Abstract
End stage renal disease (ESRD) has a four times higher incidence in African Americans compared to European Americans. This led to the hypothesis that susceptibility alleles for ESRD have a higher frequency in West African than European gene pool. We performed a genome-wide admixture scan in 1,372 ESRD cases and 806 controls and demonstrated a highly significant association between excess African ancestry and non-diabetic ESRD (LOD 5.70) but not diabetic ESRD (LOD 0.47) on chromosome 22q12. Each copy of the European ancestral allele conferred a relative risk of 0.50 (95% credible interval 0.39 – 0.63) compared to African ancestry. Multiple common SNPs (allele frequency ranging from 0.2 to 0.6) in the gene that encodes non-muscle myosin heavy chain type II isoform A (MYH9) were associated with 2-4 times greater risk of non-diabetic ESRD and accounted for a large proportion of the excess risk of ESRD observed in African compared to European Americans.
End stage renal disease (ESRD) is the near-total loss of kidney function requiring treatment of 472,000 patients with dialysis or transplantation in the US1. Diabetes and hypertension are the two leading reported causes of treated ESRD in the U.S. accounting for 44% and 27% of incident cases respectively1. African Americans have consistently had a much higher rate of ESRD than European Americans in the US. In 2005, African-Americans had a 3.7 times higher age adjusted risk of ESRD. The risk ratio by assigned primary cause of ESRD was 3.8 for hypertension, 2.6 for diabetes, 2.3 for glomerulonephritis, 2.1 for the other causes of kidney disease1. While lower socioeconomic status and poorer access to health care explains some of this excess risk2-4, African Americans appear to have greater risk than European Americans after these factors are taken into account. Family studies show clustering of ESRD independent of hypertension and diabetes5, 6 with one large study shows stronger aggregation in African Americans6.
Studies attempting to detect susceptibility genes for ESRD and other complex diseases are challenging due to the late age of onset, causing difficulty in collecting multiply-affected families, and because linkage analysis has suggested that there are no genes of high penetrance (>4-fold increased risk) in populations of European descent, the focus of most published studies7, 8. For these reasons, ESRD is an excellent phenotype for whole genome association analysis, an approach with enhanced power to detect common variants of modest penetrance, and with the further advantage that unrelated individuals can be studied.
We performed a scan for ESRD genes using a particular type of whole genome association analysis, termed admixture mapping or mapping by admixture linkage disequilibrium (MALD)9-11. Admixture mapping is particularly suitable for finding genetic risk alleles that differ in frequency between populations which we hypothesized might be the case for ESRD.
The basic principle of admixture mapping relies on the small proportion of genetic variants that differ in frequency across populations of different ancestries12. When mixing occurs between genetically heterogeneous populations, the resulting admixed population inherits chromosomal regions of either one ancestry or the other, and these regions can be identified by genotyping markers that exhibit substantially different allele frequencies between ancestral populations. Admixture mapping methodology is feasible as a result of the development of a map of admixture mapping markers in African Americans13 and appropriate analytical methods14, 15. Moreover, the admixture mapping methodology has been validated as a promising way of finding susceptibility loci for common complex conditions, such as prostate cancer16 and multiple sclerosis17.
The central hypothesis of this study is that some ESRD susceptibility alleles are present at higher frequency in African than in European Americans. Thus, the identification of ESRD susceptibility alleles is possible by screening the genome in African Americans with ESRD, searching for regions of the genome where individuals with the disease have more (or less) African ancestry than their genome-wide average. Not only do previous studies provide evidence for a genetic basis to kidney disease and ESRD7, 18, it has been hypothesized that there may exist a common set of susceptibility genes for progression to ESRD irrespective of the inciting cause, e.g. diabetic or non-diabetic19. A secondary goal of our study was to test the hypothesis that the genetic variants that confer a higher risk for ESRD in African Americans are relevant to a broad range of ESRD phenotypes.
RESULTS
Initial admixture scan using all ESRD cases identified suggestive genome-wide significance on chromosome 22
A total of 1,372 ESRD cases and 806 controls without the presence of either an elevated serum creatinine concentration or albuminuria at recruitment were included in the initial scan using 1,354 markers. The mean age of ESRD cases and non-ESRD controls was 53 (SD = 13) and 46 (SD = 12) years, respectively. Fifty-three percent of cases were male, compared to 39% of controls.
The mean age of dialysis initiation in the 841 cases was 48 years (SD = 13), and these individuals spent an average of 3 years on dialysis (information was not available in all participants). Approximately half of the ESRD cases (n=703; 51%) had diabetes (75% with known types classified as type 2 diabetes) as the etiology of ESRD, and 669 had non-diabetic causes. The mean diabetes duration was 21 years (SD = 9.6) with a mean age of onset of 36 years (SD = 13 among the 591 cases with diabetic ESRD. The majority of participants with non-diabetic ESRD had hypertension reported as the cause of ESRD (n=347; 25% of all ESRD), 87 had focal segmental glomerulosclerosis (FSGS), 69 Human Immunodeficiency Virus (HIV)-related nephropathy, 126 had other glomerulonephritis from primary or systemic diseases, and 40 had other causes. The mean duration of hypertension was 13 years among 266 hypertensive ESRD cases.
The initial scan of the 1,372 ESRD cases and 806 controls with 1,354 ancestry informative markers with 12 pre-specified European ancestry risk models ranging from 0.3 to 1.5, in increments of 0.1 and a control risk of 1. The genome-wide LOD score, which was averaged across all risk models to account for multiple comparison revealed a suggestive association with ESRD on chromosome 22 (genome wide score of 1.67, where values greater than 2 are considered genome-wide significant; Table 1). In the region of rs4417800 on chromosome 22, the single point log of the odds (LOD) score was 4.63 (highest of all chromosomes), and the highest scoring risk model was of 1 copy of the European ancestral allele conferring a 0.7-fold risk of ESRD compared to the equivalent with African ancestry (reference group).
Table 1.
Summary of results from admixture scans in African Americans with end stage renal disease (ESRD)
| ESRD phenotype |
SNPs | Cases/Controls | Genome Score |
LOD at Chr22 peak |
|---|---|---|---|---|
| All ESRD | 1354 | 1372/806 | 1.67 | 4.55 |
| DM | 1351 | 703/806 | 0.47 | −0.52 |
| Non-DM | 1354 | 669/806 | 5.70* | 8.56 |
| HTN | 1352 | 347/806 | −0.16 | 1.79 |
| FSGS | 1350 | 87/806 | 0.10 | 2.47 |
| GN | 1351 | 126/806 | 0.01 | 1.75 |
| HIV | 1348 | 69/806 | 0.11 | 2.09 |
ESRD=end stage renal disease; DM=diabetic; Non-DM = non-diabetic; FSGS=focal segmental glomerulosclerosis; HTN = hypertensive; GN = glomerulonephritis; HIV=Human immunodeficiency virus nephropathy
Non-DM includes HTN, FSGS, HIV, GN, and 40 cases with other causes
Genome scores above 2.0 are considered significant after correcting for multiple comparison on a genome-wide level; scores above 1.0 are suggestive
Significant admixture-generated signal was observed on chromosome 22 for non-diabetic ESRD but not for diabetic ESRD
To determine whether susceptibility genes for diabetic ESRD and non-diabetic ESRD were different, admixture scans limited to either diabetic (n=703) or non-diabetic ESRD (n=669) were performed as an a priori subgroup analysis. The same group of 806 non-ESRD controls was used for both scans. No genome-wide significant association was observed in the diabetic ESRD scan (genome-wide score of 0.47; Table 1), with the strongest locus-specific signal arising from chromosome 11 (LOD=2.79, which does not meet the threshold of locus-specific LOD>5 for evidence of association; Table 2 and Figure 1a).
