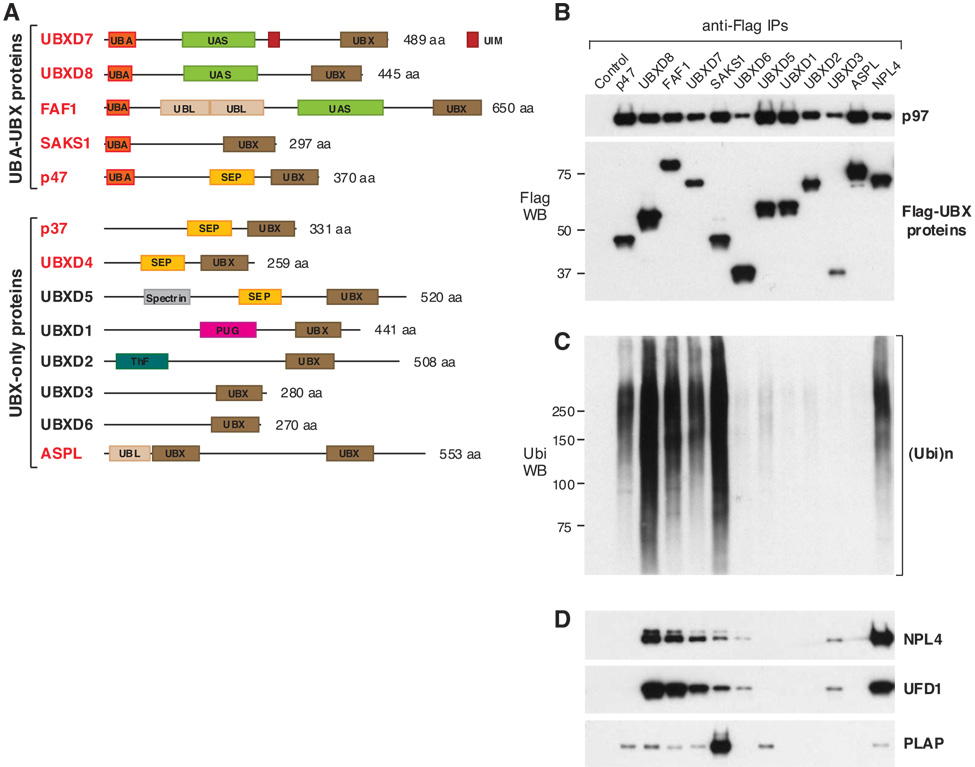
Figure 1. Mammalian UBX-Domain Proteins Interact with p97 and Some Serve as Ubiquitin Receptors.
(A) The domain composition of human UBX proteins. Those identified in p97-Myc immunoprecipitates are indicated in red. UBA – Ubiquitin-associated; UIM – Ubiquitin-interacting motif; UBL – Ubiquitin-like; ThF – Thioredoxin-like fold. Further information about the respective domains can be found at http://www.ebi.ac.uk/interpro/. (B–D) N-terminally Flag-tagged UBX proteins were expressed in 293 cells and immunoprecipitated using anti-Flag beads. Cells expressing Flag-NPL4 or no Flag-tagged protein were used as positive and negative control, respectively. Some of the endogenous proteins that were coimmunoprecipitated are shown in western blots using specific antibodies. (Ubi)n refers to ubiquitin chains of varying length.
The human proteome includes at least thirteen different UBX proteins (Fig. 1A), some of which were not identified in our initial analysis. At least three of those, UBXD1 (Carim-Todd et al., 2001), Socius/UBXD5 (Katoh et al., 2002), and Rep-8/UBXD6 (Yamabe et al., 1997), are mainly expressed in the reproductive organs and might be expressed poorly in the 293 kidney cell line used for immunoprecipitation. Upon expressing their Flag-tagged versions in 293 cells, we confirmed that eleven mammalian UBX domain-containing proteins (five of which were absent in the original p97 immunoprecipitates) coimmunoprecipitated endogenous p97 (Fig. 1B). Taken together our mass spectrometry and immunoprecipitation/western analyses confirmed that all thirteen mammalian UBX proteins bound p97. This is consistent with the observation that all seven budding yeast UBX proteins bind Cdc48 (Schuberth et al., 2004). Given that UBX proteins invariably bind p97/Cdc48, UBX emerges as a signature domain for p97 binding partners across species.
There Are Two Classes of UBX Domain-Containing Proteins Based on Their Ability to Bind Ubiquitinated Substrates
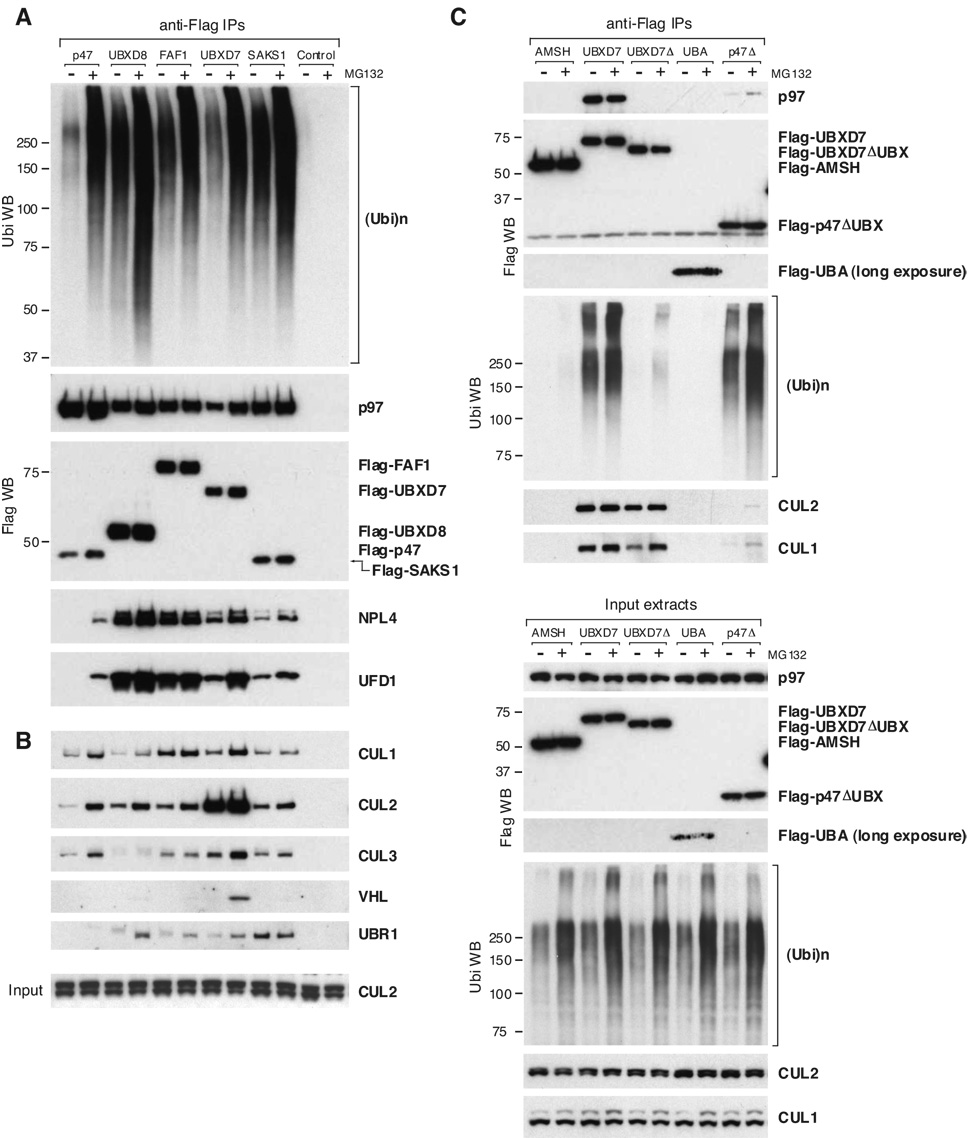
Based on their domain composition, the human UBX proteins can be divided into two main groups (Fig. 1A). The first group includes the UBA-UBX proteins (UBXD7, UBXD8, FAF1, SAKS1, and p47), characterized by the presence of an UBA (ubiquitin-associated) domain at their N-termini. The UBA domain binds ubiquitin (Hurley et al., 2006) and Flag-tagged UBA-UBX proteins coimmunoprecipitated endogenous ubiquitin conjugates (Fig. 1C). The amount of ubiquitinated proteins present in UBA-UBX protein immunoprecipitates was amplified by proteasome inhibition with MG132 (Fig. 2A), suggesting that at least some of them are UPS substrates. The second group includes the UBX-only proteins, which lack the UBA domain (Fig. 1A) and the ability to bind ubiquitinated substrates (Fig. 1C).
