Abstract
High-affinity biotinylated derivatives of 17β-estradiol (E2) are of value for isolation of various estrogen-binding proteins (including estrogen receptors) and also for studying protein-protein interactions involving these proteins. In this study, we developed a simplified route for the chemical synthesis of a biotinylated derivative of E2 (compound 7) with a side chain attached to its C-7α position. Compound 7, i.e., 7α-{7-[8-(biotinamido)-octanamido]-heptyl}-estradiol, could be readily synthesized from 6-keto-estradiol-3,17β-di-tetrahydropyranyl ether (compound 2, which can be prepared from E2), with a final yield of 36%. In vitro receptor binding assay confirmed that the synthesized affinity ligand has a high binding affinity for both human estrogen receptor α and β. When the affinity ligand (compound 7) was immobilized with avidin on an affinity column, it effectively bound human estrogen receptor α, and the receptor protein could be selectively eluted with a biotin-containing buffer. Using the same affinity ligand, prolyl 4-hydroxylase β-subunit (also known as protein disulfide isomerase) was identified as one of the high-affinity E2-binding proteins in the whole cytosolic protein mixture prepared from MCF-7 human breast cancer cells. Computational molecular modeling analysis also showed that compound 7 can bind to human estrogen receptor α in a similar manner as ICI-182,780 and raloxifene, and their binding energy values are also similar.
INTRODUCTION
High-affinity biotinylated derivatives of 17β-estradiol (E2)5 are useful for isolation of estrogen receptors (ERs) as well as other E2-binding proteins [1–4]. Also, they are useful for studying protein-protein interactions involving these estrogen-binding proteins. The most commonly-used estrogen receptor affinity ligands include two basic types: one is the C-17α substituted derivatives that are commonly synthesized from 17α-ethynylestradiol, and the other is the C-7α substituted derivatives that are usually synthesized from E2 or estrone. While the C-17α substituted derivatives have the advantage of shorter steps for chemical synthesis, ligands of this group generally have rather low binding affinity for ERs, because C-17α substitution drastically reduces their binding affinity [5, 6]. In comparison, C-7α substituted affinity ligands retain high affinity for ER binding [7, 8], just like the C-7α substituted antiestrogens such as ICI-182,780 [9, 10]. However, C-7α substituted affinity ligands are generally more difficult to make because of the long and difficult synthetic steps involved [2–4, 8, 11].
Recently, we have compared a number of methods for the chemical synthesis of C-7α substituted E2 derivatives [12]. Based on the improvements we recently made, we report here a simplified route for the chemical synthesis of a new C-7α biotinylated E2 derivative (structure shown in Scheme 1). Based on the earlier studies showing that some of the substituted E2 derivatives with a long side-chain attached to the C-7α position usually retain ERα-binding affinity, it was thus expected that the new affinity ligand we synthesized likely would also have ER-binding activity. The affinity ligand was experimentally tested in this study to determine its binding affinity for human ERα and ERβ in vitro. In addition, we also tested its usefulness for the isolation of human ERα protein or other E2-binding proteins when it was used in combination with an immobilized avidin column. To help better understand its binding interactions with human ERα, we used the computational molecular modeling tools to dock compound 7 and two of its theoretical analogs with a shorter side chain (n = 2 or 4) to the binding pocket of the receptor and compared its binding conformation with two well-known ER antagonists, ICI-182,780 and raloxifene (RAL).
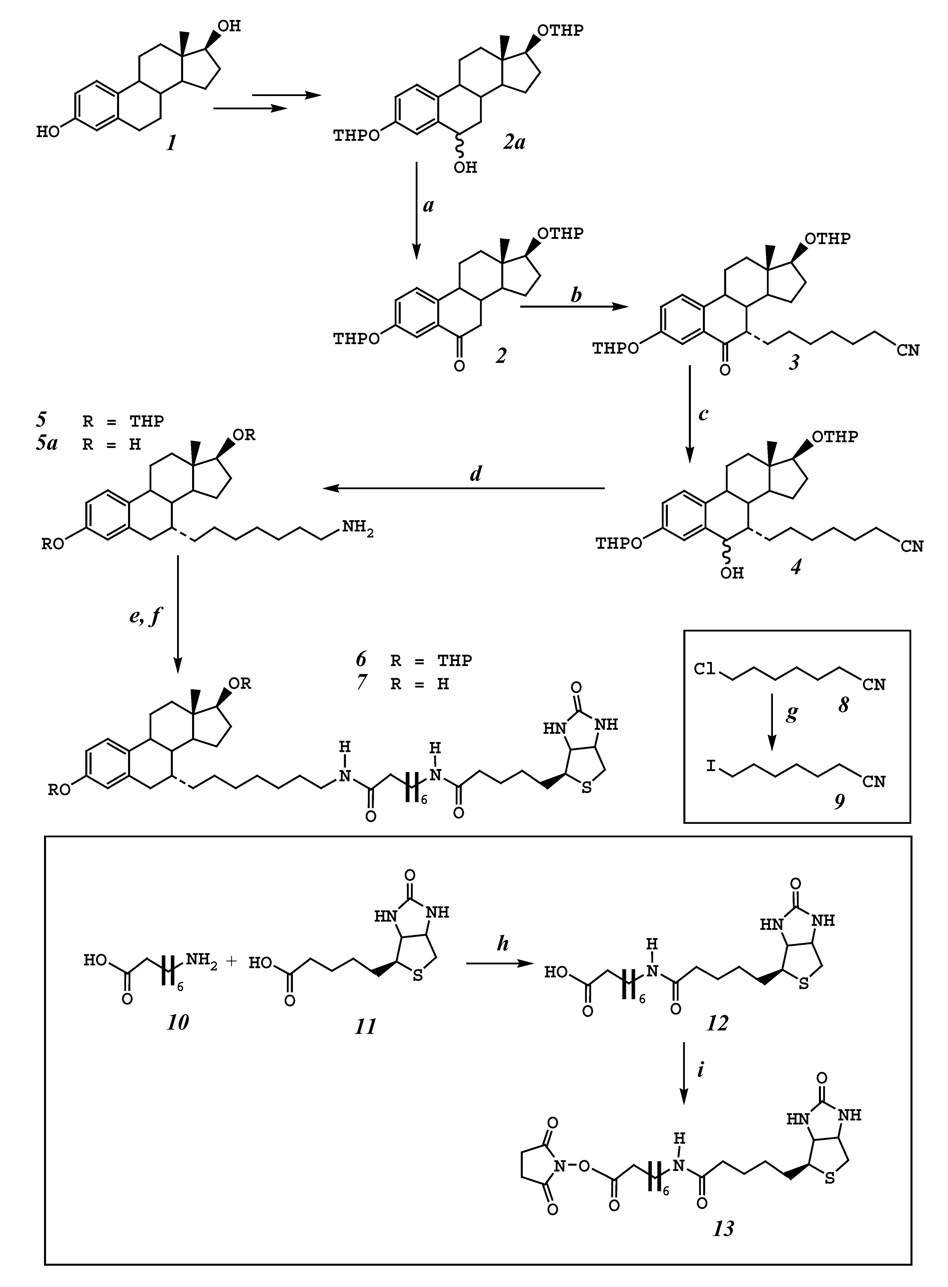
Scheme 1.
