Abstract
Eukaryotic gene expression is a complex, multistep process that needs to be executed with high fidelity and two general methods help achieve the overall accuracy of this process. Maximizing accuracy in each step in gene expression increases the fraction of correct mRNAs made. Fidelity is further improved by mRNA surveillance mechanisms that degrade incorrect or aberrant mRNAs that are made when a step is not perfectly executed[MP2]. Here, we review how cytoplasmic mRNA surveillance mechanisms selectively recognize and degrade a surprisingly wide variety of aberrant mRNAs that are exported from the nucleus into the cytoplasm.
1. Introduction
One way to increase the accuracy of gene expression is to optimize each step to ensure that only the correct gene product is made. However, despite the high fidelity of RNA polymerase, splicing etc., abnormal transcripts are still created (e.g. [1–6]). For example, transcription can generate RNAs that are non-coding [1, 5, 6], mistakes during splicing can result in transcripts with retained introns and/or skipped exons [2, 3], and mistakes during polyadenylation can result in mRNAs that have a poly(A) tail added within the coding region [4, 7] or have an abnormally long 3’UTR . Therefore, to further increase the fidelity of gene expression, mRNA surveillance pathways have evolved to selectively recognize and degrade abnormal transcripts. Messenger RNA surveillance pathways are responsible for decreasing the production of the aberrant and dominant-negative proteins encoded by abnormal mRNAs [8, 9]. Furthermore, these pathways are diverse and can occur either within the nucleus or cytoplasm. In this review, we will highlight emerging roles of the cytoplasmic degradation machinery in yeast mRNA surveillance pathways. Nonsense-mediated mRNA degradation (NMD) pathway and nuclear RNA surveillance will not be discussed in detail here because several good reviews have recently been written on these two topics, including some in this volume [6, 10–14].
1.1 Pathways of normal mRNA degradation
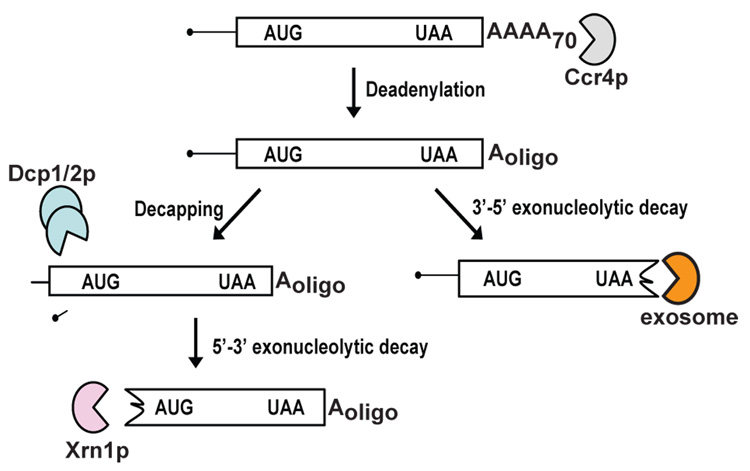
Studies using the yeast Saccharomyces cerevisiae have identified two major cytoplasmic degradation pathways for normal mRNAs (Figure 1). All of the enzymes required for these two pathways are conserved in other eukaryotes, suggesting that the pathways of mRNA decay are also conserved. Messenger RNA degradation is initiated by gradual removal of the poly(A) tail [15–18], a process which is normally carried out by the 3’ exonuclease Ccr4p, but also by Pan2p, at a slower rate [17, 19–21]. In the absence of both Ccr4p and Pan2p, poly(A) tails are stable, suggesting that there are no other enzymes that can substitute for this function [17]. Removal of the poly(A) tail triggers two mRNA degradation pathways. In the first pathway, the 5’ cap is removed by the Dcp1p/Dcp2p[MP3] complex [22–29]. Decapping of the mRNA triggers degradation of the transcript from the 5’end by Xrn1p, a 5’exoribonuclease [24, 30, 31]. In the second pathway, the body of the transcript is degraded from the 3’end by a multi-subunit 3’exoribonuclease termed the exosome [32–34]. The exosome has both nuclear and cytoplasmic functions, both requiring additional factors [14, 35]. For example, Ski2p, Ski3p, Ski7p and Ski8p are required for all cytoplasmic exosome functions [36–39]. As detailed below, mRNA surveillance utilizes the same enzymes for mRNA decay, but variations of the general mRNA decay pathway result in increased degradation rates of aberrant mRNAs.
Figure 1.
Pathways of normal mRNA degradation. Normal mRNAs are degraded by two general mRNA degradation pathways. In the first pathway, following removal of the poly(A) tail, mRNAs are decapped by the Dcp1/2p complex and degraded by Xrn1p. In a second pathway, deadenylation is followed by degradation of the body of the mRNA from the 3’end by the exosome.
2. Nonstop mRNA decay
2.1 Nonstop mRNA decay mechanism
Nonstop mRNA decay selectively degrades transcripts that lack all in-frame termination codons [4, 40]. While nonstop mRNA decay is conserved in other eukaryotes, the mechanism has been primarily studied in yeast. In the current model for nonstop mRNA decay, the translating ribosome translates to the end of the poly(A) tail of a nonstop mRNA and stalls (Figure 2). The stalled ribosome is hypothesized to be recognized by Ski7p, possibly because of the absence of a codon in the A-site of the ribosome. Recognition by Ski7p recruits the exosome to the nonstop mRNA, resulting in degradation of the transcript.