Figure 2. UBA-UBX Proteins Interact with Ubiquitinated Proteins Destined for Degradation and with Various E3 Ligases.
(A, B) Flag–(UBA-UBX) proteins were immunoprecipitated from 293 cells treated for 2 h with DMSO or MG132. Cells expressing no Flag-tagged protein were used as negative control. Some of the endogenous proteins that coimmunoprecipitated were detected by western blotting using specific antibodies. CUL2 input levels are shown at the bottom of panel B.
(C) The indicated Flag-tagged proteins were immunoprecipitated from HeLa cells treated with MG132 as above. AMSH, a protein that is not part of the p97 network, was used as negative control. UBXD7Δ and p47Δ are truncation mutants lacking the UBX domain. The UBA domain by itself was expressed at very low levels. The indicated proteins were detected using specific antibodies, in the immunoprecipitates (top) and in the input cell extracts (bottom). Ubi – ubiquitin
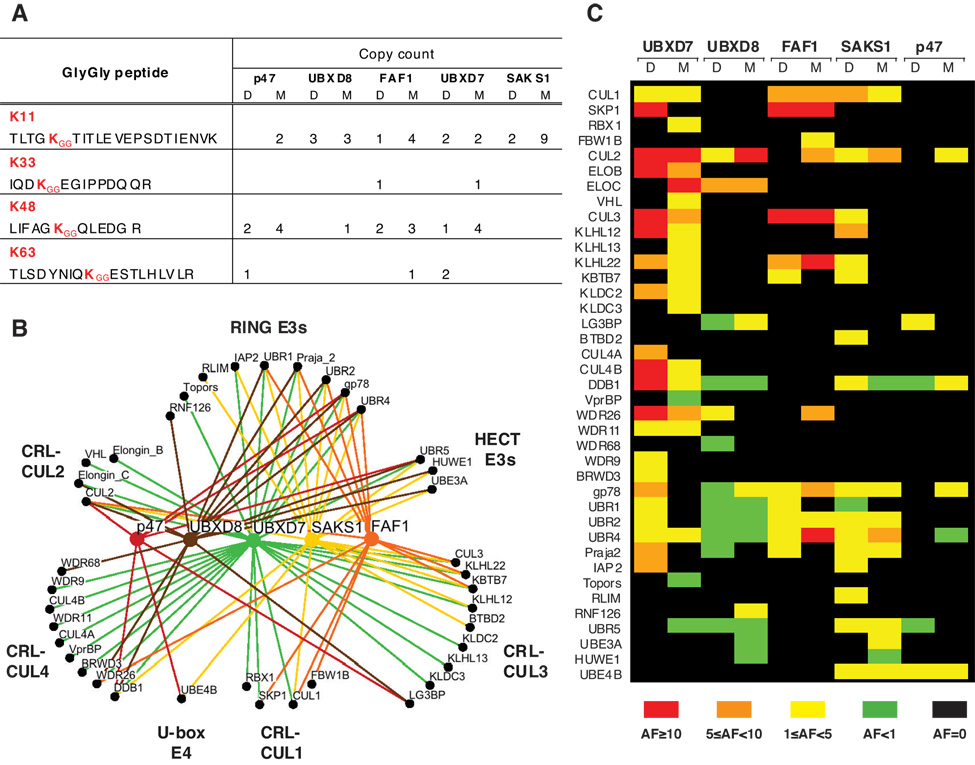
To establish what type of ubiquitin chains are recognized by mammalian UBA-UBX proteins, we searched our mass spectrometry data for ubiquitin tryptic peptides bearing GG signatures (Parker et al., 2005). Multiple spectra corresponding to ubiquitin peptides carrying a GG signature at K48 were identified, as expected for proteins targeted for proteasomal degradation (Pickart, 1997). However, it came as a surprise that a higher number of peptides carried GG signatures attached to K11. The detection of GG signature peptides by mass spectrometry is most efficient for K48 and less effective for K11 (Kirkpatrick et al., 2006), suggesting that the actual ratio of K11- to K48-linked chains could be even higher than indicated by the spectrum counts shown in Fig. 3A. While a more detailed analysis will be required to unambiguously establish ubiquitin chain specificity, it is interesting to note that K11-linked chains were detected in immunoprecipitates of all UBA-UBX proteins.
Figure 3. The E3 Interaction Network for UBA-UBX Proteins.
(A) The number and type of GG signature peptides identified by mass spectrometry in Flag–(UBA-UBX) immunoprecipitates is indicated. D – DMSO; M – MG132 (B) Osprey diagram illustrating the set of E3 ligases interacting with each UBA-UBX protein. (C) Quantitative representation of the interactions shown in B, obtained by calculating for each E3 ligase the abundance factor (AF) relative to p97. For details see Suppl. Table 2 and its legend.
General Features of the UBX Protein Interaction Networks
Because the biological functions for most UBX proteins are largely unknown (Schuberth and Buchberger, 2008), we performed a comparative MudPIT analysis of Flag-UBX protein immunoprecipitates from transiently transfected 293 cells. The resulting datasets were mined to identify interacting partners that are shared among multiple UBX protein complexes, as well as partners that are specific to a certain UBX protein.
We first focused our attention on known components of the p97 network. NPL4/UFD1 and p47 use a similar bipartite mechanism for binding the N-terminal domain of p97 and compete for p97 binding in vitro (Bruderer et al., 2004; Meyer et al., 2000). This led to the hypothesis that the interaction of NPL4/UFD1 and UBX proteins with p97 might be mutually exclusive. However, the bipartite p97-binding motif seems to be conserved only in SEP-UBX proteins like p47 (Bruderer et al., 2004) and p37 (Uchiyama et al., 2006), leaving open the possibility that other UBX-domain proteins use a different binding mode. We confirmed both by mass spectrometry (Suppl. Table 1) and by immunoblotting of Flag-p47 immunoprecipitates (Fig. 1D, Fig. 2A) that, for the most part, p47 does not form complexes with NPL4/UFD1. That seems to be an exception rather than the rule, as the other UBA-UBX proteins coimmunoprecipitated NPL4 and UFD1. Conversely, Flag-NPL4 coimmunoprecipitated UBA-UBX proteins, with most peptides identified for the UBA-UAS-UBX proteins, UBXD8, UBXD7, and FAF1 (data not shown). Similarly, the yeast UBA-UBX protein Ubx2 assembles into a Cdc48/Npl4/Ufd1/Ubx2 complex (Schuberth and Buchberger, 2005).