The flow chart for the synthesis of 7α-{7-[8-(biotinamido)-octanamido]-heptyl}-estradiol (compound 7) using E2 (compound 1) as the starting material. The reagents and the reaction conditions used are summarized below: a. Swern oxidation. b. t-BuOK, THF, 0°C, compound 9, −78°C. c. NaBH4, methanol. d. 10% Pd(OH)2/C, H2, 45 psi, ethanol, 60°C 8 h. e. compound 13, Et3N, DMF, rt, 24 h. f. AcOH:MeOH:H2O (4:2:1), rt, 24 h. g. NaI, acetone, reflux, 12 h. h. isobutyl chloroformate, tri-n-butylamine, DMF. i. N-hydroxysuccinimide, dicyclohexyl carbodiimide, DMF.
EXPERIMENTAL
1. Chemical Synthesis
Chemicals and reagents
Most of the commercially-available reagents were obtained from the Sigma-Aldrich Chemical Co. (St. Louis, MO) or ACROS (through Fisher Scientific, Atlanta, GA), and were used directly without additional steps of purification.17β-Estradiol (E2) was purchased from Steraloids, Inc. (Newport, RI). Tetrahydrofuran (THF) was distilled over sodium/benzophenone ketyl and methylene chloride was distilled over calcium hydride in our laboratory prior to use.
[2,4,6,7,16,17-3H]E2 (specific activity of 123 Ci/mmol) was obtained from Perkin Elmer (Waltham, MA) and it was re-purified using an HPLC method prior to use in the in vitro receptor binding assay. The recombinant human ERα and ERβ proteins and bovine serum albumin (BSA) were obtained from PanVera Corporation (Madison, WI). Hydroxylapatite (HAP) was obtained from Calbiochem (San Diego, CA). UltraLink immobilized monomeric avidin was obtained from Pierce Biotechnology Inc.(Rockford, IL). The primary antibody (no. sc-543) against ERα was purchased from Santa Crutz Biotechnology (Santa Cruz, CA). Human breast cancer cell line MCF-7 was obtained from ATCC (Manassas, VA). Protease inhibitor cocktail was purchased from Sigma-Aldrich Chemical Company (St. Louis, MO).
Spectrometric analyses
Mass spectra were recorded with a VG70S analytical mass spectrometer. An aliquot of the ethanol solution of the test compound was used for direct-probe mass analysis. The nuclear magnetic resonance (NMR) spectra of all test compounds in deuterochloroform solution were recorded on a Varian Mercury/VX 300 spectrometer with tetramethylsilane (TMS) as internal standard (δ = 0) unless noted otherwise.
Synthesis of 7-iodoheptanenitrile (compound 9)
Under nitrogen, 10 g (52.4 mmol) of 7-bromoheptanenitrile was added into a mixture of 39 g (262 mmol) sodium iodide and 100 mL acetone. The mixture was refluxed over night. Upon completion, the reaction mixture was allowed to cool to room temperature. The organic solvent was removed in vacuo. Then, to the residue 150 mL hexane was added. After the solid was removed by filtration, the filtrate was concentrated. Flash chromatography (hexane as an eluent) afforded compound 9 (14.5 g, 98%).
Synthesis of 8-N-biotinylaminooctanoic acid (compound 12)
8-N-Biotinylaminooctanoic acid, a known compound, was prepared according to the procedures described by Baulieu et al. [13]. Isobutyl chloroformate (160 µL) was added to a solution of biotin (250 mg) in dimethylformamide (DMF, 20 mL) containing tri-n-butylamine (320 µL). After 10 min at room temperature, the mixture was added to a suspension of 8-aminooctanoic acid (163 mg) in DMF (20 mL) at 5°C and was stirred for 2 hours. The solvent was removed in vacuo. The crude product was re-crystallized from ethanol to afford 220 mg product in 56% yield. 1H NMR (DMSO-d6, 300 MHz,): 7.71 (1H, t, J = 5.4 Hz), 6.41 (1H, s), 6.34 (1H, s), 4.28 (1H, m), 4.11 (1H, m), 3.06 (1H, m), 2.97 (2H, q, J = 12.6, 6.3 Hz), 2.80 (1H, dd, J = 12.3, 5.1 Hz), 2.55 (1H, d, J = 12.3 Hz), 2.15 (2H, t, J = 7.5 Hz), 2.02 (2H, t, J = 7.2 Hz), 1.58 ~ 1.22 (17H, m). MS (TOF MS ES+,C18H31N3O4S, M: 385): 225, 386 (M+H+, base peak), 408 (M+Na+). HRMS: 386.2113 (calculated), 386.2121 (observed).
Synthesis of N-hydroxysuccinimide ester of 8-N-biotinylaminooctanoic acid (compound 13)
N-Hydroxysuccinimide ester of 8-N-biotinylaminooctanoic acid was prepared from 8-N-biotinylaminooctanoic acid and N-hydroxysuccinimide by coupling with dicyclohexylcarbodiimide according to the procedures of Bayer and Wilchek [14]. Dicyclohexyl carbodiimide (0.8 g) is added to a solution of 8-N-biotinylaminooctanoic acid (1.54 g) and N-hydroxysuccinimide (0.6 g) in 20 mL of DMF. The suspension was stirred at room temperature for 24 hours. The precipitate was filtered, and the filtrate was maintained at 0°C overnight. Any additional precipitates were filtered, and the resulting filtrates were evaporated under reduced pressure. The residue was washed extensively with ether. The crude product was used in next step without further purification.
Synthesis of 6-keto-7α-(6-cyanohexyl)-estradiol-3,17β-di-tetrahydropyranyl ether (compound 3)
Using E2 (compound 1 in Scheme 1) as the starting material, 6-hydroxy-estradiol-3,17β-di-tetrahydropyranyl ether (compound 2a) was first prepared according to an earlier study [11].
After a solution of methylene chloride (70 mL) and oxalyl chloride (3.6 mL, 41.9 mmol) in a 250-mL three-neck round-bottom flask was cooled to −78°C, DMSO (5.9 mL, 83 mmol, in 15 mL methylene chloride) was added slowly to the stirred oxalyl chloride solution and the reaction mixture was stirred further for 5 min. Note that the addition of the DMSO solution needs to be relatively slow so that the temperature of the reaction mixture would not rise above −50°C (usually controlled between −50°C and −60°C), which is critical for this reaction. After that, compound 2a (15.7 g, 34.5 mmol, in 30 mL methylene chloride) was slowly added over 15 min, and the mixture was stirred for an additional 25 min. Then trimethylamine (TEA, 24 mL) was added and the reaction mixture was stirred for 5 min and allowed to warm to room temperature. Water (100 mL) was added and the aqueous phase was extracted with methylene chloride (50 mL). The organic phase was combined, washed with a saturated sodium chloride solution (100 mL), and dried over anhydrous sodium sulfate. Flash chromatography afforded compound 2 (14.6 g, 94% yield).