Table 2.
Summary of genome wide scan results among African Americans with end stage renal disease by subtypes (highest LOD score by chromosome)
| Ch | DM | Non-DM | HTN | FSGS | GN | HIV |
|---|---|---|---|---|---|---|
| 1 | 0.64 | 1.5 | 0.95 | 0.16 | 0.39 | 0.8 |
| 2 | 1.7 | 1.45 | 0.86 | 0.4 | 1 | 0.56 |
| 3 | −0.16 | −0.37 | −0.03 | 0.39 | 1.45 | 0.28 |
| 4 | 0.47 | 0.25 | −0.11 | 0.26 | 0.37 | 0.14 |
| 5 | 0.31 | −0.26 | −0.17 | 0.45 | 0.16 | 0.52 |
| 6 | 0.55 | 2.41 | 1.23 | 0.58 | 0.4 | 0.39 |
| 7 | −0.07 | −0.25 | 0.69 | 0.9 | 0.91 | 0.29 |
| 8 | 1.09 | 0.75 | 1.03 | 0.21 | 0.67 | 0.53 |
| 9 | −0.28 | 0.14 | −0.17 | 0.1 | 0.42 | 0.23 |
| 10 | 0.12 | 1.49 | 0.81 | 0.32 | 0.61 | 0.72 |
| 11 | 2.79 | −0.37 | 0.76 | 0.03 | 0.4 | 0.06 |
| 12 | 1.17 | 0.97 | −0.05 | 0.37 | 1.3 | 0.31 |
| 13 | 1.37 | 0.7 | 0.3 | 0.61 | 0.25 | 1.34 |
| 14 | 0.9 | 0.33 | 0.16 | 0.22 | −0.11 | 0.81 |
| 15 | 1.06 | −0.28 | 0.33 | 0.41 | −0.15 | 0.25 |
| 16 | 0.34 | −0.53 | −0.23 | 0.38 | 0.01 | 0.47 |
| 17 | 0.37 | 0.18 | 0.3 | 0.1 | 0.8 | 0.35 |
| 18 | 0.41 | 0.03 | −0.28 | 0.57 | 0.06 | 0.42 |
| 19 | −0.23 | 0.46 | −0.3 | 0.54 | 0.41 | 0.01 |
| 20 | 0.5 | −0.28 | −0.36 | 0.18 | 0.08 | 0.92 |
| 21 | 0.68 | 0.15 | 0.67 | 0.1 | 0.62 | 0.01 |
| 22 | −0.52 | 8.56 | 1.79 | 2.47 | 1.75 | 2.09 |
| X | 0.82 | 0.32 | 0.15 | 0.11 | 0.74 | 0.94 |
DM= diabetic; Non-DM= non-diabetic; HTN = hypertensive; FSGS = focal segmental glomerular sclerosis; GN = glomerulonephritis; HIV=Human immunodeficiency virus nephropathy
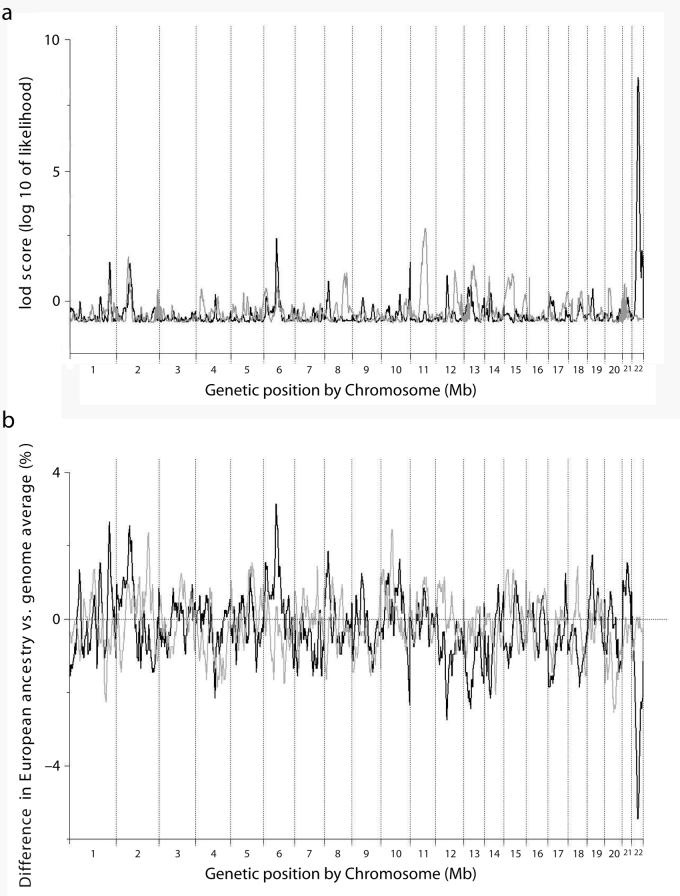
Figure 1.
(a) LOD score from genome-wide scan for diabetic (gray) and non-diabetic (black) ESRD. The genome-wide score for non-diabetic ESRD was 5.70 and 0.47 for diabetic ESRD. The highest locus-specific LOD for diabetic ESRD was 2.79 on chromosome 11 and 8.56 on chromosome 22 for non-diabetic ESRD.
(b) Difference between estimated European ancestry at different loci of the genome and the genome-wide average for 669 non-diabetic ESRD (black) and 806 controls (gray). The estimated European ancestry on chromosome 22 was lower than the average across the entire genome among non-diabetic ESRD cases but not controls.
In contrast, a highly significant genome-wide score of 5.70 was observed for the non-diabetic ESRD subset. The highest single point LOD score (LOD 8.56) was detected on chromosome 22 for the non-diabetic ESRD analysis (Table 2 and Figure 1a). The risk model with the strongest score corresponded to 1 copy of the European ancestral allele conferring a risk of 0.50 (95% credible interval 0.39 – 0.63) compared to African ancestry (reference group). Upon further narrowing of the risk models (ranging from 0.4 to 0.6 in increments of 0.02), we obtained a genome-wide score of 9.31 in this region. In addition, the estimated European ancestry in this region on chromosome 22 was lower than the average across the entire genome among non-diabetic ESRD cases but not controls (Figure 1b). The case for the strong difference between the diabetic and the non-diabetic ESRD cohorts was further strengthened by a case-control analysis in which the non-diabetic ESRD patients were used as cases and the diabetic ESRD cases were used as controls at the locus on chromosome 22. This analysis also showed a highly significant increase in African ancestry in non-diabetic ESRD, compared to diabetic ESRD cases at chromosome 22 peak (T-statistic = −10.6, P=2.94×10−15).