We also compared the ability of UBX proteins to interact with substrateprocessing cofactors of p97. VCIP135 seems to interact preferentially with SEP-UBX proteins like p47 and UBXD5 (Suppl. Table 1). Indeed, two SEP-UBX proteins, p37 and p47, both require VCIP135 for their function (Uchiyama et al., 2006). PLAP, known as Ufd3/Doa1 in budding yeast, has a strong preference for co-assembling with SAKS1 (Fig. 1D). Intriguingly, even though yeast Npl4/Ufd1 and Ufd3 bind to distinct regions of Cdc48 (Rumpf and Jentsch, 2006), in our analysis the complexes that are richest in NPL4/UFD1 are poorest in PLAP and vice versa (Fig. 1D, Suppl. Table 1). With the exception of a few peptides identified in SAKS1 and p47 immunoprecipitates UBE4B, the human ortholog of yeast Ufd2, was largely absent from our UBA-UBX protein and p97 immunoprecipitates (Suppl. Table 2).
It has been proposed that yeast Cdc48 functions in series with other targeting factors like Rad23 to mediate processing of ubiquitin conjugates and their eventual presentation to the proteasome (Medicherla et al., 2004; Richly et al., 2005). Although this model contemplates the formation of ternary complexes, we found that RAD23 and ubiquilins were largely absent from our UBX protein and p97 immunoprecipitates. However, we did identify multiple proteasome subunits, most frequently the proteasome base subunits PSMC3, PSMC4, and PSMD1 (data not shown), which made us speculate that in human cells p97–substrate complexes might directly dock onto the proteasome base without another targeting factor acting as an intermediary.
Notably, there was little cross-interaction between different UBX proteins (data not shown), suggesting that they form homomeric complexes with p97.
UBA-UBX Proteins Interact with a Large Variety of E3 Ubiquitin Ligases
A striking observation from the comparative MudPIT analysis of Flag–(UBA-UBX) protein immunoprecipitates was their ability to interact with numerous E3 ligases as indicated qualitatively in Fig. 3B. We identified multiple components of cullin-RING E3 ligase (CRL) complexes, but also single subunit RING- and HECT-domain E3s. Of these 38 ubiquitin ligases, more than a third were also identified in p97 immunoprecipitates (marked a in Suppl. Table 2), confirming they belong to the p97 network.
Individual UBA-UBX proteins did not exhibit strict specificity for particular E3 ligases, but at least some E3s seemed to be enriched in certain UBA-UBX protein immunoprecipitates (Fig. 3C). Most notably, UBXD7 showed a remarkable ability to coimmunoprecipitate CUL2. Moreover, we also identified RBX1, elongin B, elongin C, and VHL in UBXD7 immunoprecipitates. In general, UBXD7 was the UBA-UBX protein that showed the most extensive interaction with CRL subunits (Fig. 2B, Fig 3C). An UBXD7 mutant lacking the UBX domain lost the ability to interact not only with p97, but also with ubiquitinated substrates (Fig. 2C). Despite that, truncated UBXD7 largely retained its capacity to bind CUL1 and CUL2. In contrast, a p47 mutant lacking the C-terminal region could still pull down ubiquitinated proteins, but did not exhibit significant binding of cullins. This lack of correlation between ubiquitin and E3 binding, together with semi-quantitative analysis of the MudPIT data (Fig. 3C), suggest that the interaction between UBA-UBX proteins and E3 ligases is specific and not simply mediated by the ubiquitinated substrate binding to the UBA domain.
UBXD7 Interacts with HIF1α in a Manner that Is Largely Independent of p97
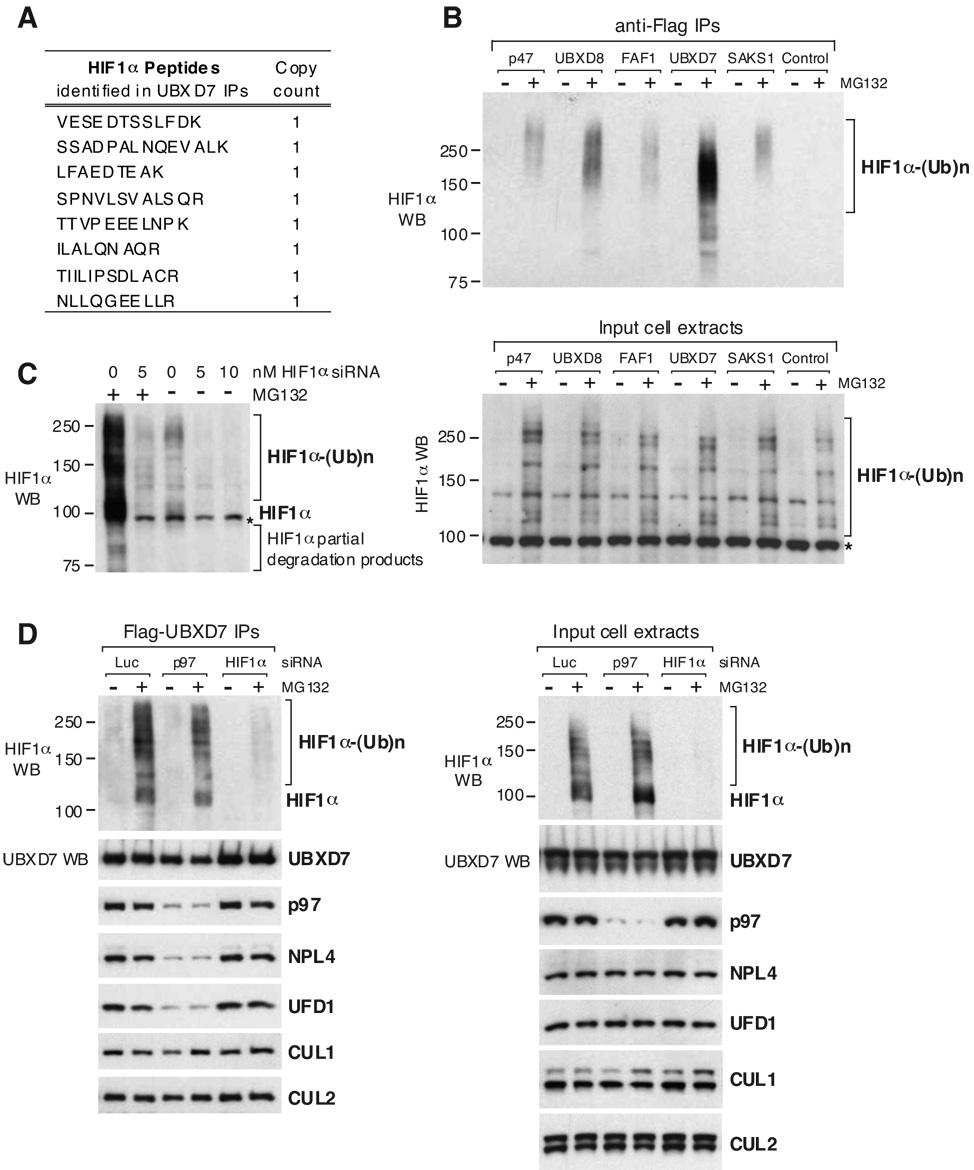
p97 cofactors like p47 and NPL4/UFD1 mediate the interaction between p97 and its ubiquitinated targets (Ye, 2006). By MudPIT analysis of individual UBX-protein immunoprecipitates, we sought to identify p97 targets specific for these cofactors, and thereby unravel which p97 functions they regulate. Therefore, we were intrigued to identify eight distinct HIF1α peptides in Flag-UBXD7 immunoprecipitates from cells in which the proteasome activity was inhibited with MG132 (Fig. 4A). HIF1, a heterodimeric transcription factor that consists of HIF1α and HIF1β subunits, regulates transcription in response to changes in O2 concentration. O2-dependent degradation of HIF1α is mediated by prolyl-hydroxylase, the CUL2/VHL ubiquitin ligase, and the proteasome (Ivan and Kaelin, 2001). Thus, we decided to pursue HIF1α as a potential p97/UBXD7 substrate.