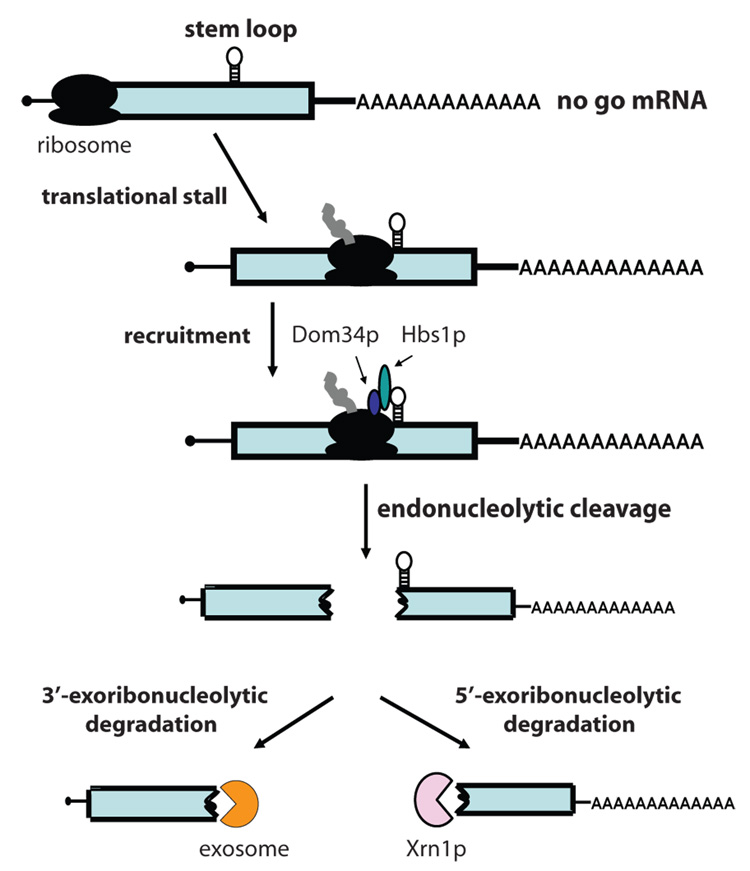
Figure 2.
Model of nonstop mRNA degradation in yeast. Transcripts that lack all in-frame termination codons (nonstop mRNAs) cause the translating ribosome to stall at the 3’end of the message. The stalled ribosome is recognized by the C-terminal domain of Ski7p. The N-terminal domain of Ski7p recruits the exosome, providing a physical link between the nonstop mRNA and the exosome, which facilitates rapid degradation of the transcript. Nonstop mRNAs may also be translationally repressed, or the encoded protein may be targeted to the proteasome; however, the significance and mechanisms of these two aspects are not yet clear.
Several lines of evidence support the current model. First, it is likely that the translating ribosome stalls at the end of a nonstop transcript. Nonstop mRNAs remain physically associated with ribosomes when used in in vitro translation reactions, while ribosomes dissociate from mRNAs that contain a stop codon [41]. Stalled ribosome-mRNA complexes have been useful tools in studying the sorting of nascent proteins, and are surprisingly stable, as they can be purified by sucrose gradient centrifugation or gel filtration [41–43]. Thus, unlike DNA or RNA polymerases, ribosomes do not simply dissociate when they reach the end of a template, which implies that a specific factor (perhaps Ski7p) is necessary for disassembly of the ribosome. Similarly, the bacterial ribosome needs trans-acting factors to dissociate when it reaches the end of a nonstop mRNA (i.e. tmRNA and SmpB; [44]).
Second, nonstop mRNA decay requires active translation. Nonstop mRNAs are stabilized in wild type yeast cells treated with a translational inhibitor, and in mutant cells depleted of charged tRNAs [4]. More importantly, translation of the nonstop mRNA itself is needed: nonstop mRNA decay is prevented when a stop codon is inserted close to the poly(A) tail [4], and when a stable structure in the 5’UTR prevents its translation (unpublished data).
Third, a nonstop PGK1pGreporter mRNA was stabilized in yeast strains lacking cytoplasmic exosome function, suggesting that the exosome degrades nonstop mRNAs [40]. In contrast, mutations inactivating the decapping enzyme or the 5’ to 3’ exoribonuclease, Xrn1p, have large effects on the degradation of normal mRNAs [22, 24, 30, 31], but do not detectably affect the stability of the nonstop PGK1pG mRNA [4].
Fourth, based on sequence similarity, Ski7p is a likely candidate for recognizing the stalled ribosome at the end of a nonstop mRNA. The C-terminal domain of Ski7p is homologous to translation factors eEF1A and eRF3 [36, 45]. Because eEF1A interacts with the ribosome when the A-site contains a sense codon and eRF3 interacts with the ribosome when the A-site contains a stop codon, it is proposed that the homologous domain of Ski7p interacts with the ribosome when the A-site is empty. This hypothesis is supported by the observation that deletion of the C-terminal domain of Ski7p inactivates the nonstop mRNA decay pathway without affecting other exosome functions [40].
Fifth, the N-terminal domain of Ski7p interacts with the exosome, and with additional exosome cofactors [36, 46]. This domain is required for both nonstop mRNA decay and other cytoplasmic exosome functions [40]. Importantly, a point mutation in the exosome that disrupts the interaction with Ski7p also blocks nonstop mRNA decay [40], confirming that the interaction between Ski7p and the exosome is functionally important.