To assess the robustness of the chromosome 22 peak, we further evaluated whether the evidence for association was dependent on a particular marker. We divided the data into even and odd-numbered SNPs. Each set of SNPs independently revealed evidence of association (genome-wide score = 6.32 for even-numbered SNP scan and genome-wide score = 5.57 for odd-numbered SNP scan).
To assess whether the significant signal from the non-diabetic ESRD scan was generated by one particular subtype of ESRD, we performed additional admixture scans for the major subtypes of non-diabetic ESRD: hypertensive ESRD and FSGS (case numbers for glomerulonephritis and HIV-related ESRD were smaller). At the same locus on chromosome 22q (rs5999762), the best risk models, as indicated by relative risks, and the corresponding locus-specific LOD scores were: 0.5 (LOD 8.467) for all non-diabetic, 0.8 (LOD 1.70) for hypertensive, and 0.35 (LOD 2.48) for focal and segmental glomerulosclerosis (FSGS) ESRD. Although the risk models were not identical for different ESRD types, with the strongest effect observed for FSGS, the signals of association were consistent across all ESRD types, with the strongest association signals generated in the vicinity of rs5999762 from the entire admixture SNP panel. The evidence of admixture signal was present for HIV nephropathy but much weaker for secondary glomerol glomerulonephritis (Table 1).
The 95% credible interval of the admixture-generated signal on chromosome 22q12 contained a biological positional candidate – MYH9
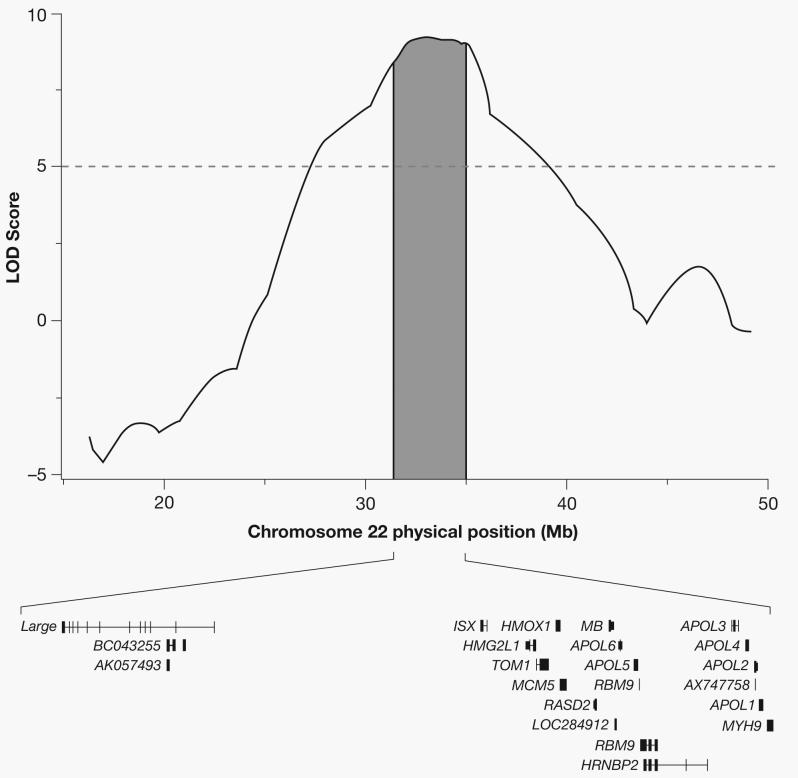
Given the consistency of a signal across different groups of non-diabetic ESRD cases, we calculated a confidence interval for the position of the underlying genetic variant(s) contributing to the increased risk for non-diabetic ESRD in African Americans. By identifying the region where the LOD score is within 0.86 of its maximum, we estimate that an approximate 95% credible interval for the region is 31.388650 – 35.039798 Mb (Figure 2). The region contains 22 genes (17 known). While many are expressed in the kidney only one, the gene encoding the non muscle myosin heavy chain 9 (MYH9), is the most highly expressed compared to the other 16 known genes in this region20. Moreover, we chose to study MYH9 as a biological positional candidate gene based on the existing knowledge of the importance of MYH9 mutations in a number of kidney diseases21-25 and ongoing collaborations between authors of the present study and a concurrent study of FSGS in African Americans. They also identified a region of interest on chromosome 22 and further established MYH9 as a risk factor for FSGS and subsequently hypertensive ESRD26. Fourteen SNPs in MYH9 known to have high allele frequency differences between the Caucasian samples (CEU) from Utah and the African samples from Yoruba (YRI) from the International HapMap Project and significantly associated with FSGS and hypertensive ESRD in the Kopp et al., study were genotyped. All fourteen MYH9 SNPs were highly significantly associated with non-diabetic ESRD using either the conservative general genetic model parameterizing three genotype-specific risks (Supplementary Tables 1 and 2) or a recessive genetic model (p values as low as 3.63×10−15after correcting for global ancestry, Table 3) at a Bonferroni-corrected alpha level of 0.003 (0.05/14). As anticipated, none of the SNPs were associated with diabetic ESRD (lowest p value 0.03 for rs16996677 after adjustment for global ancestry, Table 3). The ORs for the MYH9 SNPs between diabetic and non-diabetic ESRD were quite different. In a case-case comparison of the odds of the at-risk genotype in diabetic ESRD cases compared to the odds of the at-risk genotype in non-diabetic ESRD cases showed that all but one SNP (rs875725) were highly associated with the cause of ESRD, with diabetic ESRD cases less likely to carry the at-risk genotype for each SNP (Supplementary Table 3). Lastly, the Breslow-Day test of heterogeneity for associations between MYH9 SNPs and ESRD case-control status by diabetes status showed significant differences in association between SNPs and ESRD by diabetes for rs735853 [p=5.38×10−4], rs4821480 [p=1.21×10−5], rs2032487 [p=1.93×10−5], and rs4821481 [p=3.32×10−6] (Supplementary Table 4).
Figure 2.
Region of association with non-diabetic ESRD on chromosome 22. The peak LOD score was 9.31, and the 95% credible interval spanned from 31.388650 to 35.039798 Mb, covering 22 genes.
Table 3.