Figure 4. UBXD7 Interacts with Endogenous HIF1α Independently of p97.
(A) HIF1α peptides identified by mass spectrometry in Flag-UBXD7 immunoprecipitates from cells treated with MG132 for 2 h.
(B) The Flag–(UBA-UBX) immunoprecipitates shown in figure 2A were separated by PAGE and blotted using HIF1α specific antibodies (top). The bottom panel shows equivalent HIF1α levels in the input cell extracts.
(C) The specificity of HIF1α antibodies was tested on total cell extracts from HeLa cells treated with the indicated concentration of HIF1α siRNA, in the presence and in the absence of a 2 h treatment with MG132. A cross-reacting band partially overlapping with full-length HIF1α is indicated with *.
(D) Flag-UBXD7 was immunoprecipitated from HeLa cells treated with 5 nM of the indicated siRNAs for 48 h. Where indicated, 20 µM MG132 was added for 2 h prior to harvesting the cells. The indicated proteins were detected using specific antibodies, in the immunoprecipitates (left) and in the input cell extracts (right). Luc – luciferase
Among the UBA-UBX proteins, UBXD7 was by far the most efficient in coimmunoprecipitating endogenous HIF1α which was detected as a ubiquitinated ladder using anti-HIF1α antibodies (Fig. 4B). HIF1α is scarce in normoxia (Huang et al., 1996), hence the interaction between UBXD7 and HIF1α was only detectable after MG132 treatment, which causes accumulation of ubiquitinated HIF1α. We confirmed the specificity of the HIF1α antibodies by comparing the signal in total cell extracts from cells treated or not with HIF1α siRNA, both in the presence and in the absence of MG132 (Fig. 4C). This indicated the presence of a cross-reacting band that partially overlaps with full length HIF1α, marked with * in all the panels showing HIF1α in total cell extracts. The cross-reacting band was absent from immunoprecipitates (Fig. 4D). Proteasome inhibition also caused accumulation of HIF1α partial degradation products (Fig. 4C) that migrated faster than expected for the full-length protein (92.7 kDa), some of which also coimmunoprecipitated with UBXD7 (Fig. 4B).
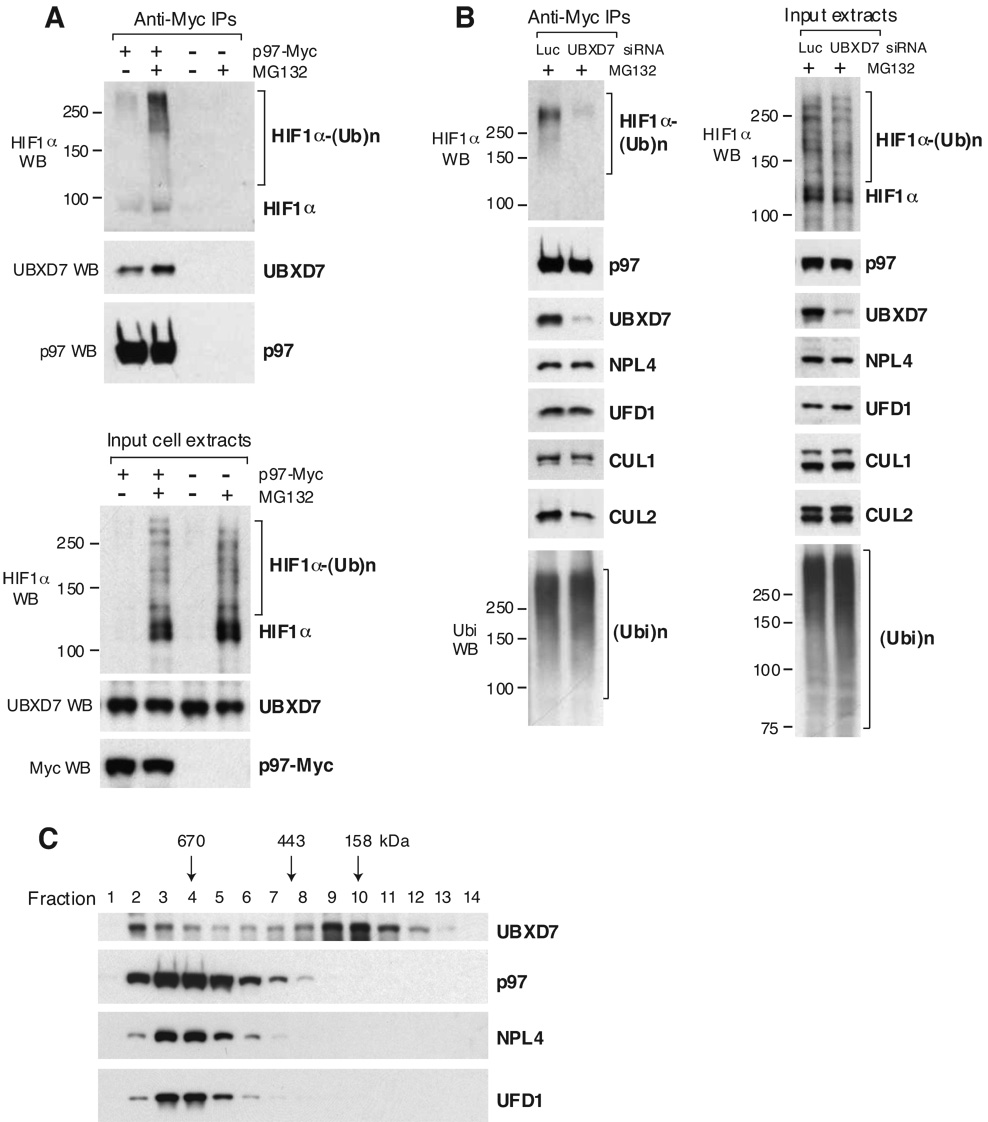
We next tested whether the interaction between UBXD7 and HIF1α depends on p97. Depletion of p97 by siRNA did not alter significantly the interaction of UBXD7 with HIF1α or cullins, but it drastically reduced the association of UBXD7 with NPL4 and UFD1 (Fig. 4D). We therefore conclude that UBXD7 interaction with the substrate and E3s does not depend on p97/NPL4/UFD1.