Interestingly, two observations suggest that degradation of nonstop mRNAs does not involve a distinct deadenylation phase by dedicated deadenylases such as Ccr4p or Pan2p. Deletion of the major deadenylase stabilizes normal mRNAs [17], but has no effect on the stability of nonstop mRNAs [4]. In addition, only fully adenylated nonstop mRNAs were detected in wild type cells, and fully adenylated mRNAs are stabilized in SKI7 mutant cells [40]. The conclusion that the exosome degrades the poly(A) tail on nonstop mRNAs is surprising since the exosome is incapable of degrading the poly(A) tail on normal mRNAs, as shown by a complete absence of deadenylation in a ccr4Δ pan2Δ double mutant [17].
Although the mechanism of nonstop mRNA decay has primarily been studied in yeast, some aspects appear to be conserved. Multiple observations suggest that human nonstop mRNAs are less stable than their wild type counterparts. First, the mRNA level of a nonstop version of mouse β-glucoronidase introduced into HeLa cells is two-fold lower than the level for the corresponding normal mRNA [4], while a nonstop version of α-globin was shown to be rapidly degraded (α-NS, t1/2=2h compared to 11.5h for WT) [47]. Second, at least some nonstop mRNAs are increased in abundance after inhibition of translation [4, 48]. Third, an α-globin reporter gene lacking all in-frame stop codons is degraded without any detectable deadenylation intermediates [47]. Thus, the dependence on translation and the independence of a distinct deadenylation phase appear to be conserved aspects of nonstop mRNA decay.
According to the model for nonstop mRNA decay (Figure 2), at least one round of translation is needed to target a nonstop mRNA for degradation by the exosome, which raises two important, partially answered questions. The first question is whether the translation rate of a nonstop mRNA is the same as for a control mRNA. Several studies have hinted that translation of nonstop mRNAs is inefficient, but a detailed molecular mechanism has not been described [49–51]. Second, the fate of the protein produced from a nonstop mRNA is also unclear. Reports from two groups indicate that nonstop proteins may be targeted for proteolysis by the proteasome. Inhibitors of the proteasome increased the levels of the protein encoded by nonstop mRNAs [50], and a screen to identify trans-acting factors in nonstop mRNA decay uncovered three mutations that inactivate the proteasome [52]. Interestingly, the effects of the proteasome and Ski7p are additive, suggesting that recognition of the stalled ribosome by Ski7p is not required for the rapid degradation of the encoded protein [52]. On the contrary, in bacterial cells recognition of the stalled ribosome by tmRNA is an integral part of targeting the encoded protein for rapid degradation [44]. One possibility is that translation of the poly(A) tail into a poly-lysine tail on the protein is the signal that targets the protein for rapid decay. Consistent with this hypothesis, a poly-lysine tail encoded at the end of an ORF can significantly reduce the amount of the protein produced [50]. However, it is not entirely clear whether this effect is specific for poly-lysine, as it seems likely that a random sequence added to the C-terminus of a protein could also destabilize the protein. Important in this respect, most of the nonstop reporters that have been studied were made by a point mutation in the stop codon. Thus, the proteins contained the amino acid sequence that is encoded in what was the 3’UTR, as well as a poly-lysine tail. Clearly, more work is needed to determine the features of nonstop mRNAs that target the encoded protein to the proteasome, and whether rapid proteolysis is a specific part of the nonstop mRNA pathway, or reflects non-specific degradation of proteins with improperly folded C-terminal extensions.
2.2. Premature polyadenylation generates nonstop mRNAs
One method of generating a nonstop transcript is by premature polyadenylation upstream of the stop codon [4, 7]. 1.2% of random yeast cDNA clones and 0.7% of human cDNAs were prematurely polyadenylated [4, 53]. Since nonstop mRNAs are less stable than normal mRNAs, this suggests that perhaps 5% to 10% of the time, the cleavage and polyadenylation machinery acts prematurely [4]. Prematurely polyadenylated mRNAs are not readily detectable by standard methods such as Northern blotting because of the instability of these transcripts, and because each individual premature polyadenylation site is used infrequently. The recent observation that truncated poly(A)+ mRNAs accumulate when the Arabidopsis exosome is inactivated is consistent with the hypothesis that exosome-mediated decay of prematurely polyadenylated mRNAs is a conserved pathway [54].
Premature polyadenylation can also serve as a method of gene-specific regulation. In S. cerevisiae, 0.8% of all open reading frames contain a cryptic polyadenylation site upstream of the normal termination codon [4]. Cryptic sites could be favored over the normal poly(A) site under various conditions to down-regulate the expression of the normal mRNA [55, 56]. This implies that premature polyadenylation, coupled with nonstop mRNA decay, could serve to down-regulate many genes.
One example of how premature polyadenylation is used to modulate gene expression occurs in the RNA14 gene in yeast. RNA14 encodes a protein required for cleavage and polyadenylation [57]. If Rna14p is abundant, the cleavage and polyadenylation machinery efficiently recognizes a polyadenylation site within the RNA14 ORF, thus down-regulating further Rna14p synthesis [58] . Similar premature polyadenylation mechanisms operate in other organisms in genes encoding cleavage and polyadenylation factors [59]. This suggests that premature polyadenylation of such genes is a common feed-back regulation mechanism. In some organisms, premature polyadenylation within an intron generates an mRNA with a stop codon in the partially retained intron. In contrast, since the premature polyadenylation occurs within a yeast ORF, a nonstop mRNA is generated. Therefore, nonstop mRNA decay likely increases the effectiveness of this feed-back regulation loop by reducing the accumulation of truncated proteins.