Adjusted* odds ratio (OR) and 95% confidence interval (95% CI) of non-diabetic (Non-DM) and diabetic (DM) ESRD for 14 MYH9 SNPs
| Non-DM | DM | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | position | alleles | MAF (%) |
ref allele | OR | 95% CI | P value1 | P value2 | OR | 95% CI | P value1 | P value2 |
| rs7078 | 35007860 | G/A | 15.3 | G | 1.97 | (1.52,2.55) | 3.41×10−7 | 1.58×10−5 | 1.28 | (1.01,1.62) | 0.04 | 0.13 |
| rs12107 | 35007928 | A/G | 9.3 | A | 1.57 | (1.16,2.12) | 3.64×10−3 | 0.01 | 0.90 | (0.69,1.18) | 0.45 | 0.76 |
| rs735853 | 35009161 | G/C | 10.2 | G | 1.90 | (1.38,2.62) | 9.05×10−5 | 0.14 | 0.91 | (0.69,1.19) | 0.47 | 0.77 |
| rs5756129 | 35014038 | C/T | 19.3 | C | 1.71 | (1.35,2.17) | 7.92×10−6 | 3.21×10−5 | 1.06 | (0.85,1.32) | 0.63 | 0.89 |
| rs5756130 | 35014277 | T/C | 12.5 | T | 1.58 | (1.2,2.08) | 1.20×10−3 | 1.00×10−3 | 1.00 | (0.78,1.28) | 0.99 | 0.99 |
| rs875725 | 35021637 | T/C | 6.0 | C | 1.17 | (0.84,1.65) | 0.35 | 0.29 | 0.91 | (0.66,1.25) | 0.55 | 0.84 |
| rs4821480 | 35025193 | T/G | 29.5 | T | 2.18 | (1.73,2.73) | 1.72×10−11 | 1.36×10−5 | 0.95 | (0.75,1.19) | 0.65 | 0.90 |
| rs2032487 | 35025374 | T/C | 33.9 | T | 2.41 | (1.94,3.01) | 4.51×10−15 | 2.78×10−8 | 1.11 | (0.89,1.38) | 0.35 | 0.64 |
| rs4821481 | 35025888 | T/C | 34.7 | T | 2.40 | (1.93,2.98) | 3.63×10−15 | 2.80×10−8 | 1.07 | (0.86,1.33) | 0.53 | 0.82 |
| rs3752462 | 35040129 | C/T | 25.6 | C | 2.11 | (1.69,2.63) | 5.19×10−11 | 1.98×10−5 | 1.08 | (0.87,1.33) | 0.47 | 0.77 |
| rs5756152 | 35042418 | A/G | 28.7 | G | 2.59 | (1.88,3.55) | 4.49×10−9 | 2.25×10−6 | 1.28 | (0.9,1.83) | 0.16 | 0.38 |
| rs1005570 | 35045220 | A/G | 44.9 | G | 2.45 | (1.93,3.12) | 2.56×10−13 | 4.33×10−9 | 1.27 | (0.98,1.63) | 0.07 | 0.19 |
| rs16996674 | 35056598 | T/C | 23.4 | C | 3.10 | (2.15,4.47) | 1.49×10−9 | 4.14×10−7 | 1.51 | (1.01,2.27) | 0.04 | 0.13 |
| rs16996677 | 35057229 | A/G | 26.8 | G | 3.03 | (2.16,4.25) | 1.61×10−10 | 1.47×10−7 | 1.50 | (1.03,2.17) | 0.03 | 0.10 |
OR and p value1 adjusted for global ancestry; P value2 adjusted for both global and local ancestry; recessive genetic model assumed
MAF = minor allele frequency in African Americans, refers to the allele listed first in the column “alleles”
Ref allele refers to allele used as the reference group in the logistic regression
After adjusting for both global and local ancestry, all but 3 of the SNPs remained highly significantly associated with non-diabetic ESRD, with P-values as low as 4.33×10−9 (Table 3). Moreover, these SNPs were also associated with both hypertensive and FSGS ESRD, with the magnitude of association between markers and FSGS ESRD being larger than that for hypertensive ESRD; however, the significance of the association was reduced due to the smaller sample size of FSGS cases (Table 4).
Table 4.
Adjusted* odds ratio (OR) and 95% confidence interval (95% CI) of hypertensive (HTN) and focal segmental glomerulosclerosis (FSGS) ESRD for 14 MYH9 SNPs
| HTN | FSGS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | position | alleles | MAF (%) |
ref allele | OR | 95% CI | P value1 | P value2 | OR | 95% CI | P value1 | P value2 |
| rs7078 | 35007860 | G/A | 15.3 | G | 1.70 | (1.24,2.33) | 1.04×10−3 | 4.03×10−3 | 3.52 | (1.73,7.16) | 5.17×10−4 | 1.85×10−3 |
| rs12107 | 35007928 | A/G | 09.3 | A | 1.34 | (0.93,1.93) | 0.12 | 0.16 | 2.95 | (1.26,6.91) | 0.01 | 0.02 |
| rs735853 | 35009161 | G/C | 10.2 | G | 2.08 | (1.36,3.16) | 6.79×10−4 | 0.02 | 1.79 | (0.86,3.7) | 0.12 | 0.82 |
| rs5756129 | 35014038 | C/T | 19.3 | C | 1.62 | (1.21,2.18) | 1.19×10−3 | 2.04×10−3 | 2.81 | (1.55,5.07) | 6.35×10−4 | 1.19×10−3 |
| rs5756130 | 35014277 | T/C | 12.5 | T | 1.69 | (1.19,2.4) | 3.45×10−3 | 3.36×10−3 | 2.42 | (1.19,4.93) | 0.01 | 0.02 |
| rs875725 | 35021637 | T/C | 06.0 | C | 1.01 | (0.67,1.51) | 0.97 | 0.90 | 3.69 | (1.14,11.94) | 0.03 | 0.03 |
| rs4821480 | 35025193 | T/G | 29.5 | T | 2.07 | (1.56,2.74) | 3.7×10−7 | 3.18×10−5 | 3.66 | (2.11,6.34) | 3.98×10−6 | 4.54×10−3 |
| rs2032487 | 35025374 | T/C | 33.9 | T | 2.28 | (1.74,2.98) | 2.38×10−9 | 5.77×10−7 | 4.85 | (2.82,8.33) | 1.09×10−8 | 6.14×10−5 |
| rs4821481 | 35025888 | T/C | 34.7 | T | 2.32 | (1.77,3.03) | 7.81×10−10 | 4.37×10−7 | 4.34 | (2.61,7.21) | 1.45×10−8 | 9.57×10−5 |
| rs3752462 | 35040129 | C/T | 25.6 | C | 2.00 | (1.52,2.64) | 6.79×10−7 | 9.52×10−5 | 3.50 | (2.01,6.07) | 8.65×10−6 | 2.53×10−3 |
| rs5756152 | 35042418 | A/G | 28.7 | G | 2.54 | (1.76,3.68) | 7.71×10−7 | 1.59×−5 | 4.63 | (2.73,7.87) | 1.45×10−8 | 2.44×10−6 |
| rs1005570 | 35045220 | A/G | 44.9 | G | 2.07 | (1.55,2.76) | 9.18×10−7 | 3.54×10−5 | 4.49 | (2.81,7.18) | 3.45×10−10 | 3.63×10−7 |
| rs16996674 | 35056598 | T/C | 23.4 | C | 2.78 | (1.81,4.27) | 2.83×10−6 | 3.56×10−5 | 6.84 | (3.91,11.97) | 1.69×10−11 | 6.82×10−9 |
| rs16996677 | 35057229 | A/G | 26.8 | G | 2.60 | (1.74,3.87) | 2.60×10−6 | 5.98×10−5 | 5.38 | (3.09,9.35) | 2.55×10−9 | 8.07×10−7 |
OR and p value1 adjusted for global ancestry; P value2 adjusted for both global and local ancestry; recessive genetic model assumed
MAF = minor allele frequency in African Americans, refers to the allele listed first in the column “alleles”
Ref allele refers to allele used as the reference group in the logistic regression
Common SNPs in MYH9 accounted for the significant admixture signal on chromosome 22
To determine the extent to which the MYH9 SNPs could account for the significant admixture signal in this region (as indicated by the highly significant association between estimates of local ancestry and non-diabetic ESRD case-control status shown in Figure 3), we performed two logistic regression analyses per marker. For the first analysis we estimated the association between local ancestry and non-diabetic ESRD case-control status. For the second analysis we estimated the ancestry association to having the disease after the inclusion of one of the fourteen MYH9 markers as covariates. The latter analysis estimates the residual risk of non-diabetic ESRD associated with admixture after controlling for each of the 14 SNPs in MYH9 gene.