UBXD7 Recruits p97 to HIF1α
To validate that the interaction of UBXD7 with HIF1α occurs within the p97 network, we showed that p97 itself coimmunoprecipitated endogenous HIF1α (Fig. 5A). Interestingly, the HIF1α that immunoprecipitated with p97 was more extensively polyubiquitinated than the pool bound to UBXD7 (compare Fig. 5A, B with Fig. 4B, D). The accumulation of ubiquitinated HIF1α in p97 immunoprecipitates after proteasome inhibition correlated with increased amounts of endogenous UBXD7 bound to p97 (Fig. 5A), suggesting that UBXD7 binding to p97 might depend on the availability of substrate. Further support for this idea came from gel filtration experiments of HeLa cell extracts in which all proteins were expressed endogenously (Fig. 5C). We observed two fractionation peaks for endogenous UBXD7, only one of which overlapped with p97. In contrast, NPL4 and UFD1 fractionation closely resembled p97. This fractionation pattern indicated that the default state for NPL4/UFD1 was p97-bound, whereas only a fraction of UBXD7 was p97-bound, possibly in response to a stimulus such as interaction with a substrate. Indeed, the accumulation of ubiquitinated substrates upon proteasome inhibition by MG132 resulted in a shift of UBXD7 towards p97-positive fractions (Suppl. Fig. 1A), a phenomenon that was reverted upon p97 depletion (Suppl. Fig. 1B).
Figure 5. UBXD7 Recruits p97 to HIF1α.
(A) p97-Myc was immunoprecipitated from HeLa cells treated or not with 20 µM MG132 for 2 h prior to harvesting the cells. Cells expressing no Myc-tagged protein were used as negative control. The indicated proteins were detected in the immunoprecipitates (top) and in the input cell extracts (bottom) using specific antibodies.
(B) p97-Myc was immunoprecipitated from HeLa cells treated for 48 h with 5 nM of the indicated siRNAs and incubated with MG132 as above. The indicated proteins were detected using specific antibodies, in the immunoprecipitates (left) and in the input cell extracts (right). Luc – luciferase
(C) HeLa cell extracts were fractionated on a Superdex 200 gel filtration column. Individual fractions were concentrated by TCA precipitation and subjected to western blotting using specific antibodies. All proteins were endogenously expressed.
If the substrate-ligase complex binds UBXD7, which in turn binds p97, HIF1α association with p97 should depend on UBXD7. Indeed, the ability of p97 to coimmunoprecipitate endogenous HIF1α-ubiquitin conjugates was lost in cells treated with UBXD7 siRNA (Fig. 5B). In contrast, UBXD7 depletion had no significant effect on NPL4, UFD1 or poly-ubiquitin binding to p97. While CUL1 binding to p97 was also unaffected by UBXD7 depletion, we observed a significant reduction of CUL2 binding (Fig. 5B), consistent with UBXD7 being the best CUL2 binder among UBA-UBX proteins (Fig. 2B). This suggests that the UBA-UBX adaptor mediates p97 interaction with the substrate and the corresponding E3 ligase.
HIF1α Is a Novel p97 Substrate
The endogenous HIF1α that interacted with both UBXD7 and p97 was mainly ubiquitinated and accumulated upon proteasome inhibition (Fig. 4B, 4D, 5A), supporting the idea that it was destined for UPS-dependent degradation.
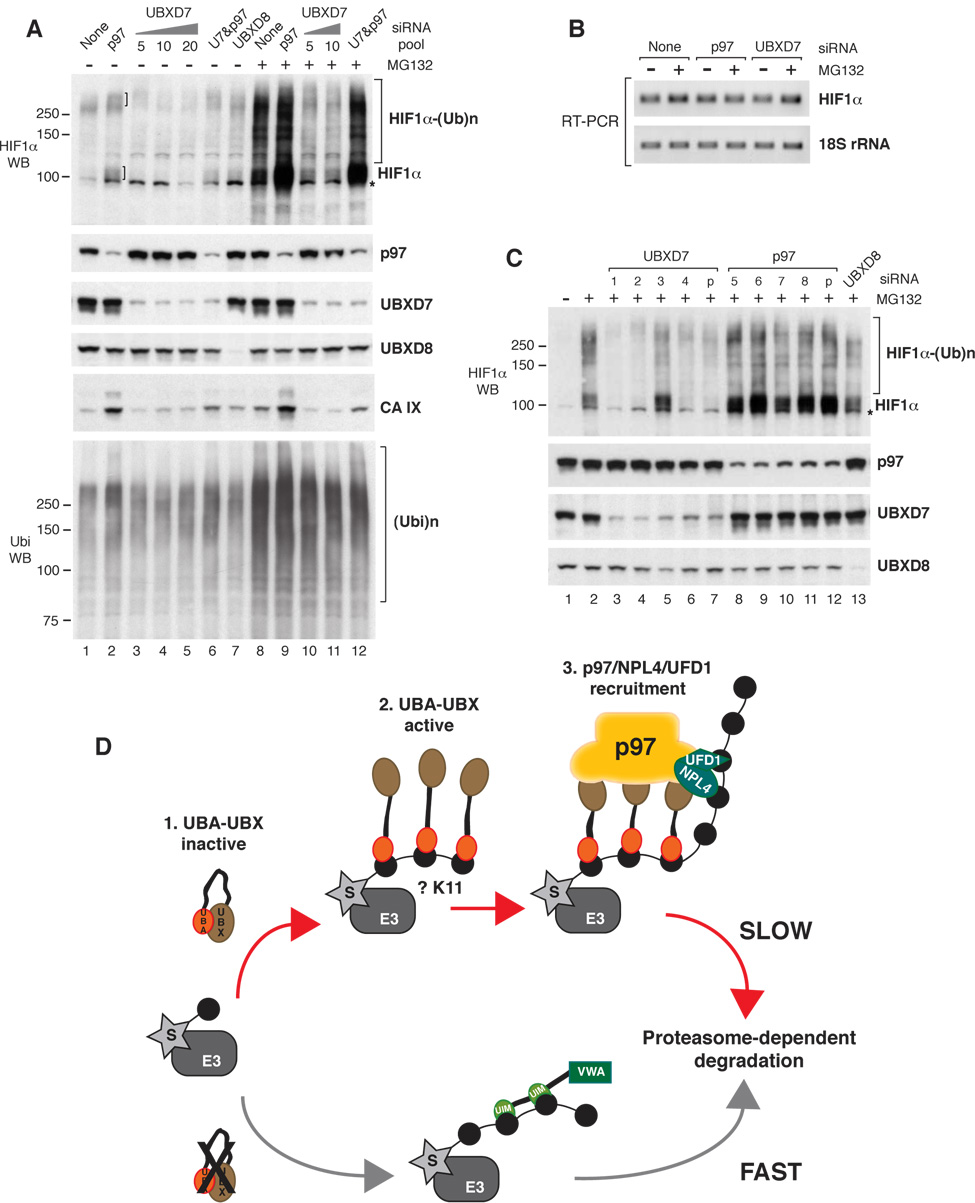
To test whether p97 regulates HIF1α degradation, we performed siRNA-mediated depletion experiments. Figure 6A shows the effect of various siRNA pools on HIF1α levels in total cell extracts. p97 depletion caused accumulation of endogenous HIF1α as species >100 kDa and ≫250 kDa, and this effect was amplified by brief exposure to MG132 (Fig. 6A, compare lane 1 with 2 and lane 8 with 9). However, p97 depletion did not promote HIF1α accumulation as effectively as proteasome inactivation. This could be due to various reasons: i) the low amounts of p97 that remain in the cell after siRNA treatment may be sufficient to promote HIF1α degradation, ii) other targeting factors, like Rpn10/PSMD4, RAD23, or ubiquilins may be able to partially compensate for the lack of p97, or iii) only a subset of ubiquitinated HIF1α molecules depend on p97 for degradation. p97 depletion also caused a mild increase in the total pool of ubiquitinated proteins (Fig. 6A, compare lane 1 with 2). The p97 siRNA pool did not alter HIF1α mRNA levels (Fig. 6B), indicating that the observed effects at the protein level were most likely due to perturbations in HIF1α degradation.