To synthesize compound 3, a 1.0 M solution of potassium t-butoxide (t-BuOK) in THF (20 mL, 20 mmol) was added to a cooled solution (at 0°C) of compound 2 (1.2 g, 2.67 mmol) in THF (20 mL). The reaction mixture was stirred at 0°C for 40 min and then cooled to −78°C. 7-Iodoheptanenitrile (compound 9, 2.0 g, 7.1 mmol) was added dropwise to the solution. The reaction mixture was allowed to warm to 0°C and stirred for 2 hours, and then to room temperature and stirred overnight. The reaction was quenched with water and extracted with ethyl acetate. The organic phase was dried over sodium sulfate and the solvent was evaporated in vacuo. Flash chromatography (hexane:ethyl acetate = 4:1, v:v) afforded compound 3 in 62% yield (0.93 g). 1H NMR (300 Hz, CDCl3): 7.61 (1H, brS), 7.15~7.25 (2H, m), 5.39 (1H, m), 4.62(1H, m), 3.80 (2H, m), 3.68 (1H, t, J = 8.4 Hz), 3.56 (1H, m), 3.42 (1H, m), 2.59 (1H, m), 2.39 (2H, m), 2.24 (2H, t, J = 7.2Hz), 1.16~2.02 (31H, m), and 0.73 (3H, s). MS (Scan ES+, C35H49O5N, M+1 564): 564 (base peak), 586 (M+Na). HRMS (M+1): 564.3689 (calculated) and 564.3682 (observed).
Synthesis of 6-hydroxy-7α-(6-cyanohexyl)-estradiol-3,17β-di-tetrahydropyranyl ether (compound 4)
Compound 3 (0.51 g, 0.91 mmol) was dissolved in 20 mL methanol. To this solution was added sodium borohydride (0.69 g, 18.1 mmol). The reaction mixture was stirred at room temperature for 4 hours. The methanol was removed in vacuo. The residue was then dissolved in water (20 mL). The aqueous solution was extracted with methylene chloride three times (3 × 20 mL). Organic fractions were combined, dried over sodium sulfate, and concentrated. The crude product was purified by flash chromatography (ethyl acetate : hexane = 1:3, v:v) to afford compound 4 (0.5 g, 98%). 1H NMR (300 Hz, CDCl3): 7.4 (1H, d, J = 2.4 Hz), 7.09 (1H, d, J = 8.8 Hz), 6.84 (1H, m), 5.35 (1H, m), 4.81 (1H, m), 4.57(1H, m), 3.84 (2H, m), 3.68 (1H, t, J = 7.6 Hz), 3.52 (1H, m), 3.42 (1H, m), 2.18~2.31 (4H, m), 1.87~2.07 (4H, m), 1.76 (4H, m), 1.20~1.65 (25H, m), and 0.73 (3H, s). MS (TOF MS ES+, C35H51O5N, M+ 565): 540, 548, 566, 583 (M + NH4 +) (base peak), 588 (M + Na+). HRMS [588 (M + Na+)]: 588.3665 (calculated) and 588.3660 (observed).
Synthesis of 7α-(7-aminoheptyl)-estradiol-3,17β-di-tetrahydropyranyl ether (compound 5)
Compound 4 (0.4 g, 0.71 mmol) and 10% palladium hydroxide on carbon (400 mg) were added to 50 mL of ethanol. The reaction vessel was evacuated of air and filled to a pressure of 45 psi with hydrogen on a Parr apparatus, and then shaken for 8 hours at 60°C. The mixture was then filtered through celite, and the filtrate was evaporated under reduced pressure to give a mixture of compound 5 and compound 5a [7α-(7-aminoheptyl)-estradiol] by a ratio of 5:1 based on 1H NMR (total 0.39 g, ~100%). This mixture was not further purified for the next synthetic step.
For spectrometric analysis, a fraction of the samples was used for separation of compounds 5 and 5a by flash chromatography (hexane:ethyl acetate = 4:1, v:v). Compound 5. 1H NMR (300 Hz, CDCl3): 7.11 (1H, m), 6.77 (1H, d, J = 8.4 Hz), 6.68 (1H, brS), 5.29 (1H, m), 4.59 (1H, m), 3.85 (2H, m), 3.67 (1H, t, J = 8.4 Hz), 3.55 (1H, m), 3.43 (1H, m), 2.81 (2H, m), 2.21 (2H, m), 1.92~1.12 (38H, m), and 0.73 (3H, s). MS (TOF MS ES+, C35H55O4N, M+ 553): 555 (M + 1) (base peak). HRMS (M + 1): 554.4209 (calculated) and 554.4208 (observed). Compound 5a. 1H NMR (300 Hz, DMSO-d6): 9.15 (1H, brs), 8.20 (3H, brs), 7.07 (1H, d, J = 8.4 Hz), 6.57 (1H, d, J = 8.4 Hz), 6.51 (1H, s), 4.94 (1H, brs), 3.59 (1H, t, J = 7.8 Hz), 2.77 (3H, m), 2.65 (1H, d, J = 16.8Hz), 2.27 (2H, m), 1.97~1.11 (21H, m), and 0.72 (3H, s). MS (C25H39NO2, M 385): 385 (base peak). HRMS (M): 385.2981 (calculated) and 385.2984 (observed).
Synthesis of 7α-{7-[8-(biotinamido)-octanamido]-heptyl}-estradiol (compound 7)
The crude preparation containing mostly compound 5 (42 mg, 0.11 mmol) was dissolved in 10 mL DMF. Then 0.053 g (0.11 mmol) of N-hydroxysuccinimide ester of 8-N-biotinylaminooctanoic acid (compound 13) and 47 µL of TEA were added. The mixture was stirred at room temperature for 24 hours. The solvents were removed under high vacuum, and the raw product was dissolved in a mixture of acetic acid, methanol and water (4:2:1, v:v:v) and stirred for 24 hours at room temperature. The solvents were removed in vacuo, and the raw product was purified by HPLC to afford compound 7.
1H NMR (300 Hz, DMSO-d6): 8.98 (1H, s), 7.72 (2H, m), 7.04 (1H, d, J = 8.7 Hz), 6.49 (1H, dd, J = 8.7, 2.7 Hz), 6.43 (1H, s), 6.41 (1H, d, J = 2.7 Hz), 6.36 (1H, s), 4.48 (1H, d, J = 4.8 Hz), 4.29 (1H, m), 4.13 (1H, m), 3.53 (1H, m), 3.09 (1H, m), 3.0 (4H, m), 2.80 (2H, m), 2.62(2H, m), 2.26 (2H, m), 2.0 (4H, m), 190~1.05 (38H, m), and 0.66 (3H, s). MS (TOF MS ES+, C43H68N4O5S, M+ 752.5): 753.5 (M + 1) (base peak). HRMS (M + 1): 753.4988 (calculated) and 753.4982 (observed).