2.3. Nonstop mRNA decay and human disease
It is estimated that ~30% of all genetic disorders result from the generation of a transcript that is targeted for NMD. Although nonstop mRNA decay has not been extensively studied in human cells, some data indicate that nonstop mutations contribute to disease. For example, mutations in the stop codons of APRT and GPR54 cause 2,8-dihydroxyadenine urolithiasis [60] and idiopathic hypogonadotrophic hypogonadism [61], respectively. In both cases, the normal stop codon is mutated, which results in nonstop mRNAs that are reduced in abundance when compared to unaffected human subjects. Importantly, the mutant GPR54 protein is partially functional after being overexpressed in mammalian cell lines [61], suggesting that drugs that block nonstop mRNA decay could provide effective treatment. In other disease alleles, nonstop mRNAs are generated by mutations within the coding region. In nonstop Factor X and α-GalA, small deletions within the coding region move the normal stop codon out of frame, resulting in hemophilia [62] and Fabry disease [63], respectively. A mutation of a splice site in RPS19 generates a nonstop mRNA by skipping the exon that contains the stop codon, which causes Diamond Blackfan anemia [48]. These cases demonstrate that nonstop mRNAs can be generated by a variety of mutations. Importantly, disease appears to be caused by a reduction in the mRNA and protein level, rather than an altered protein, and thus nonstop mRNA decay likely contributes to disease.
3. No-go mRNA decay
Pseudoknots, rare codons, or stem-loops within the open reading frame cause the translating ribosome to stall. Transcripts with stalled ribosomes are degraded by the no-go mRNA surveillance pathway. In the current model for no-go mRNA decay, a stalled ribosome within the coding region of the no-go transcript is thought to recruit Dom34p and Hbs1p (Figure 3;[64]). Dom34p and Hbs1p are homologous to eRF1 and eRF3, respectively, suggesting that they may function to recognize stalled ribosomes. Interaction of Dom34p and Hbs1p with the stalled ribosome is thought to trigger endonucleolytic cleavage of the no-go mRNA. The resulting 5’ cleavage product is degraded by the cytoplasmic exosome, while the 3’ cleavage product is degraded by the 5’ exoribonuclease, Xrn1p. Consistent with this model, an xrn1Δ strain accumulates the expected 3’ cleavage product, but deleting DOM34 from this strain prevents this accumulation [64]. Likewise, a strain lacking cytoplasmic exosome activity accumulates the expected 5’ cleavage product, and deleting DOM34 from this strain prevents this accumulation [64]. Similarly, mutants in HBS1 show drastically reduced levels of no-go mRNA decay intermediates, but hbs1Δ is not as effective as dom34Δ, implying that Hbs1p may not play as great a role as Dom34p [64]. Consistent with this observation, recent structural work suggests that Dom34p is responsible for endonucleolytic cleavage of no-go mRNAs [65]. The X-ray crystal structure of archaeal and yeast Dom34p revealed that it is composed of three domains [65, 66]. While domains two and three were similar to eRF1, domain one shared no structural similarity with eRF1, but instead is similar to the Sm-[MP4]fold [65, 66]. Interestingly, the key residues of eRF1 that recognize the stop codon are in domain one, and the absence of this domain readily explains why Dom34p does not recognize ribosomes with a stop codon in the A-site. Instead, domain one of Dom34p contains four conserved acidic amino acid residues, reminiscent of some nucleases. The observation that Dom34p copurifies with endoribonuclease activity that is dependent on one of the acidic residues [65] suggests that Dom34p may be the endonuclease in no-go mRNA decay. However, it has not yet been shown that no-go decay is abolished if the putative active site of Dom34p is mutated.
Figure 3.
Model for no-go mRNA decay in yeast. Stem-loop structures, pseudoknots, and rare codons cause the translating ribosome to stall within the coding region of an mRNA. The stalled ribosome is recognized by the Hbs1p/Dom34p complex. Subsequently, the mRNA is cleaved endonucleolytically, generating 5’ and 3’ fragments that are degraded by the exosome and Xrn1p, respectively.
Ribosomes can also be stalled because of aberrancies within the ribosome. Mutations in ribosomal RNAs result in the assembly of nonfunctional ribosomes that are preferentially degraded through the nonfunctional ribosome decay pathway (NRD) [67]. Thus, in the case of no-go decay, a stalled ribosome leads to mRNA decay, while in NRD, a stalled ribosome leads to rRNA decay. Yet in other cases of ribosomal stalling, yeast mRNAs are stabilized. (e.g. treatment with cycloheximide or depletion of charged tRNA in a cca1-1 strain [68, 69]). Thus, an important question yet to be addressed is how the cellular machinery distinguishes between whether the mRNA or rRNA in a stalled complex is defective, or whether in both cases both mRNA and rRNA are degraded.