Figure 6. p97 Promotes HIF1α Degradation.
(A) Total cell extracts were prepared from cells treated with 5 nM of the indicated siRNA pools unless other siRNA concentration is specified. The siRNA treatment was 48 h and it was combined or not with 2 h of MG132 treatment. The indicated proteins were detected using specific antibodies. U7 – UBXD7
(B) HIF1α mRNA was amplified by RT-PCR using specific primers. 18S rRNA was amplified as control.
(C) Total cell extracts were prepared from cells treated with 5 nM of the indicated siRNA oligonucleotides or pools (p) for 48 h and incubated with 20 µM MG132 for 2 h. The indicated proteins were detected using specific antibodies.
A non-specific band cross-reacting with the HIF1α antibodies is indicated with *.
(D) UBXD7 Recruits p97/NPL4/UFD1 to the Ubiquitinated Substrate and Prevents Its Interaction with Other Proteasome Targeting Factors.
Top: 1 - UBA and UBX domains inactivate each other when the protein is not bound to the substrate. 2 - Substrate oligo-ubiquitination or attachment of multiple monoubiquitin allows recruitment of several UBA-UBX molecules per substrate. UBA domains mask the emerging ubiquitin-chain and prevent substrate degradation. 3 - Substrate binding frees the UBX domains that become available to recruit p97/NPL4/UFD1. The ubiquitin chain is elongated and the substrate is delivered to the proteasome for degradation.
Bottom: In the absence of UBXD7, other proteasome targeting factors mediate accelerated substrate degradation. The Rpn10/PSMD4 subunit of the proteasome is depicted as an alternative ubiquitin receptor. S – substrate, E3 – ubiquitin ligase
As a measure of HIF1α activity, we analyzed the levels of carbonic anhydrase IX (CA IX), an established target of HIF1α transcriptional activity (Wykoff et al., 2000). CA IX protein levels were very low in normoxia (Fig. 6A, lane 1), but accumulated in cells depleted of p97 (Fig. 6A, lanes 2 and 9). It has been reported by several groups that HIF1α that accumulates in the presence of MG132 is transcriptionally inactive (Kaluz et al., 2007) and this explains why MG132 had a major effect on HIF1α levels, but little effect on CA IX levels.
We next confirmed that individual p97 siRNA oligonucleotides behaved similar to the p97 siRNA pool (Fig. 6C, lanes 8–12). Even if the amplitude of the effect varied among siRNAs, the general trend was the same — p97 depletion led to HIF1α accumulation.
Taken together our results suggest that efficient HIF1α degradation depends on p97, thereby establishing HIF1α as the first endogenous substrate of mammalian p97 that is not associated with the ER.
Given that UBXD7 recruits HIF1α to p97 (Fig. 5B), we thought that UBXD7 depletion would phenocopy p97 depletion. Thus, it was unexpected to see that UBXD7 depletion caused a reduction in both full length and ubiquitinated HIF1α (Fig. 6A). The contrast between p97 and UBXD7 depletion was most obvious upon brief treatment with MG132 (Fig. 6A, lanes 9–11). This result was confirmed by three of the four siRNA oligonucleotides in the UBXD7 siRNA pool (Fig. 6C, compare lane 2 with lanes 3, 4, 6, 7). Moreover, when the cells were treated with a combination of UBXD7 and p97 siRNAs, UBXD7 depletion seemed to partially offset the lack of p97 (Fig. 6A, Suppl. Fig. 2). As elaborated in the discussion section we speculate that in the absence of UBXD7, HIF1α does not engage the p97 network and is more readily available to alternative proteasome receptors.
The protein levels of CA IX perfectly mimicked those of HIF1α (Fig. 6A); they were highest in cells depleted of p97 (lanes 2 and 9), lowest in cells depleted of UBXD7 (lanes 3–5 and 10, 11), and intermediate in cells depleted of both UBXD7 and p97 (lanes 6 and 12). As for p97, the UBXD7 siRNA pool did not have a significant effect on HIF1α mRNA levels (Fig. 6B).
DISCUSSION
The role of the p97/Cdc48 ATPase in ubiquitin-dependent proteolysis has been studied intensively over the past several years, mainly from the perspective of the important contribution that it makes to ERAD. Despite the prevalent ER-centric view of p97 function, hints have emerged that p97 plays a broader role in regulating the turnover of UPS substrates. For example, p97/Cdc48 has been implicated in the turnover of the protein kinase Cdc5 (Cao et al., 2003), the Cdk inhibitor Far1 (Fu et al., 2003), and the myosin chaperone UNC45 (Janiesch et al., 2007). However, it remains unknown how pervasive p97's non-ER functions are.
In the work reported here, we sought to gain greater insight into p97 biology by performing a focused, ‘network proteomics’ analysis (Graumann et al., 2004) of p97 and its UBX-domain cofactors. Two major insights have emerged from this effort. First, we found that UBA-UBX proteins associate with an unexpectedly broad range of ubiquitin ligases, including cullins 1 through 4, nine RING ligases, and three HECT domain enzymes. Among these, p97 has been linked previously only to the ERAD-related ubiquitin ligase gp78 (Zhong et al., 2004). Given the great number of CRLs expressed in human cells and their intimate connection to a broad range of regulatory processes, our findings suggest that the substrate repertoire of p97 is far more diverse than previously appreciated and nominate p97 as a candidate regulator of numerous processes in which it has not previously been implicated. The second major finding, which flows directly from the first, is that we have forged a direct and unexpected functional connection between p97 and HIF1α which is the key governor of cellular and organismal responses to oxygen tension.
Network Proteomics Provides Novel Insights into p97 Biology
Our analysis of the p97 proteome has unearthed a trove of observations that challenge some current assumptions about p97/Cdc48. First, our findings imply that ERAD may comprise but a small fraction of p97's role in the UPS. This is consistent with CDC48 being an essential gene in yeast, whereas ERAD is dispensable (Swanson et al., 2001). Second, we challenged the notion that UBX proteins and NPL4/UFD1 form mutually exclusive complexes with p97. This view is based on molecular analysis of p47, which our results reveal may be the exception, rather than the rule. Other UBX-domain proteins including UBXD7, UBXD8, and FAF1 clearly form higher-order complexes that contain p97 and NPL4/UFD1. Third, our proteomic findings suggest that substrate-processing cofactors such as VCIP135, PLAP, and UFD2 may be restricted to specific UBX-protein/p97 complexes. Fourth, we noted an unexpected prevalence of K11-linked ubiquitin chains in pull-downs of UBA-UBX proteins.
Taken together, our findings demonstrate how network proteomics can provide a broad view that is not accessible from the analysis of a single component.
The Role of p97 and UBXD7 in HIF1α Turnover
In the second half of the work described here, we took one observation from the proteomic analysis and investigated it in depth. Specifically, we delved into the finding that in cells treated with MG132, UBXD7 coimmunoprecipitated all components of the CUL2/VHL ubiquitin ligase as well as its most prominent substrate, HIF1α. Although HIF1α metabolism has been the focus of intensive investigation, it has not been previously linked to p97 in any way.