3. ERα and ERβ binding assays
The ERα and EṚβ competitive binding assays were performed according to the method described in our recent study [15]. The ER binding buffer used for dilution of the receptor preparations consisted of 10% glycerol, 2 mM dithiothreitol, 1 mg/mL BSA, and 10 mM Tris–HCl (pH 7.5). The ERα washing buffer contained 40 mM Tris-HCl and 100 mM KCl (pH 7.4), but the ERβ washing buffer contained only 40 mM Tris-HCl (pH 7.5). The 50% (v/v) hydroxylapatite (HAP) slurry was prepared by using the 50 mM Tris-HCl solution (pH 7.4). The reaction mixture contained 50 µL of varying concentrations of the test compound in the ER binding buffer, and 45 µL of [3H]E2 solution (at 22.22 nM). Then 5 µL of ERα or ERβ protein was added and mixed gently. Nonspecific binding by [3H]E2 was determined by addition of a 400-fold concentration of the nonradioactive E2 (8 µM). The binding mixture was incubated at room temperature for 2 hours. At the end of the incubation, 100 µL of the HAP slurry was added and the tubes were incubated on ice for 15 minutes with three times of brief vortexing. An aliquot (1 mL) of the washing buffer was added, mixed, and centrifuged at 10,000 g for 1 min, and the supernatants were discarded. This wash step was repeated twice. The HAP pellets were then resuspended in 200 µL ethanol, and the content was transferred to scintillation vials for measurement of the 3H-radioactivity in a liquid scintillation counter (Packard Tri-CARB 2900 TR; Downers Grove, IL). The data obtained from duplicate measurements were expressed as the % specific binding of [3H]E2 versus the log molar concentration of the competing compound. The IC50 value (calculated using the SigmaPlot software) represents the concentration of a test compound required to reduce the [3H]E2 binding by 50%.
4. Effectiveness of the affinity ligand (compound 7) for isolation of human E2-binding proteins
In the first experiment, the human ERα protein was incubated with compound 7 or biotin alone (at 10 µM) at 4°C overnight, and then the mixture was loaded onto a small column pre-packed with UltraLink immobilized monomeric avidin. After a washout with five-bed volume of 10 mM sodium phosphate buffer (pH 7.4), a sequential elution using three-bed volume of 10 mM sodium phosphate buffer (pH 7.4) and two-bed volume of 2 mM biotin-containing sodium phosphate was performed. The fractions were then subject to 13% trichloroacetic acid precipitation, and the western blotting of the precipitated ERα protein was performed.
In the second sets of experiments, MCF-7 cells were cultured in the presence of 10 nM of E2, harvested in 10 mM sodium phosphate buffer, and sonicated in the presence of 1 mM EDTA and a protease inhibitor cocktail. The whole cytosolic protein extracts were then incubated with compound 7 (at 10 µM) at 4°C over-night before loaded onto a small column pre-packed with UltraLink immobilized monomeric avidin. After a washout with five-bed volume of 10 mM sodium phosphate buffer (pH 7.4, containing 0.5 M NaCl), a sequential elution using three-bed volume of 10 mM sodium phosphate buffer (pH 7.4) and two-bed volume of E2-containing (20 µM) sodium phosphate buffer was performed. After incubation at 4°C for 6 hrs, continuous elution with two-bed volume E2-containing buffer was carried out to elute the tightly-bound proteins. The fractions were then subject to 13% trichloroacetic acid precipitation and SDS-PAGE analysis. The bands of interest were cut and sent to ProtTech (Norristown, PA) for protein identification using the NanoLC-MS/MS peptide sequencing technology.
5. Molecular docking of ligands into the binding pocket of human ERα
Docking study was performed with InsightII modeling software (Version 2005, Accelrys Inc. San Diego, CA) installed in a Dell Precision 690 workstation with Red Hat Enterprise Linux WS4.0 operating system (Red Hat Inc. Raleigh, NC). Energy minimization and molecular dynamics simulation were performed with Discovery Studio modeling program (Version 1.7, Accelrys Inc. San Diego, CA) installed on the same computer. CHARMm force field was used by DiscoveryStudio for molecular dynamics simulation.
Because the crystal structure of human ERα in complex with a full antagonist is not available, the crystallographic structure of ERα in complex with RAL (PDB code: 1ERR [16]), which is a partial agonist/antagonist, was thus used as a template for the docking study of ICI-182,780 and our affinity ligand (i.e., compound 7, structure shown in Fig. 3). The structures of ICI-182,780 and compound 7 were first built using the Builder module and then minimized with the Discover module of the InsightII. The flexible docking procedures were carried out using the Simulated Annealing Docking method in Affinity module of InsightII. The binding pocket was defined to include all residues within the 7–Å reach of the original ligand RAL. A combination of Monte Carlo and simulated annealing methods was used to explore all possible conformations of ICI-182,780 and compounds 7. One hundred conformations were obtained and the one with the lowest potential energy was chosen for further minimization using the Standard Dynamics Cascade protocol in the Discovery Studio. The backbone of the protein and its key residues in the binding pocket (namely, E353, R394 and H524) were fixed during the docking procedures.
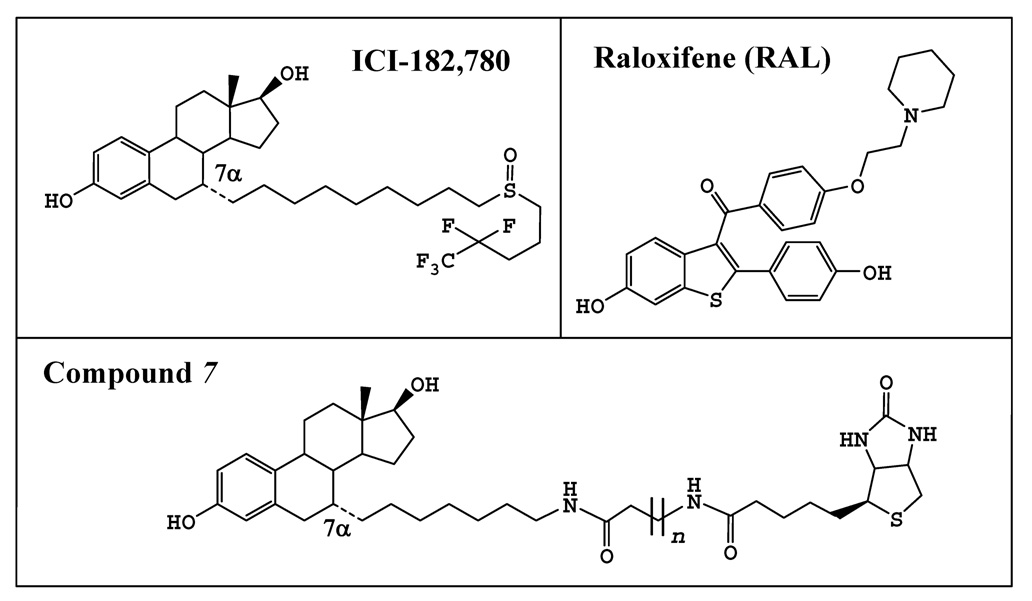
Figure 3.
Chemical structures of ICI-172,780, raloxifene (RAL), and compounds 7. For compound 7, n = 6. Note that for the two putative analogs of compound 7, namely, compounds 7a and 7b, the length of their side chains were reduced to n = 4 and 2, respectively.
For energy minimizations, the steepest descent method was employed first to 10 kcal/(molÅ) root mean square (RMS) energy gradient and followed by the Polak and Ribiere conjugate gradient method until the final convergence criterion reached 0.01 kcal/(molÅ) RMS energy gradient. Then the whole system was heated from 100 to 300 K in 2 ps and equilibrated in 300 K for 100 ps. One hundred conformations were collected in 20 ps production phase at 300 K. The conformation with the lowest potential energy was further minimized and used for Binding Energy Calculation protocol. The backbone of the protein and its key residues in the binding pocket (namely, E353, R394 and H524) were constrained during the simulation process.