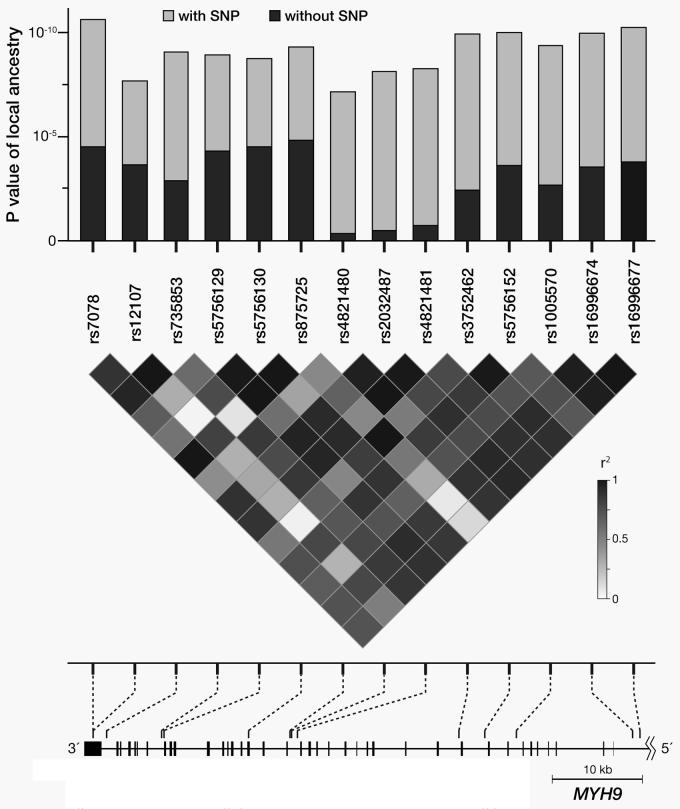
Figure 3.
Effect of MYH9 SNPs on the association between local ancestry in the region of chromosome 22q12 and non-diabetic ESRD. The results of the local ancestry estimates were obtained from 14 different ANCESTRYMAP runs, each containing one of the 14 SNPs at a time. The light gray bars indicate the highly significant associations between local ancestry at the locus of interest on chromosome 22q12 and case-control status. The dark gray bars show the greatly reduced associations between local ancestry and case-control status after the inclusion of MYH9 SNP in the logistic regression model. Three SNPs (rs4821480, rs2032487, and rs4821481), in particular, accounted for almost the entire significance of the association between local ancestry and case-control status. The LD pattern of 14 SNPs in MYH9 shown is that of the African-American FIND and CHOICE participants. In addition, known rare mutations Mendelian conditions are shown for the region of MYH9 containing the 14 SNPs studied.
Figure 3 shows that any of the three SNPs rs4821480, rs2032487, and rs4821481, with pair-wise r2 > 0.96 (Figure 3), is sufficient to account for all of the association between the excess African ancestry observed on chromosome 22q12 and non-diabetic ESRD. More specifically, when the markers are included in the logistic regression, the p-value for the association between local ancestry and non-diabetic ESRD fell to 0.34, 0.47, and 0.71 after adjusting for rs4821480, rs2032487, and rs4821481, respectively.
For rs2032487, the frequency of the at-risk allele (C) was 0.66 in African Americans. Based on the International HapMap Project, the frequency of the allele is estimated to be 75.8% in West Africans from Yoruba (YRI) and 0% in Asians from both Tokyo Japan (JPT) and Beijing China (HCB) but was not typed in the Caucasians from Utah (CEU). However, the frequencies of rs4821481 and rs4821480 (both in almost perfect LD with rs2032487 in our population) were known for both YRI and CEU. Both the G allele of rs4821480 and the C allele of rs4821481 had a frequency of 75.8% in YRI but only 6.1% in CEU. These three SNPs represent highly unusual difference (above the 99th percentile of autosomal SNPs in terms of allele frequency differentiation). Thus, the strong correlation of the allele to ancestry explains the strength of the admixture signal.
Multiple MYH9 SNPs remained significantly associated with non-diabetic ESRD after controlling for ancestry
As expected, not only did MYH9 SNPs account for the admixture signal on chromosome 22 but they were significantly associated with non-diabetic ESRD even after accounting for difference in global ancestry. To understand the contribution of alleles to the phenotype16, we estimated the odds ratio associated with each SNP for non-diabetic ESRD after controlling for only for global, or genome-wide, ancestry. Multiple clusters of SNPs remained highly significant. For example, the odds ratio associated with two copies of the C allele compared to those with either one or two copies of the T allele for rs2032487 (one of the SNP that accounted for the admixture generated signal) is 2.41 (95% CI: 1.94, 3.01; p= 4.51×10−15, Table 3) for all non-diabetic ESRD, 2.28 (95%CI: 1.74, 2.98; p= 2.38×10−9) for hypertensive ESRD, and 4.85 (95% CI: 2.82, 8.33; p= 1.09×10−8) for FSGS (Table 4). Sixty-four percent of all non-diabetic ESRD cases had the CC genotype compared to 42% in the controls.