An interesting aspect of the relationship between nonstop and no-go decay is that Ski7p and Hbs1p are a pair of duplicated genes in yeast [70], whereas most other eukaryotes have only one homolog. This raises the question whether this single homolog in other eukaryotes functions in nonstop decay, no-go decay, or both. The single Ski7p/Hbs1p homolog from the related yeast Saccharomyces kluyveri could complement the growth phenotypes of an HBS1 mutant and a SKI7 mutant, suggesting that it can do both nonstop mRNA decay and no-go mRNA decay [71]. The human homolog of Hbs1p/Ski7p (eRFS; [72]) is unable to complement either an HBS1 mutant or a SKI7 mutant (unpublished data). The most likely explanation is that human eRFS is nonfunctional when expressed in yeast, leaving the question whether eRFS can function in nonstop and/or no-go decay unanswered. The evolutionary history of Hbs1p and Ski7p suggests the possibility that in most eukaryotes the single Ski7p/Hbs1p protein recognizes stalled ribosomes within the coding region and at the end of an mRNA. This also suggests that in other eukaryotes, nonstop and no-go mRNA decay may be a mixture of endonucleolytic decay and exosome-mediated decay.
4. Antiviral activities of mRNA surveillance
Most eukaryotic cells are invaded by RNA viruses and have evolved various innate immunity mechanisms to specifically recognize and degrade viral RNAs. Innate immune systems recognize general features of the viral mRNA, such as dsRNA, RNA with a triphosphate 5’ end, or mRNAs with unadenylated 3’ ends. Plants [73], worms [74], flies [75, 76], fungi [77], and mammals [78] have evolved to selectively recognize viral RNAs as foreign and target them to the RNAi machinery. In humans, several proteins recognize viral RNA. RIG-I recognizes the 5’ triphosphate-containing RNAs found on some uncapped viral RNAs [79, 80], while dsRNA is recognized by MDA5 [81], PKR [82], TLR3 [83] and 2’5’ A synthetase [84]. None of these antiviral activities have obvious homologs in yeast, but the 5’ and 3’ cytoplasmic mRNA decay machineries exhibit antiviral functions by degrading uncapped and unadenylated viral RNAs [39, 85, 86]. This process is best characterized in yeast but may also occur in other eukaryotes.
The majority of studies of antiviral mechanisms in yeast utilized the yeast dsRNA L-A virus and its toxin-producing satellite M1 virus to identify host antiviral factors that control levels of the M1 toxin [87]. These studies identified eight “superkiller” (or ski) genes which are involved in cytoplasmic mRNA decay [88, 89]. The SKI gene products have also been implicated in controlling viral RNA levels of the positive-strand, unadenylated Brome Mosaic Virus, suggesting that this is a general antiviral mechanism [90].
The Ski2-8 gene products are each required for exosome-mediated mRNA decay (Figure 1), suggesting the possibility that viral mRNAs are especially susceptible to exosome-mediated mRNA decay. This was confirmed by showing that unadenylated mRNAs are rapidly degraded by the cytoplasmic exosome [86]. In addition, the Ski2-8 gene products may also control translation of unadenylated mRNAs. This possibility is based on a series of studies that showed that levels of protein translated from unadenylated transcripts were increased in ski- strains when compared to wild type strains without a detectable difference in mRNA stability [45, 85, 91–93]. One caveat of these experiments is that they all used electroporation to introduce adenylated or unadenylated mRNAs and, consequently, may not mimic what normally occurs in vivo. Indeed, it was subsequently shown that unadenylated mRNAs that were not susceptible to 3’ to 5’ decay were equally translated when electroporated into wild type or ski mutant cells [39]. Thus, while it is clear that the SKI gene products control mRNA stability and preferentially degrade unadenylated mRNAs, it is not entirely clear whether they also control their translation.
Currently, it is not known what targets unadenylated transcripts for rapid decay by the cytoplasmic exosome. One simple explanation would be that the poly(A) binding protein, Pab1p, normally protects mRNAs from exosome-mediated mRNA decay. However, in a pab1Δ strain, normal transcripts do not appear to be rapidly degraded by the cytoplasmic exosome [94]. The main function of the preferential decay of unadenylated mRNAs is not entirely clear. One possibility is that unadenylated mRNAs are normally produced by a mistake in gene expression, and the preferential decay of unadenylated mRNAs has evolved as an mRNA surveillance pathway that has fortuitous antiviral activity. A second possibility is that the function of this pathway is purely antiviral, and that an uninfected cell does not contain any substrates for this pathway. Yet another possibility is that the main function of exosome-mediated mRNA decay of unadenylated mRNAs is to degrade cleavage products of endoribonucleases, such as those generated during RNAi and no-go mRNA decay [64, 95].