RESULTS AND DISCUSSION
Chemical synthesis
Compound 2 was readily obtained in a high yield from E2 according to a previously described procedure [11, 12, 17]. Compound 9 was prepared from compound 8 by reacting with sodium iodide refluxing in acetone overnight in 90% yield. Compound 2 was then reacted with compound 9 to give compound 3 in 60% yield using potassium tert-butoxide in THF [11, 18]. Compound 3 was reduced to compound 4 using sodium borohydride in methanol, then hydrogenolysis of compound 4 under the conditions of 45 psi hydrogen, heating (to 60°C), and palladium hydroxide (used as catalyst) in ethanol was carried out to give compounds 5 and 5a (with a ratio of approximately 5 to 1). Here it is optional to separate compound 5a from 5 since the protecting groups could be removed later. This mixture was then reacted with N-hydroxysuccinimide ester of 8-N-biotinylaminooctanoic acid (compound 13) to give 7α-{7-[8-(biotinamido)-octanamido]-heptyl}-estradiol-3,17β-di-tetrahydropyranyl ether (compound 6), followed by removal of the THP protecting groups in a solution of acetic acid, methanol and water (4:2:1, v:v:v) to give the final compound 7 in 81% yield over two steps. In the hydrogenolysis step, if the reaction time was prolonged to 24 hours, compound 5 could be transformed to 5a completely. The advantage of this process is that the removal of hydroxyl on the C-6 position, the reduction of the nitrile to amine at the end of the side chain and deprotection of THP at the C-3 and C-17β positions could be achieved in one single step under the mild conditions.
Compared to other methods [2–4, 7, 8, 11], this is a relatively shorter route for the chemical synthesis of 7α-(7-aminoheptyl)-E2 and also its analogs with different lengths of the side chain. The procedures also appear to be environmental-friendly and rather economic with regard to the chemical reagents required and waste products generated. From this compound, with one more step, C-7α-biotinylated E2 affinity ligands can be readily synthesized. It is of note that this intermediate is not just limited to the synthesis of biotinylated E2 affinity ligands, it can also be used for the synthesis of other useful chemicals such as fluorescent probes for estrogen-binding proteins [19].
In vitro ER binding assays
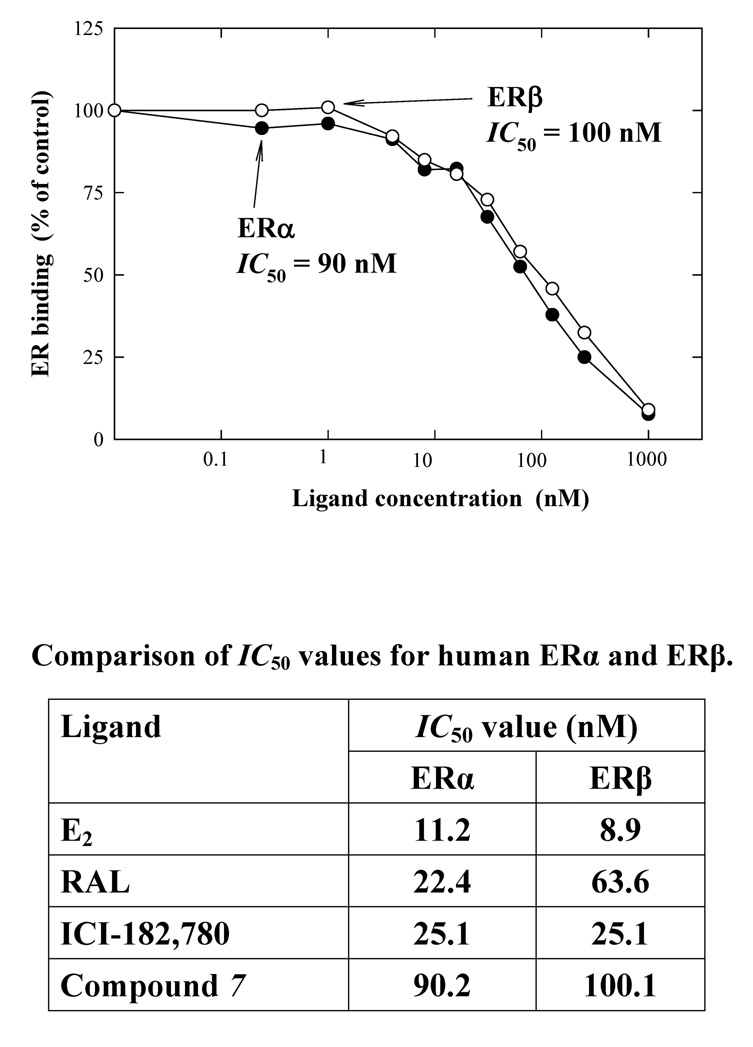
In the present study, we also determined the relative binding affinity of compound 7 for the recombinant human ERα and ERβ proteins (data summarized in Fig. 1). The IC50 values of ICI-182,780 and RAL for human ERα are quite similar to each other (25.1 and 22.4 nM, respectively). The IC50 values of the synthesized affinity ligand (compound 7) for ERα and ERβ were 90 and 100 nM, respectively. The apparent affinities of this ligand for human ERs were slightly lower than those of ICI-182,780.
Figure 1.
Comparison of the relative binding affinity (IC50) of compound 7 for human ERα and ERβ. The binding affinity was also compared with those of E2, RAL and ICI-182,780 (summarized in the accompanying Table below). The relative binding affinity of each chemical was determined by measuring its inhibition of the binding of 10 nM [3H]E2 to the recombinant human ERα and ERβ. Eight concentrations (0.06, 0.24, 0.98, 3.9, 15.6, 62.5, 250, and 1000 nM) of each competing estrogen were tested. The IC50 values for the competing ligands were calculated according to the sigmoid inhibition curves.