Interestingly, further upstream of rs2032487 were two other common SNPs (rs169966774 with a frequency of 23.4% for the high-risk allele [T] and rs16996677 with a frequency of 26.8% for the high-risk allele [A]) in intron 3 that were in almost perfect LD with each other and were even more significantly associated with non-diabetic ESRD after adjusting for global and local ancestry. Assuming a three-genotypic risk model, those with either the AG or AA genotype at rs16996677 were about 1.63 (95% CI: 1.29 to 2.05) and 3.82 (95% CI: 2.67 to 5.47) more likely to have non-diabetic ESRD than their counterparts with the GG genotype (Supplementary Table 1). The frequency of the AA and AG genotypes were 19% and 43%, respectively, in non-diabetic ESRD cases but only 7% and 39% in the controls. The excess risk of non-diabetic ESRD associated with the A allele was also observed using a recessive model where individuals with the AA genotype were about 3.03 times more likely to have non-diabetic ESRD than their counterparts who had either the GG or AG genotype (95%CI: 2.16 to 4.25; p= 1.61×10−10; Table 3). Upon cross tabulation of rs2032487 and rs16996677, both SNPs remained independently associated with non-diabetic ESRD. Although the two SNPs are correlated, they are not in complete LD(r2 ∼ 0.88), suggesting that these two SNPs are capturing a haplotype that contains the causal variant. Lastly, both adjusted and stratified analyses to account for the effect of ancestry on rs2032487 and rs16996677 were performed. Both SNPs remained highly significantly associated with non-diabetic ESRD even after adjusting for both global and local ancestry (Table 4 for the odds ratios that adjusted for only global ancestry and Supplementary Table 2 for the odds ratios that adjusted for both global and local ancestry). In analyses that was stratified by ancestral background of this region of the chromosome (i.e. 0, 1, or 2 copies of the African ancestral chromosome), both SNPs remained associated with non-diabetic ESRD on either the background of 1 or 2 copies of the African ancestral chromosome (odds ratios were not estimated for those with zero copies of the African ancestral chromosome because only about 1% of the population had zero copies of the African ancestral chromosome in this region and about 84% of the population had two copies of the African ancestral chromosome; Supplementary Table 7).
DISCUSSION
In this genome-wide admixture scan of 1,372 ESRD cases and 806 controls, we have identified a locus on chromosome 22q12 that demonstrated a significant excess of African ancestry among non-diabetic ESRD cases compared to controls without nephropathy. Moreover, we showed that multiple SNPs in MYH9 can account for the admixture-generated signal in this region. Although the evidence for MYH9 harboring one or more susceptibility alleles for non-diabetic ESRD in African Americans is exceedingly strong, it is not clear that the genetic variant responsible (the causal variant) has yet been identified since none of the SNPs typed in the present study are known to impact either gene transcription or translation directly. Moreover, multiple MYH9 SNPs, including rs2032487 and rs16996677, were correlated with each other and were highly significantly associated with non-diabetic ESRD. Thus, it is likely that our results have simply identified genetic variants that are in strong linkage disequilibrium with the causal variant.
An intriguing result of the admixture scan is that independent of the knowledge of the causal SNP, the results allow us to estimate how much of the epidemiologically increased risk for non-diabetic ESRD in African Americans as compared with European Americans can be accounted for by this locus. Using estimates and the assumptions made in the admixture scan, we calculate that compared with individuals who have entirely African ancestry, individuals with 1 European chromosome at the locus are estimated to have 0.50 times the prevalence (95% credible interval 0.39-0.63), and individuals with 2 European chromosomes have 0.5×0.5 = 0.25 the prevalence. We estimate from this that if it were possible to reduce the prevalence of non-diabetic ESRD in African Americans (assuming 16.9% European ancestry on average), to the rate that would be expected if all the African Americans had inherited European ancestry at the locus, the prevalence of the non-diabetic ESRD in African Americans would decrease to 30% (95% credible interval of 19-45%) of its current estimate (Supplementary Table 5 contains details of this calculation).
The lack of association between MYH9 SNPs and diabetic ESRD is unlikely the result of low statistical power. With 703 diabetic ESRD cases and 806 controls, this study would have had close to 100% power to detect an odds ratio of 2.4 (the odds ratio associated with both rs2032487 and rs4821481, which has an r2 ∼ 1 with rs4821480) assuming that the frequency of the at-risk genotype is 36% in the controls (percent of individuals who are homozygous for the at-risk allele at either SNP) and an alpha of 0.003. The marked contrast in results for diabetic versus non-diabetic ESRD suggests that mutations in MYH9 are not strongly associated with diabetic ESRD, especially ESRD associated with type 2 diabetes. In diabetic ESRD, unlike other types of ESRD, the initiation of renal damage is due to hyperglycemia with an increase in glomerular filtration rate and mesangial proliferation. The inexorable progression of renal disease, once renal damage occurs, led to the hypothesis that there are common underlying pathogenic pathways leading to ESRD19. While the lack of association of MYH9 in this study with diabetic kidney disease does not disprove the hypothesis that there are common mechanisms for progression of all forms of chronic kidney disease, it shows that there are at least some mechanisms of ESRD progression that are unique to specific diseases. In particular, the results indicate that MYH9, while important in non-diabetic ESRD, is not important for progression of diabetic ESRD, although the present study was not able to test whether MYH9 may have differential effects on progression to ESRD depending on the type of diabetes (type 1 versus type 2). Thus, genetic variation in MYH9 may only be important in the kidney's response to injury not caused by hyperglycemia.
MYH9 encodes for the protein nonmuscle myosin heavy chain, class II, and isoform type A in eukaryotic cells. The gene is approximately 110 kb with 41 exons and is highly conserved among a number of mammalian species and very similar to other nonmuscle myosin isoforms27. The protein is abundantly expressed in the kidney, liver, and platelets. These proteins have a variety of cellular functions, such as cellular polarity, architecture, and trafficking27, 28. Rare mutations in MYH9 are associated with several Mendelian conditions, including an autosomal dominant form of deafness (OMIM 160775), Epstein syndrome (OMIM 153650), Fechtner syndrome (OMIM 153640), May-Hegglin anomaly (OMIM 155100), and Sebastian syndrome (OMIM 605249); moreover, many of these conditions may include renal disease (Figure 3).
Within the kidney, the expression occurs in the glomerulus, specifically the podocyte, peritubular capillaries and tubules. Aggregation of abnormal myosin and damage to the cytoskeleton of the podocyte and tubular cells could lead to progressive kidney disease; however, it remains to be determined how sequence variations in MYH9 directly result in development and progression of non-diabetic kidney disease.
In summary, common genetic variations in the MYH9 locus on chromosome 22q can account for much of the excess risk of non-diabetic ESRD observed in African Americans compared to European Americans, although environmental risk factors and interactions between genes and environment undoubtedly play an additional role as well. Moreover, there is also a distinct genetic susceptibility to non-diabetic ESRD among African Americans that is not evident among cases with diabetic ESRD, suggesting that the mechanisms leading from the initiation of chronic kidney disease to progression to ESRD may differ based on the inciting cause. Thus, drug therapies that are often targeted at common pathways to prevent kidney disease progression may need more specificity for effective treatment.
Finally, the identification of susceptibility alleles that exist in different frequencies between African Americans and European Americans has potential implications for future screening strategies for African Americans with non-diabetic kidney disease. Our finding suggests that many African-American individuals reported as having hypertensive ESRD may actually have an identifiable etiology other than hypertension. This may explain why strict blood pressure control often fails to slow the progression of purported hypertensive kidney disease in many African Americans29. The MYH9 gene suggests a novel pathway for kidney disease progression with major potential implications. Experiments should identify the causal variants, elucidate the pathophysiologic mechanisms, and evaluate strategies to influence this novel pathway that may decrease risk of progression to ESRD among African Americans. This can ultimately lead to identification of high risk individuals and the development of new approaches to combat the epidemic rise of ESRD in the US and other nations.