5. Cytoplasmic decay of Splice-defective mRNAs
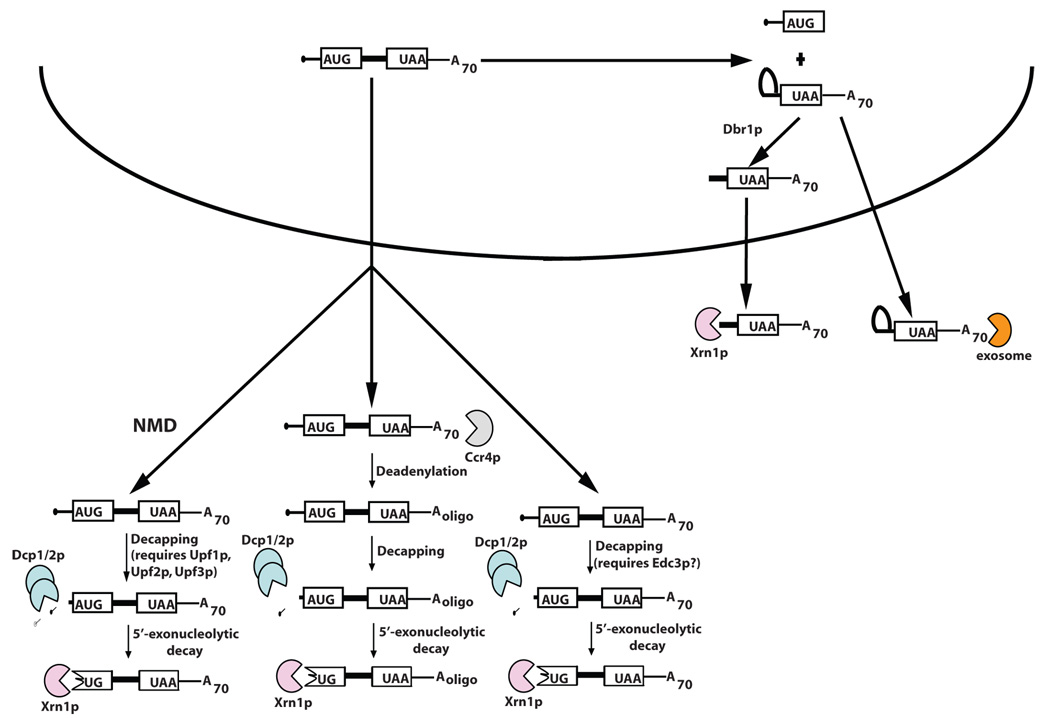
Incorrectly processed pre-mRNAs can be exported to the cytoplasm [96–98], where they can be translated, and ultimately degraded. Introns in pre-mRNAs typically contain premature termination codons that can target these pre-mRNAs to the NMD pathway [99]; however, not all exported pre-mRNAs are degraded by NMD. Reporter pre-mRNAs with mutations in the 5’ splice site or branchpoint site are deadenylated, decapped, and degraded from the 5’ ends [100]. Interestingly, this decay is independent of the NMD pathway, despite the presence of multiple premature termination codons within the intron. Other studies also implicate the 5’ cytoplasmic mRNA decay pathway in the down-regulation of transcripts with retained introns. For example, in the absence of Mer1p, mRNAs from the MER2, MER3, and AMA1 genes are incorrectly processed, exported to the cytoplasm, and degraded by the 5’ decay machinery. Despite retained introns containing premature termination codons in all three transcripts, only MER2 and MER3 were substrates for the NMD pathway [101]. Export of pre-mRNA is also a feature of the feedback regulation of YRA1 expression [102]. When levels of Yra1p reach a certain threshold, the YRA1 pre-mRNA is exported, and degraded by the 5’ mRNA decay machinery. Like the examples mentioned above, the YRA1 pre-mRNA contains a premature termination codon but is not degraded by the NMD pathway. Instead, it first undergoes Edc3p-mediated activation of decapping, followed by 5’ decay by Xrn1p [103]. It is not yet known if Edc3p is involved in degradation of only YRA1 pre-mRNA, or of a wider variety of exported pre-mRNAs[MP5]. If the latter is true, this could define a new role for Edc3p in the cell and begin to describe how some pre-mRNAs are recognized and degraded in the cytoplasm. Cumulatively, these data show that exported pre-mRNAs are not always degraded by the NMD pathway, but can be degraded by other pathways that may be dependent upon Edc3p (Figure 4).
Figure 4.
Unspliced and partially spliced mRNAs are exported and degraded in the cytoplasm. Pre-mRNAs can be exported into the cytoplasm where they are degraded either by nonsense-mediated decay, deadenylation-dependent decapping, or deadenylation-independent decapping. A 3’ splice-defective pre-mRNA containing a lariat structure at its 5’end is debranched in the nucleus by Dbr1p and exported to the cytoplasm where it is degraded by Xrn1p. Alternately, lariat structures can be exported to the cytoplasm and degraded by the exosome.
Two studies show that pre-mRNAs that can not perform the second step of splicing, and therefore contain lariat introns, can also be exported to the cytoplasm for decay. Lariat intermediates from a reporter mRNA with a mutant 3’ splice site are debranched by the Dbr1p debranching enzyme and then predominantly degraded from their 5’ ends by cytoplasmic Xrn1p [100]. In a second study, cleavage sites for the Rnt1p endonuclease were identified in a subset of yeast introns. Lariat intermediates from these genes are first cleaved by Rnt1p and then degraded in the cytoplasm by Xrn1p [104].These two studies show that lariat intermediates that fail to complete splicing are subject to cytoplasmic mRNA decay from their 5’ ends by Xrn1p. Alternatively, such incorrectly processed mRNAs could also be targeted to the cytoplasmic exosome (Figure 4) [100,104].
6. Cytoplasmic decay of transcribed intergenic sequences
In recent years it has become evident that large parts of mammalian genomes are transcribed, however the functions of many unconventional transcripts are unknown [105, 106]. In yeast, similar RNAs have been named cryptic unstable transcripts (CUTs), which are capped and polyadenylated yet unstable in vivo [1, 107]. The degradation of CUTs is thought to occur by polyadenylation by the poly(A) polymerase Trf4p and subsequent degradation by the nuclear exosome [1]. However, recent evidence suggests that some CUTs are exported out of the nucleus and degraded by the cytoplasmic mRNA decay machinery [108].