Use of biotin-E2 ligand (compound 7) for the isolation of E2-binding proteins
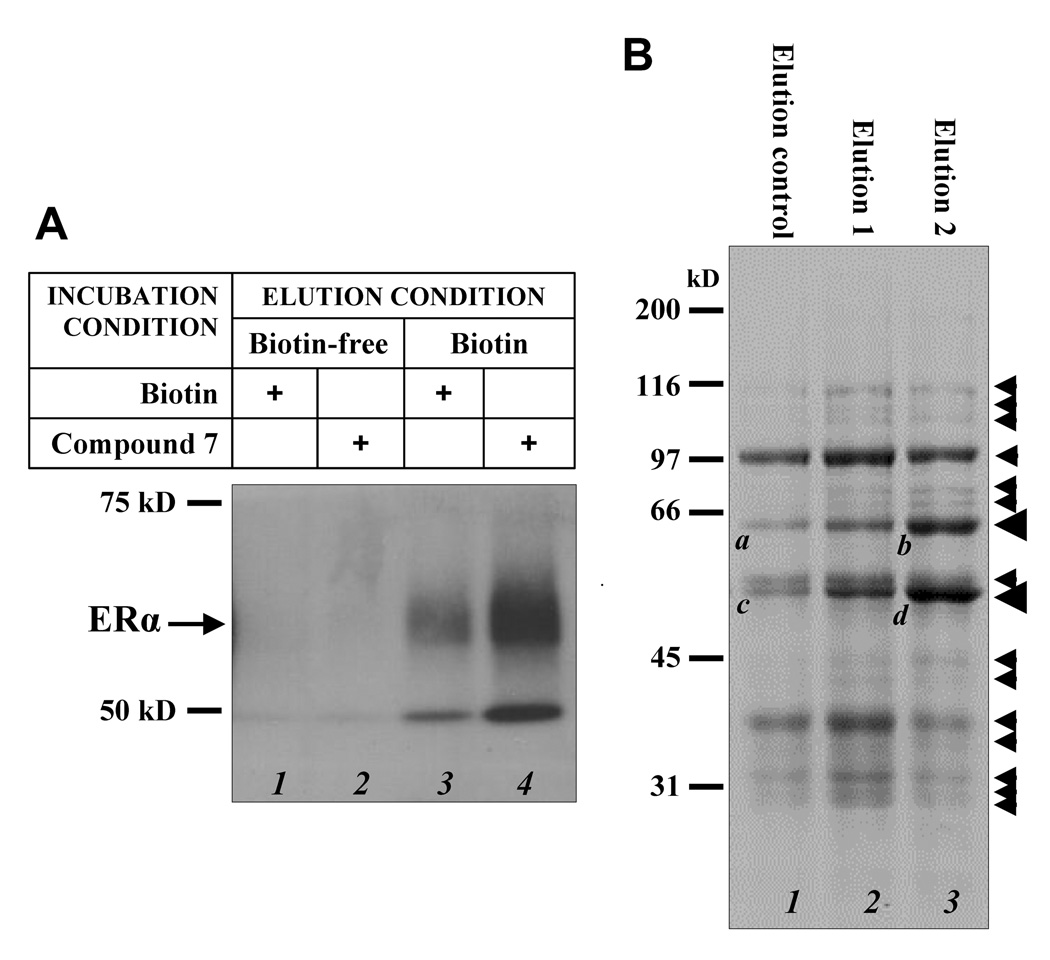
To determine the usefulness of the synthesized affinity ligand for isolation of E2-binding proteins, we first determined whether it could isolate the recombinant human ERα protein. The ERα protein was incubated with compound 7 or biotin was then loaded onto an avidin-immobilized sepharose affinity column. We found that the receptor protein could be selectively eluted off the affinity column by a biotin-containing buffer (compare lane 4 with lane 2 in Fig. 2A). These data suggest the ability of the affinity ligand to specifically bind human ERα and the affinity ligand-bound ERα can be selectively eluted off the avidin-immobilized affinity column with a biotin-containing buffer. It is of interest to note that biotin itself (in the absence of an affinity ligand) also appeared to increase ERα binding to the column, and the bound ERα was selectively eluted off the column with a biotin-containing buffer (compare lane 3 with lane 1 in Fig 2A). This observation is intriguing, and it may suggest that human ERα protein may have a weak binding affinity for biotin. This possibility merits further investigation. The lower bands with a molecular weight of approximately 50 kD in lane 3 and lane 4 are probably a degradation product of ERα based on the fact that this protein could be selectively washed out by a biotin-containing buffer (compare lane 4 and lane 2) and could be stained with anti-ERα antibodies.
Figure 2.
Use of compound 7 for the isolation of human ERα and other E2-binding proteins from human MCF-7 cells. A. Western blot analysis of the eluted fractions from affinity chromatography of human recombinant ERα protien after incubation with either biotin alone (lanes 1 and 3) or compound 7 (lanes 2 and 4). The avidin-immobilized affinity column was first eluted with biotin-free solution (lanes 1 and 2) and then followed by 2 mM biotin-containing solution (lanes 3 and 4). The proteins in the collected fractions were separated by SDS-PAGE electrophoresis. B. SDS-PAGE analysis of potential E2-binding proteins isolated from cytosolic proteins of MCF-7 cells. After incubation of crude cytosolic protein mixtures with compound 7 or biotin, the mixture was loaded onto the affinity column and eluted sequentially with an E2-free elution buffer (elution control), 20 µM E2-containing elution buffer (elution 1), or after incubation at 4°C for 6 hrs with 20 µM E2 and then eluted with the E2-containing buffer (elution 2). The small arrows label the proteins that appear to be preferentially eluted off with an E2-containing buffer either during the first elution or during the second elution. The two quantitatively-major bands with a fat arrowhead were further analyzed with LC-MS/MS for structural compositions. The data are shown in Table 1.
Following the above test which confirmed that the newly-synthesized affinity ligand is capable of binding human ERα and the affinity ligand-ERα complex can be selectively washed off with a biotin-containing elution buffer, we then used this affinity ligand to isolate various E2-binding proteins or those proteins that may tightly bind to the E2-binding proteins from the cytosolic protein extracts of MCF-7 human breast cancer cells. The mixture containing crude cytosolic lystates and the affinity liand was incubated at 4°C overnight and then directly applied to an avidin-immobilized affinity column. The column was eluted with a buffer with or without 20 µM E2. The eluted proteins were analyzed using SDS-gel electrophoresis. Fig. 2B showed that several protein bands were stronger during the two sequential elutions with an E2-containing buffer. The first elution was done directly with the E2-containing buffer without incubation, and the subsequent second elution was done after incubation with the E2-containing buffer for 6 hrs at 4°C.
Among the protein bands identified in the second elution, two quantitatively-major ones (bands b and d labeled with a fat arrowhead in Fig. 2B) were further analyzed in the present study for their structural compositions by LC-MS/MS analysis (Table 1). The corresponding bands in the elution control, i.e. bands a and c, were also analyzed for comparison. Prolyl 4-hydroxylase β-subunit, a protein commonly known as protein disulfide isomerase (PDI), was identified to be present in band b with 33 peptide fragments compared to 14 peptides identified in band a. Similarly, 11 peptide fragments of PDI were identified in band d compared to none in band c. It is of note that since the theoretical molecular weight of PDI is larger than proteins in band d, it is likely that either a quantitatively-major degradation fragment of PDI or a truncated spicing variant of PDI that still retained E2-binding activity was present in band d. In addition, several heat shock protein-70 kD isoforms or their truncated splicing variants were identified in the lower bands (c and d). Some of these proteins are known to be tightly associated with ERα.
Table 1.
LC/MS-MS identification of potential proteins in the cytosol of MCF-7 human breast cancer cells that can directly bind E2 or bind to E2-bidning proteins.
| Protein band in Fig. 3B | Number of peptides | Proteins identified |
|---|---|---|
| a | 14 | prolyl 4-hydroxylase β-subunit (PDI) [190384|gb|AAC13652.1]* |
| 11 | pyruvate kinase [35505|emb|CAA39849.1] | |
| 6 | chaperonin containing TCP1, subunit 6A [76827911|gb|AAI06943.1] | |
| 5 | CCT7 [48145555|emb|CAG33000.1] | |
| b | 33 | prolyl 4-hydroxylase β-subunit (PDI) [190384|gb|AAC13652.1] |
| 12 | pyruvate kinase [35505|emb|CAA39849.1] | |
| 10 | 60 kDa chaperonin (Protein Cpn60) (groEL protein) [2493643|sp|P95800] | |
| 6 | chaperonin containing TCP1, subunit 6A [76827911|gb|AAI06943.1] | |
| c | 18 | heat shock 70 kDa protein 8 isoform 2variant [62896815|dbj|BAD96348.1] |
| 10 | thyroid autoantigen 70 kDa [57165052|gb|AAW34364.1] | |
| 3 | heat shock 70 kDa protein 9B precursor [24234688|ref|NP_004125.3] | |
| d | 23 | heat shock 70 kDa protein 8, isoform 1 [16741727|gb|AAH16660.1] |
| 11 | prolyl 4-hydroxylase β-subunit (PDI) [190384|gb|AAC13652.1] | |
| 6 | thyroid autoantigen 70 kDa (Ku antigen) [57165052|gb|AAW34364.1] |
The protein access number is provided in the bracket following the protein name.