Using UBXD7 as a prototype UBA-UBX protein and HIF1α as a model substrate, we gained new insights into the role of UBA-UBX adaptors within the p97 network. Three observations led us to suggest that substrate binding to UBXD7 precedes the formation of UBXD7 complexes with p97/NPL4/UFD1 and may be a prerequisite for it: i) the interaction of UBXD7 with the substrate and E3 does not depend on p97/NPL4/UFD1, whereas the interaction of p97/NPL4/UFD1 with substrate and E3 requires UBXD7, ii) UBXD7 binding to p97 increases upon proteasome inhibition and accumulation of substrate, as shown by immunoprecipitation and gel filtration experiments, iii) in contrast to NPL4/UFD1, a considerable pool of endogenous UBXD7 is not bound to p97. By analogy to the idea that UBL and UBA domains can undergo intramolecular association (Lowe et al., 2006; Ryu et al., 2003), it is conceivable that UBX and UBA domains interact with each other, intra- or inter-molecularly, thereby maintaining UBXD7 in an inactive state (Fig. 6D-1). Only after the UBA domain engages an ubiquitinated substrate, would the UBX domain become available to recruit the p97/NPL4/UFD1 complex through interaction with the N-terminal domain of p97 (Fig. 6D-2,3).
An important complement to the binding studies that linked HIF1α to UBXD7 and p97 was a series of siRNA knockdown experiments. We found that endogenous HIF1α accumulates in cells depleted of p97, while the opposite is seen when cells are depleted of UBXD7. To explain this apparent paradox, we propose a two-step function for UBXD7 in mediating HIF1α degradation via the p97 pathway. Binding of UBXD7 to ubiquitinated HIF1α commits it to the p97 pathway and shields it from other proteasome targeting factors. A protective role of UBXD7 that precedes its role in recruiting p97/NPL4/UFD1 would explain the observed discrepancy between the UBXD7 and p97 siRNA results. In cells depleted of p97, ubiquitinated HIF1α becomes trapped in non-productive complexes with UBXD7. However, in cells depleted of UBXD7, ubiquitinated HIF1α cannot be guided into the p97 pathway and is free to engage other targeting factors or the proteasome itself through its Rpn10/PSMD4 or Rpn13 subunits (Fig. 6D bottom). This would provide a more expeditious route for degradation than the pathway gated by UBXD7, hence the observed reduction in HIF1α levels.
In contrast to the results we obtained for UBXD7 and HIF1α, deletion of yeast UBX2 causes stabilization of ERAD substrates (Neuber et al., 2005; Schuberth and Buchberger, 2005). The difference between the behavior of HIF1α and that of ERAD substrates upon depletion of the respective UBA-UBX adaptor protein might be due to the fact that ERAD includes an ER retro-translocation step. In the case of ER-associated substrates, p97 provides the driving force for their retro-translocation through the ER membrane (Ye, 2006), which makes it irreplaceable by other targeting factors lacking the ATPase activity. As a soluble substrate, HIF1α relinquishes such a requirement.
HIF1α is the first known UBA-UBX protein ligand that is not associated with the ER, hence the generality of its behavior in the absence of UBXD7 must await the identification of other receptor-ligand pairs. While elucidating the exact role played by UBXD7 in HIF1α degradation will require further studies, the p97 siRNA results clearly indicate a role for p97 in HIF1α degradation. Taken together, our results highlight the complexity of the substrate targeting and processing pathways that operate downstream of ubiquitin ligases and upstream of the proteasome and call attention to the need for further studies to elucidate these pathways in greater detail.
Why Choose p97 as a Proteasome Targeting Factor?
Depletion of p97 causes less accumulation of HIF1α than does inhibition of the proteasome, suggesting the possibility that p97 is involved in the degradation of only a subset of HIF1α molecules. Perhaps p97 is required to degrade only those molecules that exist in a particular assembly state, i.e. bound to HIF1β, promoter DNA, and/or general transcription machinery. Indeed, p97 is known to function as an ‘unfoldase’ of protein aggregates (Kobayashi et al., 2007), as a ‘dislocase’ of ERAD substrates (Bar-Nun, 2005) or as a ‘separase’ in Spt23/Mga2 activation (Rape et al., 2001; Shcherbik and Haines, 2007). In the absence of UBXD7, HIF1α degradation would occur without benefit of the functions provided by p97. We suggest that p97-independent degradation may be less processive, or possibly less selective for the ubiquitin-conjugated subunit of a protein complex.
K11 versus K48
As indicated above, K11 linkages of ubiquitin were unexpectedly prominent in UBA-UBX immunoprecipitates. The UBA domains of RAD23 interact with a surface of ubiquitin that includes K48 (Ryu et al., 2003) and they inhibit assembly of K48-linked chains in vitro (Ortolan et al., 2000; Raasi and Pickart, 2003). If UBA-UBX proteins employed a similar binding mode, their UBA domains would be masking K48 of ubiquitin, thereby favoring modification of alternative lysine residues such as K11. The unexpected prominence of K11-linked chains reported here could explain why these linkages were estimated to be equi-abundant with K63-linked chains in budding yeast cells (Peng et al., 2003).
Moreover, K11-linked ubiquitin chains accumulate in neurodegenerative disorders associated with protein aggregation, like Alzheimer’s (Cripps et al., 2006) or Huntington’s disease (Bennett et al., 2007). Mutations in p97 are the underlying cause for the syndrome of inclusion body myopathy with Paget’s disease of the bone and frontotemporal dementia – IBMPFD (Watts et al., 2004) and p97 colocalizes with protein aggregates in Huntington’s, Machado-Joseph, and Parkinson’s disease (Hirabayashi et al., 2001; Mizuno et al., 2003). K11 linkages can be generated by the ubiquitin ligase APC/C working in concert with the E2 enzymes Ubc4 and UbcH10 (Kirkpatrick et al., 2006). Very recently, Rape and colleagues reported that K11-linkages are required for the turnover of APC/C substrates (Jin et al., 2008). Taken together these observations suggest an unexpected connection between APC/C, p97, and human disorders rooted in defective protein homeostasis.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents and Plasmids
The following antibodies have been used: anti-Flag, anti-ubiquitin (Sigma), anti-p97 (Research Diagnostics), anti-NPL4 (Abnova), anti-UFD1, anti-CUL3 (BD Transduction Laboratories), anti-PLAP (Epitomics), anti-CUL1, anti-CUL2 (Zymed), anti-VHL (Santa Cruz Biotechnology), anti-UBR1 (courtesy of A. Varshavsky lab), anti-HIF1α (Novus), anti-UBXD7 (courtesy of Millipore), anti-UBXD8 (Imgenex), and anti-CA IX (courtesy of J. Pastorek and S. Pastorekova). MG132 was purchased from Biomol. The truncation mutants were obtained by site directed mutagenesis. STOP codons were placed at the corresponding position in the wild-type plasmid such that the Flag-UBXD7ΔUBX construct expresses UBXD7(1–400), Flag-UBA expresses UBXD7(1–62), and Flagp47ΔUBX expresses p47(1–232). See Suppl. Table 3 for the list of wild-type plasmids used in this study.
Mass Spectrometry Analysis
Mass spectrometrical sample analysis was performed as described previously (Graumann et al., 2004) using multidimensional capillary chromatography in line to a LCQ DecaXP electrospray ion trap mass spectrometer (ThermoFinnigan). Data analysis was performed using Sequest (Eng et al., 1994) and DTASelect (Tabb et al., 2002) against the IPI human database (Kersey et al., 2004) version 3.15.1. using the parameters indicated in Graumann et al. (2004).