METHODS
Participants were recruited as part of the Family Investigation of Nephropathy and Diabetes (FIND) and the Choices for Healthy Outcomes In Caring for ESRD (CHOICE) study. The FIND study populations and design have been described in detail30. Briefly, participants were recruited from eleven participating centers. The African American MALD study recruited both diabetic and non-diabetic ESRD cases and spouse/partner controls. Subjects were recruited and samples collected according to the declaration of Helsinki principles, and a certificate of confidentiality was filed at the National Institutes of Health. The CHOICE study is a national prospective cohort study of 1,041 ESRD participants (294 were African American) from 81 dialysis clinics associated with Dialysis Clinic, Incorporated, New Haven CAPD and the Hospital of St. Raphael which has been described previously31. The study was approved by the Institutional Review Boards at Johns Hopkins University, and the associated clinical sites, and participants provided written informed consent.
From both studies, a total of 1,427 ESRD cases (883 from FIND African-American MALD study; 299 from FIND Family study; 245 from the CHOICE study) and 839 controls (only from the African-American FIND MALD study) were included. Of these 2,256 cases and controls, 36 individuals were dropped during routine data check in ANCESTRYMAP, 17 were dropped for having potential gender inconsistencies between self-reported gender and probability of heterozygosity states on markers on the X chromosome, and 25 were dropped for having low genotype calling rates.
Diabetes (primarily type 2 diabetes) was defined as self-reported and/or prevalent treatment with insulin and/or oral hypoglycemic agents or documented in the medical history. Diabetes, hypertension and kidney disease duration were obtained from the medical history and by medical record review. Information on specific type of diabetes obtained from medical chart abstraction was available on 160 of the diabetic ERSD cases: 118 (74%) of them had type 2 diabetes, 22 had type 1 diabetes (14%), and 20 (12%) had unknown type of diabetes listed on their medical records. Only cases recruited from the Johns Hopkins FIND site had medical chart abstraction review to classify diabetes. Age of onset for diabetes, hypertension and kidney disease was by self report. ESRD due to diabetic nephropathy was defined by having 2 of the following 3 clinical criteria (a) onset of diabetes ≥5 years prior to renal replacement therapy (b) documented diabetic retinopathy or (c) proteinuria (>3.0 g protein/g creatinine). ESRD due to nondiabetic nephropathy was defined as biopsy proven non-diabetic kidney disease or by cause of ESRD abstracted from medical chart charts obtained during physician visits and hospitalizations. ESRD from hypertension was defined by one of the following: 1) evidence of hypertensive nephropathy based on previous biopsy findings or 2) the exclusion of all other causes of ESRD by the participants' primary nephrologist. In addition, in those cases of hypertensive ESRD without a biopsy, less than 6% had evidence of significant proteinuria of >3.0 g protein/g creatinine at the time of presentation. We also specifically excluded all congenital, known monogenic forms, and cancer-related renal diseases.
Genotyping
We attempted 1,536 single nucleotide polymorphisms (SNPs) using the Illumina BeadLab platform for the initial admixture mapping scan. Details of the genotyping for this experiment are described by Nalls et al.32 This panel was constructed mainly by using the panel of ancestry informative markers from previously published map by Smith et al.13 and then improving this panel by mining new ancestry informative markers from the data sets of supplemented with >1,500 new markers identified by Hinds et al.33 and the Phase 2 International Haplotype Map34, and validating them to confirm that they were indeed ancestry informative. These SNPs were then prioritized as most informative about West African versus European ancestry, according to their predicted usefulness for determining ancestry.After quality checks, including the requirement that at least 85% of SNPs were successfully genotyped for each sample and that none of the SNPs were in linkage disequilibrium in the ancestral populations, 182 SNPs were excluded from analysis (call rates for each of the SNPs kept are shown in Supplementary Table 6). The physical genome positions used in this study are based on build 35 of the public genome reference. To obtain the genetic positions, we used the Rutgers Integrated Map35 as previously described by Reich et al.17. Using the TaqMan pre-designed genotyping assays (Applied Biosystems, Foster City, CA) under standard conditions, we genotyped an additional 14 SNPs in MYH9, selected for having a pronounced frequency difference between reference African (YRI) and European (CEU) populations.
Admixture scan
ANCESTRYMAP was used to perform admixture mapping analyses. This program uses a Markov Chain Monte Carlo-based (MCMC) methodology14. To accumulate evidence of association in these models, we averaged the Bayes factors emerging from each model at each point in the genome, taking the log-base-10 of this number to produce a LOD score. As reported in Reich and Patterson (2005), a LOD score for association at a particular locus of >5 is approximately genome-wide significant. To obtain a formal assessment of statistical significance on a genome wide level, we calculated an additional statistic that averaged the risks specified in the models as genome wide Bayes factors and took the log-base-10; a value >2 indicates statistically significant association to the phenotype. For the initial scan, we ran ANCESTRYMAP for a burn-in period of 100 iterations with 200 follow-on iterations and averaged scores obtained from 12 pre-specified European ancestry risk models ranging from 0.3 to 1.5 in increments of 0.1. For example, the model testing a risk of 1.5 assumed a 1.5-fold increased risk due to inheritance of one copy of European ancestral allele for cases, with a control risk of 1. To calculate the 95% credible interval for the position of the disease locus once we found an association, we first summarized the evidence for association by taking the sum of the likelihood ratios across the entire locus. Then, starting at the peak of the locus, we moved in both directions until the region included 95% of the value of the sum.
In order to test for association between SNPs and ESRD above and beyond the confounding effect of ancestry association, we obtained local and global estimates of ancestry by using ANCESTRYMAP. Global ancestry was obtained using all of the markers from the initial scan. Estimate of local ancestry on MYH9 was obtained fourteen times, each time using one of the MYH9 SNPs along with the other SNPs from the original scan. Estimates obtained from these 14 SNPs were used to adjust for the effect of ancestry locally on the association between MYH9 SNPs and case-control status definition.
Association between SNPs in MYH9 and various case-control status definitions was determined by the means of odds ratios, 95% confidence intervals, and p values obtained from logistic regression models (SAS V.10). A general model parameterizing three genotypic risks was assumed for initial association analyses (Supplementary Tables 1 and 2). Based on ORs derived from the general models, we further tested and presented the results for the recessive model. A Bonferroni-corrected alpha of 0.003 (0.05/14 SNPs) was used to declare significance. The logistic regression was used to perform a case-case comparison to determine whether MYH9 SNPs were associated with diabetic ESRD (as opposed to non-diabetic ESRD). The Breslow-Day test for heterogeneity was used to determine with the associations between MYH9 SNPs and ESRD status differed significantly by diabetes status.