The SRG1 RNA is one example of a non-coding regulatory RNA that is degraded in the cytoplasm. Transcription of SRG1 produces three transcripts of different sizes, all of which are polyadenylated. The two smaller transcripts are exported to the cytoplasm, decapped, and degraded from their 5’ ends by Xrn1p. Similar to some of the pre-mRNAs mentioned above, these transcripts are degraded independent of deadenylation and of the NMD pathway. The longer transcript is also exported into the cytoplasm but is degraded by the NMD pathway [108].
Microarray analysis of intergenic sequences in various mRNA decay mutants provided a global idea of how CUTs are degraded in yeast [108]. Here, the majority of CUTs tested accumulated in cytoplasmic mRNA decay mutants. The majority showed higher transcript levels in decapping and cytoplasmic 5’ mRNA decay mutants, with a subset of these affected by mutants in the NMD machinery. In addition, levels of some CUTs were increased in nuclear and cytoplasmic exosome mutant strains. These data suggest that some CUTs can be degraded in the nucleus, but that many unstable transcripts are exported to the cytoplasm where they are degraded by the cytoplasmic mRNA decay machinery. Interestingly, cytoplasmic degradation of many CUTs is independent of both deadenylation and NMD, suggesting a yet to be discovered mRNA decay pathway for these transcripts.
7. Conclusions
Previously, it was believed that the majority of mutant transcripts were retained and degraded in the nucleus. However, it is becoming apparent that a significant fraction of aberrant transcripts are exported to the cytoplasm where they are degraded by various surveillance pathways. Normal, and most abnormal, transcripts are degraded by the same RNases, although at very different rates: normal mRNAs are generally relatively stable, while aberrant mRNAs are rapidly degraded, in part, because they do not need to be deadenylated before they are degraded. This suggests that factors exist to provide the mRNA degradation machinery with specificity for aberrant mRNAs. Some of these factors have been identified: for example, Upf1p, Upf2p and Upf3p function to recognize mRNAs with early stop codons [109–112], Ski7p functions to recognize mRNAs without stop codons [40], and Dom34p and Hbs1p function to recognize mRNAs with stalled ribosomes [64]. For some factors, (i.e. Ski7p, Hbs1p and Dom34p), hypotheses have been formulated that explain how they recognize aberrant mRNAs and how they target them for decay. However, the molecular roles of other factors (i.e. Upf1-3 and Edc3p) are still unclear. Once the functions of these proteins are uncovered, the method by which aberrant mRNAs are distinguished from normal mRNAs in a cell should be uncovered. Furthermore, the intricacies of how aberrant mRNAs are targeted to degradative enzymes should provide insight into how unstable normal mRNAs can be targeted for rapid decay.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 2.He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci U S A. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 6.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 7.Sparks KA, Dieckmann CL. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cali BM, Anderson P. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol Gen Genet. 1998;260:176–184. doi: 10.1007/s004380050883. [DOI] [PubMed] [Google Scholar]
- 9.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 10.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 12.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 13.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 15.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 16.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 17.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 18.Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boeck R, Tarun S, Jr, Rieger M, Deardorff JA, Muller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Chiang YC, Denis CL. CCR4, a 3'-5' poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 23.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 24.Hsu CL, Stevens A. Yeast cells lacking 5'-->3' exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5' cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 31.Larimer FW, Hsu CL, Maupin MK, Stevens A. Characterization of the XRN1 gene encoding a 5'-->3'exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 32.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs Anderson JS, Parker RP. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: conserved eukaryotic RNA processing complex containing multiple 3'-->5'exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 35.Lorentzen E, Conti E. The exosome and the proteasome: nano-compartments for degradation. Cell. 2006;125:651–654. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3'-to-5' mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p)and Ski7p proteins in 3'-to-5' degradation of mRNA. Mol Cell Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown JT, Johnson AW. A cis-acting element known to block 3' mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA. 2001;7:1566–1577. [PMC free article] [PubMed] [Google Scholar]
- 40.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 41.Perara E, Rothman RE, Lingappa VR. Uncoupling translocation from translation: implications for transport of proteins across membranes. Science. 1986;232:348–352. doi: 10.1126/science.3961485. [DOI] [PubMed] [Google Scholar]
- 42.Wilson C, Connolly T, Morrison T, Gilmore R. Integration of membrane proteins into the endoplasmic reticulum requires GTP. J Cell Biol. 1988;107:69–77. doi: 10.1083/jcb.107.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilmore R, Collins P, Johnson J, Kellaris K, Rapiejko P. Transcription of full-length and truncated mRNA transcripts to study protein translocation across the endoplasmic reticulum. Methods Cell Biol. 1991;34:223–239. doi: 10.1016/s0091-679x(08)61683-0. [DOI] [PubMed] [Google Scholar]
- 44.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 45.Benard L, Carroll K, Valle RC, Masison DC, Wickner RB. The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae. J Virol. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Lewis MS, Johnson AW. Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA. 2005;11:1291–1302. doi: 10.1261/rna.2060405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong J, Liebhaber SA. A cell type-restricted mRNA surveillance pathway triggered by ribosome extension into the 3' untranslated region. Nat Struct Mol Biol. 2007;14:670–676. doi: 10.1038/nsmb1256. [DOI] [PubMed] [Google Scholar]
- 48.Chatr-Aryamontri A, Angelini M, Garelli E, Tchernia G, Ramenghi U, Dianzani I, Loreni F. Nonsense-mediated and nonstop decay of ribosomal protein S19 mRNA in Diamond-Blackfan anemia. Hum Mutat. 2004;24:526–533. doi: 10.1002/humu.20117. [DOI] [PubMed] [Google Scholar]