Cell Extracts and Immunoprecipitation
For immunoprecipitation experiments, the cells were lysed in buffer A (50 mM HEPES/KOH, pH 7.5; 5 mM Mg(OAc)2; 70 mM KOAc; 0.2% Triton X-100; 10% glycerol; 0.2 mM EDTA; protease inhibitors) and incubated with anti-Flag agarose beads (Sigma) or anti-Myc sepharose beads (Covance).
Total extracts of cells treated with siRNA were prepared using buffer B (50 mM HEPES/KOH, pH 7.2; 400 mM NaCl; 1% NP-40; 0.2 mM EDTA; 10% glycerol; protease inhibitors) to enable extraction of nuclear HIF1α.
siRNA-Mediated Protein Depletion
Various siRNA oligonucleotides purchased from Dharmacon were transfected into HeLa cells using Oligofectamine (Invitrogen) and the protocol suggested by the manufacturer. The cells were lysed 48 hours after siRNA transfection.
RT-PCR
RNA was isolated with the RNeasy Mini kit (Qiagen). cDNA was synthesized using Omniscript reverse transcriptase (Qiagen) and specific primers. PCR amplification was performed using the HotStarTaq DNA polymerase (Qiagen) and specific primers chosen to yield short DNA fragments of 200–300 base pairs. The protocols were those suggested by Qiagen. Primer sequence is available upon request.
Gel Filtration
HeLa cell lysates in buffer C (50 mM HEPES/KOH, pH 7.2; 5 mM Mg(OAc)2; 70 mM KOAc; 0.2% Triton X-10; 5% glycerol; 0.2 mM EDTA; protease inhibitors) were fractionated on a Superdex 200 column (GE Healthcare). The collected fractions were concentrated by TCA precipitation prior to western blot analysis.
The molecular weight standards were Thyroglobulin (670 kDa; Bio-Rad), Apoferritin (443 kDa; Sigma), and γ-globulin (158 kDa; Bio-Rad).
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Kleiger and T. Chou for help with gel filtration, J. Quimby for help with DNA cloning, S. Schwarz for advice on cell culture, R. Oania for technical assistance, and S. Buonomo for experimental protocols. We also thank T. Nagase, G. Warren, H. Katoh, S. Buonomo for plasmids and Millipore, J. Pastorek, S. Pastorekova, Z. Xia for antibodies. We are grateful to A. Varshavsky and members of the Deshaies’ lab for critical reading of the manuscript. G.A. was supported by DRG-1745-02 of the Damon Runyon Cancer Research Foundation and the Howard Hughes Medical Institute (HHMI). R.J.D. is an HHMI Investigator and this work was funded by HHMI. R.J.D. is a founder, shareholder, and consultant for Proteolix.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bar-Nun S. The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast. Curr Top Microbiol Immunol. 2005;300:95–125. doi: 10.1007/3-540-28007-3_5. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bruderer RM, Brasseur C, Meyer HH. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J Biol Chem. 2004;279:49609–49616. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- Carim-Todd L, Escarceller M, Estivill X, Sumoy L. Identification and characterization of UBXD1, a novel UBX domain-containing gene on human chromosome 19p13, and its mouse ortholog. Biochim Biophys Acta. 2001;1517:298–301. doi: 10.1016/s0167-4781(00)00248-7. [DOI] [PubMed] [Google Scholar]
- Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J Biol Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- Doolman R, Leichner GS, Avner R, Roitelman J. Ubiquitin is conjugated by membrane ubiquitin ligase to three sites, including the N terminus, in transmembrane region of mammalian 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for sterol-regulated enzyme degradation. J Biol Chem. 2004;279:38184–38193. doi: 10.1074/jbc.M405935200. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fu X, Ng C, Feng D, Liang C. Cdc48p is required for the cell cycle commitment point at Start via degradation of the G1-CDK inhibitor Far1p. J Cell Biol. 2003;163:21–26. doi: 10.1083/jcb.200307025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. Embo J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR, 3rd, Wold BJ, Deshaies RJ. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol Cell Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi M, Inoue K, Tanaka K, Nakadate K, Ohsawa Y, Kamei Y, Popiel AH, Sinohara A, Iwamatsu A, Kimura Y, et al. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8:977–984. doi: 10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kaelin WG., Jr The von Hippel-Lindau tumor suppressor protein. Curr Opin Genet Dev. 2001;11:27–34. doi: 10.1016/s0959-437x(00)00152-0. [DOI] [PubMed] [Google Scholar]
- Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G, Krause S, Hoppe T. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluz S, Kaluzova M, Stanbridge EJ. Does inhibition of degradation of hypoxia-inducible factor (HIF) alpha always lead to activation of HIF? lessons learnt from the effect of proteasomal inhibition on HIF activity. J Cell Biochem. 2007 doi: 10.1002/jcb.21644. [DOI] [PubMed] [Google Scholar]
- Katoh H, Harada A, Mori K, Negishi M. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol. 2002;22:2952–2964. doi: 10.1128/MCB.22.9.2952-2964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Manno A, Kakizuka A. Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells. 2007;12:889–901. doi: 10.1111/j.1365-2443.2007.01099.x. [DOI] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Lowe ED, Hasan N, Trempe JF, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. Embo J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. Embo J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Hori S, Kakizuka A, Okamoto K. Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci Lett. 2003;343:77–80. doi: 10.1016/s0304-3940(03)00280-5. [DOI] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Parker CE, Mocanu V, Warren MR, Greer SF, Borchers CH. Mass spectrometric determination of protein ubiquitination. Methods Mol Biol. 2005;301:153–173. doi: 10.1385/1-59259-895-1:153. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Peters JM, Walsh MJ, Franke WW. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. Embo J. 1990;9:1757–1767. doi: 10.1002/j.1460-2075.1990.tb08300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Targeting of substrates to the 26S proteasome. Faseb J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Pye VE, Dreveny I, Briggs LC, Sands C, Beuron F, Zhang X, Freemont PS. Going through the motions: the ATPase cycle of p97. J Struct Biol. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay MQ, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Richly H, Rumpf S, Buchberger A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 2004;5:818–824. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol Cell. 2007;25:385–397. doi: 10.1016/j.molcel.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, Beuron F, Zhang X, Freemont P, Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- Yamabe Y, Ichikawa K, Sugawara K, Imamura O, Shimamoto A, Suzuki N, Tokutake Y, Goto M, Sugawara M, Furuichi Y. Cloning and characterization of Rep-8 (D8S2298E) in the human chromosome 8p11.2-p12. Genomics. 1997;39:198–204. doi: 10.1006/geno.1996.4480. [DOI] [PubMed] [Google Scholar]
- Ye Y. Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung HO, Kloppsteck P, Niwa H, Isaacson RL, Matthews S, Zhang X, Freemont PS. Insights into adaptor binding to the AAA protein p97. Biochem Soc Trans. 2008;36:62–67. doi: 10.1042/BST0360062. [DOI] [PubMed] [Google Scholar]
- Yuan X, Shaw A, Zhang X, Kondo H, Lally J, Freemont PS, Matthews S. Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J Mol Biol. 2001;311:255–263. doi: 10.1006/jmbi.2001.4864. [DOI] [PubMed] [Google Scholar]
- Zhong X, Shen Y, Ballar P, Apostolou A, Agami R, Fang S. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J Biol Chem. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.