It is of note that earlier studies have reported that protein disulfide isomerase (PDI) has considerable E2-binding affinity [20]. Recently, we have also selectively expressed the human PDI and pyruvate kinase in E. coli. We confirmed that the recombinant PDI has selective E2-binding activity, but pyruvate kinase was found to have no specific E2-binding activity (unpublished data). Currently, we are in the process of characterizing other protein bands shown in Fig. 2B that were isolated by the affinity ligand, such as the 60-kD chaperonin by using the same approach. Lastly, it is of note that among the proteins isolated by the affinity ligand, the amount of ERα protein was very low or barely detectable. The main reason was that the ERα-positive MCF-7 cells were cultured in a medium supplemented with excessive amount of E2 (at 10 nM), and under this estrogen-rich condition, ERα was expected to be mostly translocated into the nuclei and the amount of the ERα present in the cytosolic compartment would be low or undetectable.
Computational modeling
According to earlier studies of ICI-182,780 and its analogs [9, 10], it was empirically predicted that the designed affinity ligands (compounds 7) most likely would function as ER antagonists owing to their long C-7α side chains. To provide a more scientific prediction of the ERα-binding ability of the affinity ligand, we conducted computational molecular modeling analysis of its binding interactions with human ERα ligand binding domain compared with ICI-182,780.
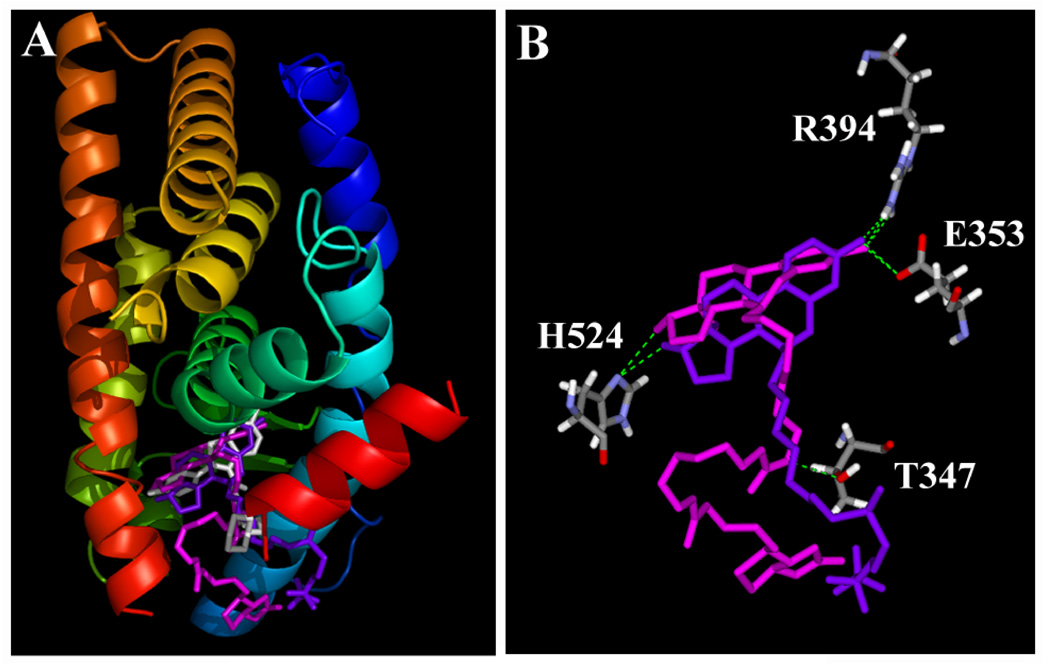
Our docking models showed that compounds 7 and ICI-182,780 can fit rather tightly inside the binding pocket in a very similar manner (Fig. 4A), with their steroidal rings and especially their C-3 and C-17β hydroxyl groups overlapping with each other extensively (Fig. 4B). Their hydroxyl groups form hydrogen bonds with the amino acid residues E353, R394 and H524 of ERα nearly in the same positions as those of E2. In addition to the hydrogen bonds formed between the C-3 or C-17β hydroxyl groups of compound 7 and ERα, its side chain forms an additional hydrogen bond with T347 of ERα. We also calculated the relative binding energy values (∆Ebinding) for these binding models (data summarized in Table 2). In addition, we also docked two putative analogs of compound 7 (compounds 7a and 7b) with a shorter side chain (structures shown in Fig. 3). The binding energy values for these two analogs did not vary significantly compared to the value for compound 7 (Table 2).
Figure 4.
Interactions of raloxifene (RAL), ICI-182,780 and compound 7 with the ligand binding domain (LBD) of human ERα. A. The three-dimensional structures of ERα LBD in complex with RAL (magenta), ICI-182,780 (purple) and compound 7 (white). The secondary structure of ERα (in ribbon) is colored from blue (N-terminus) to red (C-terminus). This figure is drawn using the PyMOL software. B. Key amino acid residues in ERα LBD that interact with ICI-182,780 and compound 7. Amino acids are colored for their elements (white for hydrogen, red for oxygen, and blue for nitrogen). Compound 7 is colored in magenta and ICI-182,780 in purple. The green dashes show hydrogen bonds between ligands and ERα. This figure is drawn using the DiscoveryStudio software.
Table 2.
The binding energy values (ΔEbinding, in kcal/mol) of raloxifene (RAL), ICI-182,780, and compounds 7 with the ligand binding domain of human ERα.
| Ligand | ΔEbinding (kcal/mol) |
|---|---|
| Raloxifene (RAL) | −218.7 |
| ICI-182,780 | −191.5 |
| Compound 7 | −284.9 |
| Compound 7a (n = 4) | −281.9 |
| Compound 7b (n = 2) | −282.1 |
Note: The binding energy value (ΔEbinding) was calculated with the following equation: ΔEbinding = Ecomplex − (Eligand + EERα), where Ecomplex is the potential energy for the complex of human ERα with the ligand, Eligand is the potential energy of the ligand itself, and EERα is the potential energy of the receptor itself.
Our docking structure of ICI-184,780 in complex with ERα revealed that this pure antagonist adopts a unique binding mode inside the binding pocket by flipping their core steroidal structure 180° along the C-3 and C-17β hydroxyl axis. In this way, the long C-7α side chain is positioned toward the C-11β channel and then exits the ligand binding pocket in a similar manner as RAL. Interestingly, in this altered binding mode, the characteristic hydrogen bonds of C-3 and C-17β hydroxyl groups with the receptor binding pocket basically remain unchanged. Notably, although the crystallographic structure of human ERα in complex with ICI-184,780 is not available at present, the docking conformation of ICI-184,780 as developed in this study matched the reported crystal structure of human ERβ in complex with ICI-164,384 (an analog of ICI-184,780) [21], which provides support for the proposed binding structure based on our computational models. Our modeling data also suggest that the C-11β-substituted ER antagonists could be more ideal than the C-7α-substituted antagonists.