- 49.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3'-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A)tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akimitsu N, Tanaka J, Pelletier J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 2007;26:2327–2338. doi: 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177:773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3'-end-processing sequences in diverse species. Proc Natl Acad Sci U S A. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, Alonso J, Brukhin V, Grossniklaus U, Ecker JR, Belostotsky DA. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 55.Mayer SA, Dieckmann CL. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3'-end formation. Mol Cell Biol. 1989;9:4161–4169. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayer SA, Dieckmann CL. Yeast CBP1 mRNA 3' end formation is regulated during the induction of mitochondrial function. Mol Cell Biol. 1991;11:813–821. doi: 10.1128/mcb.11.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minvielle-Sebastia L, Preker PJ, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3'-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 58.Mandart E. Effects of mutations in the Saccharomyces cerevisiae RNA14 gene on the abundance and polyadenylation of its transcripts. Mol Gen Genet. 1998;258:16–25. doi: 10.1007/s004380050702. [DOI] [PubMed] [Google Scholar]
- 59.Pan Z, Zhang H, Hague LK, Lee JY, Lutz CS, Tian B. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. Gene. 2006;366:325–334. doi: 10.1016/j.gene.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 60.Taniguchi A, Hakoda M, Yamanaka H, Terai C, Hikiji K, Kawaguchi R, Konishi N, Kashiwazaki S, Kamatani N. A germline mutation abolishing the original stop codon of the human adenine phosphoribosyltransferase (APRT) gene leads to complete loss of the enzyme protein. Hum Genet. 1998;102:197–202. doi: 10.1007/s004390050677. [DOI] [PubMed] [Google Scholar]
- 61.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 62.Ameri A, Machiah DK, T.T T, Channell C, Crenshaw V, Fernstrom K, Khachidze M, Duncan A, Fuchs S, Howard TE. A nonstop mutation in the factor (F)X gene of a severely hemorrhagic patient with complete absence of coagulation factor (F)X. Thrombosis and Haemostasis. 2007 doi: 10.1160/th07-02-0125. in press. [DOI] [PubMed] [Google Scholar]
- 63.Yasuda M, Shabbeer J, Osawa M, Desnick RJ. Fabry disease: novel alpha-galactosidase A 3'-terminal mutations result in multiple transcripts due to aberrant 3'-end formation. Am J Hum Genet. 2003;73:162–173. doi: 10.1086/376608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee HH, Kim YS, Kim KH, Heo I, Kim SK, Kim O, Kim HK, Yoon JY, Kim HS, Kim do J, Lee SJ, Yoon HJ, Kim SJ, Lee BG, Song HK, Kim VN, Park CM, Suh SW. Structural and functional insights into Dom34, a key component of No-Go mRNA decay. Mol Cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 66.Graille M, Chaillet M, van Tilbeurgh H. Structure of yeast Dom34: a protein related to translation termination factor eRF1 and involved in No-Go decay. J Biol Chem. 2008 doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 67.LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell. 2006;24:619–626. doi: 10.1016/j.molcel.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 68.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peltz SW, Donahue JL, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 71.van Hoof A. Conserved functions of yeast genes support the Duplication, Degeneration and Complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wallrapp C, Verrier SB, Zhouravleva G, Philippe H, Philippe M, Gress TM, Jean-Jean O. The product of the mammalian orthologue of the Saccharomyces cerevisiae HBS1 gene is phylogenetically related to eukaryotic release factor 3 (eRF3) but does not carry eRF3-like activity. FEBS Lett. 1998;440:387–392. doi: 10.1016/s0014-5793(98)01492-6. [DOI] [PubMed] [Google Scholar]
- 73.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 74.Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 75.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 76.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci U S A. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matskevich AA, Moelling K. Dicer is involved in protection against influenza A virus infection. J Gen Virol. 2007;88:2627–2635. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
- 79.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 80.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 81.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 82.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 83.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 84.Gribaudo G, Lembo D, Cavallo G, Landolfo S, Lengyel P. Interferon action: binding of viral RNA to the 40-kilodalton 2'-5'-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masison DC, Blanc A, Ribas JC, Carroll K, Sonenberg N, Wickner RB. Decoying the cap- mRNA degradation system by a double-stranded RNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meaux S, van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wickner RB. Prions and RNA viruses of Saccharomyces cerevisiae. Annu Rev Genet. 1996;30:109–139. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 88.Ridley SP, Sommer SS, Wickner RB. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toh EA, Guerry P, Wickner RB. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benard L, Carroll K, Valle RC, Wickner RB. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990;87:7628–7632. doi: 10.1073/pnas.87.19.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Widner WR, Wickner RB. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 95.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 97.Rain JC, Legrain P. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5' to 3' mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 100.Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 101.Scherrer FW, Jr, Spingola M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA. 2006;12:1361–1372. doi: 10.1261/rna.2276806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 105.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 106.Bickel KS, Morris DR. Silencing the transcriptome's dark matter: mechanisms for suppressing translation of intergenic transcripts. Mol Cell. 2006;22:309–316. doi: 10.1016/j.molcel.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 107.Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 110.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 111.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 112.Lee BS, Culbertson MR. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci U S A. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]