Supplementary Material
Acknowledgements
The authors thank the patients, staff, laboratory, and physicians who participated in the FIND and the CHOICE Study at Dialysis Clinic, Inc and at Johns Hopkins University. This study was also supported by the following research grants: U01DK070657, U01DK57304, K01DK067207 (WHLK), U01DK57292, and DK07024 (the CHOICE study) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and, in part, by the Intramural Research Program of the NIDDK. The CHOICE study was also supported in part by HS08365 from the Agency for Healthcare Research and Quality, Rockville, MD and HL62985 from the National Heart Lung and Blood Institute, Bethesda, MD. This work was supported by the National Center for Research Resources for the General Clinical Research Center (GCRC) grants: Case Western Reserve University M01-RR-000080; Wake Forest University M01-RR-07122; Harbor-UCLA Medical Center M01-RR-00425; College of Medicine - University of California Irvine M01-RR-00827-29; Frederic C. Bartter M01-RR-01346. DR was supported by a Burroughs Wellcome Career Development Award in the Biomedical Sciences.
This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Reference List
- 1.U.S.Renal Data System . In: USRDS 2007 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. National Institute of Diabetes and Digestive and Kidney Disease, editor. National Institute of Diabetes and Digestive and Kidney Disease; Bethesda, MD: 2007. [Google Scholar]
- 2.Klag MJ. End-stage renal disease in African-American and white men - 16-year MRFIT findings. JAMA. 1997;277:1293–1298. [PubMed] [Google Scholar]
- 3.Tarver-Carr ME, et al. Excess risk of chronic kidney disease among African-American versus white subjects in the United States: A population-based study of potential explanatory factors. Journal. of. the. American. Society. of. Nephrology. 2002;13:2363–2370. doi: 10.1097/01.asn.0000026493.18542.6a. [DOI] [PubMed] [Google Scholar]
- 4.Perneger TV, Whelton PK, Klag MJ. Race and end-stage renal disease. Socioeconomic status and access to health care as mediating factors. Arch Intern Med. 1995;155:1201–1208. [PubMed] [Google Scholar]
- 5.Lei HH, Perneger TV, Klag MJ, Whelton PK, Coresh J. Familial aggregation of renal disease in a population-based case- control study. J. Am. Soc. Nephrol. 1998;9:1270–1276. doi: 10.1681/ASN.V971270. [DOI] [PubMed] [Google Scholar]
- 6.Freedman BI, Spray BJ, Tuttle AB, Buckalew VM. The familial risk of end-stage renal disease in African Americans. Am J Kidney Dis. 1993;21:387–393. doi: 10.1016/s0272-6386(12)80266-6. [DOI] [PubMed] [Google Scholar]
- 7.Fox CS, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study 3188. J Am Soc. Nephrol. 2004;15:2457–2461. doi: 10.1097/01.ASN.0000135972.13396.6F. [DOI] [PubMed] [Google Scholar]
- 8.Hunt SC, et al. Linkage of serum creatinine and glomerular filtration rate to chromosome 2 in Utah pedigrees. Am. J. Hypertens. 2004;17:511–515. doi: 10.1016/j.amjhyper.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens JC, Briscoe D, O'Brien SJ. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am. J. Hum. Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 11.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am. J. Hum. Genet. 1998;63:241–251. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 13.Smith MW, et al. A high-density admixture map for disease gene discovery in african americans. Am J Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson N, et al. Methods for high-density admixture mapping of disease genes. Am J Hum. Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann. Hum. Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 16.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich D, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nature Genetics. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 18.Iyengar SK, et al. Linkage analysis of candidate loci for end-stage renal disease due to diabetic nephropathy. J. Am. Soc. Nephrol. 2003;14 doi: 10.1097/01.asn.0000070078.66465.55. [DOI] [PubMed] [Google Scholar]
- 19.Remuzzi G, Benigni A, Remuzzi A. Mechanisms of progression and regression of renal lesions of chronic nephropathies and diabetes. J. Clin. Invest. 2006;116:288–296. doi: 10.1172/JCI27699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunishima S, Matsushita T, Hamaguchi M, Saito H. Identification and characterization of the first large deletion of the MYH9 gene associated with MYH9 disorders. Eur. J. Haematol. 2008 doi: 10.1111/j.1600-0609.2008.01046.x. [DOI] [PubMed] [Google Scholar]
- 22.Kunishima S, et al. Mapping of a gene for May-Hegglin anomaly to chromosome 22q. Hum. Genet. 1999;105:379–383. doi: 10.1007/s004390051119. [DOI] [PubMed] [Google Scholar]
- 23.Saito H, Kunishima S. Historical hematology: May-Hegglin anomaly. Am. J. Hematol. 2008;83:304–306. doi: 10.1002/ajh.21102. [DOI] [PubMed] [Google Scholar]
- 24.Kelley MJ, Jawien W, Ortel TL, Korczak JF. Mutation of MYH9, encoding non-muscle myosin heavy chain A, in May-Hegglin anomaly. Nat. Genet. 2000;26:106–108. doi: 10.1038/79069. [DOI] [PubMed] [Google Scholar]
- 25.Seri M, et al. Epstein syndrome: another renal disorder with mutations in the nonmuscle myosin heavy chain 9 gene. Hum. Genet. 2002;110:182–186. doi: 10.1007/s00439-001-0659-1. [DOI] [PubMed] [Google Scholar]
- 26.Kopp JB, et al. Genome wide admixture mapping identifies MYH9 as a major effect risk gene for focal segmental glomerulosclerosis. Nature Genetics. 2008 doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Apolito M, Guarnieri V, Boncristiano M, Zelante L, Savoia A. Cloning of the murine non-muscle myosin heavy chain IIA gene ortholog of human MYH9 responsible for May-Hegglin, Sebastian, Fechtner, and Epstein syndromes. Gene. 2002;286:215–222. doi: 10.1016/s0378-1119(02)00455-9. [DOI] [PubMed] [Google Scholar]
- 28.Marini M, et al. Non-muscle myosin heavy chain IIA and IIB interact and co-localize in living cells: relevance for MYH9-related disease. Int. J. Mol. Med. 2006;17:729–736. [PubMed] [Google Scholar]
- 29.Wright JT, Jr., et al. Effect of blood pressure lowering and antihypertensive drug class on progression of hypertensive kidney disease: results from the AASK trial. JAMA. 2002;288:2421–2431. doi: 10.1001/jama.288.19.2421. [DOI] [PubMed] [Google Scholar]
- 30.Knowler WC, et al. The Family Investigation of Nephropathy and Diabetes (FIND); Design and methods. J. Diabetes Complications. 2005;19:1–9. doi: 10.1016/j.jdiacomp.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Powe NR, et al. Choices for healthy outcomes in caring for end stage renal disease. Semin. Dial. 1996;9:9–11. [Google Scholar]
- 32.Nalls MA, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am. J. Hum. Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 34.The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong X, et al. A combined linkage-physical map of the human genome. Am. J. Hum. Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.