CONCLUSIONS
In the present study, we have synthesized a new 7α-biotinylated E2 analog as affinity ligand for human ERα and ERβ from 6-keto-estradiol-3,17β-di-tetrahydropyranyl ether (compound 2). The key step involves the hydrogenolysis of compound 4 using palladium hydroxide on carbon as catalyst to remove the 6-hydroxyl group and the simultaneous reduction of nitrile to amine to yield 7α-(7-aminoheptyl)-estradiol-3,17β-di-tetrahydropyranyl ether (compound 5). The overall yield for the synthesis is approximately 36%. In vitro receptor binding assay confirmed that the synthesized affinity ligand has a rather high binding affinity for both human ERα and ERβ. Using this affinity ligand, protein disulfide isomerase was identified as one of the high-affinity E2-binding proteins present in the cytosolic fraction of human MCF-7 cells. Computational molecular modeling analysis also showed that compound 7 can bind to human ERα in a similar way as ICI-182,780 and RAL, and their binding energy values are comparable.
Footnotes
The study was supported, in part, by a grant from the NIH (CA97109). Note that part of the work described in this paper was completed when the authors were at the University of South Carolina, Columbia, SC 29208, USA.
Abbreviations used: E2, 17β-estradiol; ERα and ERβ, estrogen receptor alpha and beta subtypes, respectively; NMR, nuclear magnetic resonance; THP, tetrahydropyran; DMF, dimetylformamide; TEA: triethylamine; TMS: tetramethylsilane; t-BuOK, potassium t-butoxide; RAL, raloxifene, PDI, protein disulfide isomerase.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kaiser T, Gudat P, Stock W, Pappert G, Grol M, Neumeier D, Luppa PB. Biotinylated steroid derivatives as ligands for biospecific interaction analysis with monoclonal antibodies using immunosensor devices. Analytical Biochem. 2000;282:173–185. doi: 10.1006/abio.2000.4596. [DOI] [PubMed] [Google Scholar]
- 2.Hauptmann H, Paulus B, Kaiser T, Herdtweck E, Huber E, Luppa PB. Concepts for the syntheses of biotinylated steroids. Part II: 17β-Estradiol derivatives as immunochemical probes. Bioconjugate Chem. 2000;11:537–548. doi: 10.1021/bc9901651. [DOI] [PubMed] [Google Scholar]
- 3.Redeuilh G, Secco C, Baulieu EE. Biotinylestradiol for purification of estrogen receptor. Methods Enzymol. 1990;184:292–300. [PubMed] [Google Scholar]
- 4.Hussey SL, Muddana SS, Peterson BR. Synthesis of a β-estradiol-biotin chimera that potently heterodimerizes estrogen receptor and streptavidin proteins in a yeast three-hybrid system. J Am Chem Soc. 2003;125:3692–3693. doi: 10.1021/ja0293305. [DOI] [PubMed] [Google Scholar]
- 5.Bucourt R, Vignau M, Torelli V. New biospecific adsorbents for the purification of estradiol receptor. J Biol Chem. 1978;253:8221–8228. [PubMed] [Google Scholar]
- 6.Gao H, Katzenellenbogen JA, Garg R, Hansch C. Comparative QSAR analysis of estrogen receptor ligands. Chem Rev. 1999;99:723–744. doi: 10.1021/cr980018g. [DOI] [PubMed] [Google Scholar]
- 7.French AN, Wilson SC, Welch MJ, Katzenellenbogen JA. A synthesis of 7α-substituted estradiols: synthesis and biological evaluation of a 7α-pentyl-sbustituted BODIPY fluorine-18-labeled 7α-pentylestradiol analog. Steroids. 1993;58:157–169. doi: 10.1016/0039-128x(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 8.Redeuilh G, Secco C, Baulieu EE. The use of the biotinyl estradiol-avidin system for the purification of "nontransformed" estrogen receptor by biohormonal affinity chromatography. J Biol Chem. 1985;260:3996–4002. [PubMed] [Google Scholar]
- 9.Howell A, Osborne CK, Morris C, Wakeling AE. ICI 182,780 (Faslodex™): Development of a novel, ȁCpure” antiestrogen. Cancer. 2000;89:817–825. doi: 10.1002/1097-0142(20000815)89:4<817::aid-cncr14>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Jordan VC. Antiestrogens and selective estrogen receptor modulators as multifunctional medicines. 1. Receptor interactions. J Med Chem. 2003;46:883–908. doi: 10.1021/jm020449y. [DOI] [PubMed] [Google Scholar]
- 11.Skaddan MB, Wust FR, Katzenellenbogen JA. Synthesis and binding affinities of novel re-containing 7α-substituted estradiol complexes: Models for breast cancer imaging agents. J Org Chem. 1999;64:8108–8121. doi: 10.1021/jo990641g. [DOI] [PubMed] [Google Scholar]
- 12.Jiang XR, Sowell JW, Zhu BT. Synthesis of 7α-substituted derivatives of 17β-estradiol. Steroids. 2006;71:334–342. doi: 10.1016/j.steroids.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Redeuilh G, Secco C, Baulieu E. The use of the biotinyl estradiol-avidin system for the purification of ”Cnontransformed” estrogen receptor by biohormonal affinity chromatography. J Biol Chem. 1985;260:3996–4002. [PubMed] [Google Scholar]
- 14.Bayer E, Wilchek M. Insolubilized biotin for the purification of avidin. Methods Enzymol. 1974;34:265–267. doi: 10.1016/s0076-6879(74)34023-2. [DOI] [PubMed] [Google Scholar]
- 15.Zhu BT, Han GZ, Shim JY, Wen Y, Jiang XR. Quantitative structure-activity relationship of various endogenous estrogen metabolites for human estrogen receptor α and β subtypes: Insights into the structural determinants favoring a differential subtype binding. Endocrinology. 2006;147:4132–4150. doi: 10.1210/en.2006-0113. [DOI] [PubMed] [Google Scholar]
- 16.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engström O, Öhman L, Greene JL, Gustafsson J-Å, Carlquist M. Molecular basis of agonism and antagonismin the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 17.Tedesco R, Fiaschi R, Napolitano E. 6-Oxoestradiols from estradiols: Exploiting site selective mtalation of aralkyl systems with superbases. Synthesis. 1995:1493–1495. [Google Scholar]
- 18.Tedesco R, Katzenellenbogen JA, Napolitano E. An expeditious route to 7α-substituted estradiol derivatives. Tetrahedron Lett. 1997;38:7997–8000. [Google Scholar]
- 19.Muhlenbruch B, Kirmeier F, Roth HJ. Synthese und eigenschaften fluoreszierender estradiol-derivate. Arch Pharm (Weinheim) 1986;319:196–203. doi: 10.1002/ardp.19863190215. [DOI] [PubMed] [Google Scholar]
- 20.Primm TP, Gilbert HF. Hormone binding by protein disulfide isomerase, a high capacity hormone reservoir of the endoplasmic reticulum. J Biol Chem. 2001;276:281–286. doi: 10.1074/jbc.M007670200. [DOI] [PubMed] [Google Scholar]
- 21.Pike AC, Brzozowski AM, Walton J, Hubbard RE, Thorsell AG, Li YL, Gustafsson JA, Carlquist M. Structural insights into the mode of action of a pure antiestrogen. Structure. 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]